Abstract
Iron is an essential element for almost all living organisms, actively involved in a variety of cellular activities. To acquire iron from soil, strategy I plants such as Arabidopsis (Arabidopsis thaliana) must first reduce ferric to ferrous iron by Fe(III)-chelate reductases (FROs). FRO genes display distinctive expression patterns in several plant species. However, regulation of FRO genes is not well understood. Here, we report a systematic characterization of the AtFRO6 expression during plant growth and development. AtFRO6, encoding a putative FRO, is specifically expressed in green-aerial tissues in a light-dependent manner. Analysis of mutant promoter-β-glucuronidase reporter genes in transgenic Arabidopsis plants revealed the presence of multiple light-responsive elements in the AtFRO6 promoter. These light-responsive elements may act synergistically to confer light responsiveness to the AtFRO6 promoter. Moreover, no AtFRO6 expression was detected in dedifferentiated green calli of the korrigan1-2 (kor1-2) mutant or undifferentiated calli derived from wild-type explants. Conversely, AtFRO6 is expressed in redifferentiated kor1-2 shoot-like structures and differentiating calli of wild-type explants. In addition, AtFRO7, but not AtFRO5 and AtFRO8, also shows a reduced expression level in kor1-2 green calli. These results suggest that whereas photosynthesis is necessary but not sufficient, both light and cell differentiation are necessary for AtFRO6 expression. We propose that AtFRO6 expression is light regulated in a tissue- or cell differentiation-specific manner to facilitate the acquisition of iron in response to distinctive developmental cues.
All living organisms except lactobacilli have an absolute requirement for iron that is involved in a variety of cellular activities, including in respiration, chlorophyll biosynthesis, photosynthetic electron transfer, nitrogen assimilation, and DNA synthesis. In addition, numerous proteins, especially enzymes, require iron as an essential component in the form of heme or iron-sulfur (Marschner, 1995). Although abundant in soil, iron is one of the most common nutrients limiting plant growth and development, largely due to its extremely low solubility under aerobic environments of high pH (Guerinot and Yi, 1994). To acquire iron from soil, higher plants mainly utilize two different strategies, namely, strategy I and strategy II (Römheld and Marschner, 1986). All plants, with the exception of the grasses, employ the strategy I mechanism to effectively acquire iron from soil under iron deficiency stress.
Over the past several years, knowledge about the molecular basis of iron acquisition from soil in strategy I plants has greatly increased. Based on the sequence homology with the yeast (Saccharomyces cerevisiae) Fe(III) reductase1 (FRE1) and FRE2 that have been shown to be involved in iron acquisition (Dancis et al., 1990; Roman et al., 1993), AtFRO2, a Fe3+-chelate reductase, was identified from Arabidopsis (Arabidopsis thaliana; Robinson et al., 1999). Subsequently, PsFRO1 from pea (Pisum sativum) and LeFRO1 from tomato (Lycopersicon esculentum) were also identified and characterized (Waters et al., 2002; Li et al., 2004). Recent studies indicated that expression of LeFRO1 in tomato and FRO2 in Arabidopsis was regulated by FER (Ling et al., 2002; Li et al., 2004) and FIT1 (Colangelo and Guerinot, 2004; Jakoby et al., 2004), respectively. Tomato FER and Arabidopsis FIT1 (also named as FRU), which encode bHLH-type proteins, are functional orthologs in controlling iron acquisition (Yuan et al., 2005). In addition, Chloronerva, a gene encoding a nicotianamine synthase, is required in the transcriptional down-regulation of LeFRO1 under the iron sufficiency condition in tomato (Ling et al., 1999; Li et al., 2004). Moreover, substantial progress has also been made in efforts to explicate the mechanism of iron transport. For example, IRT1 is initially characterized as an iron-regulated metal transporter in Arabidopsis (Eide et al., 1996). Subsequent studies demonstrated that IRT1 is an iron transporter essential for plant growth and development (Henriques et al., 2002; Varotto et al., 2002; Vert et al., 2002). Expression of IRT1 is regulated at the transcriptional and posttranscriptional levels in response to iron deficiency (Connolly et al., 2002). In addition, members of the NRAMP family of divalent cation transporters, initially identified in bacteria, appear to be highly conserved across different kingdoms (Fleming et al., 1997; Gunshin et al., 1997). Three NRAMP-like genes in Arabidopsis, AtNramp1, AtNramp3, and AtNramp4, were shown to be involved in iron homeostasis. Similar to other iron metabolism-related genes, expression of these three Arabidopsis genes is subjected to the regulation of iron availability (Curie et al., 2000; Thomine et al., 2000).
In Arabidopsis, a typical strategy I species, iron is first reduced on the root surface from ferric to ferrous iron by Fe(III)-chelate reductases (FROs) and then transferred across the rhizodermal plasmalemma into root cells. Subsequently, iron is oxidized and transported as Fe3+-citrate complex for long-distance transport in the xylem from roots to shoots (Hell and Stephan, 2003). For assimilation in leaves and other tissues, iron is again reduced by FROs (Brüggemann et al., 1993; Gonzalez-Vallejo et al., 2000). However, it is not clear whether different genes are involved in the Fe(III) reduction process in different tissues. In the Arabidopsis genome, a FRO gene family with eight putative members was recently identified (Wu et al., 2005). These genes display distinctive expression patterns, including in roots (AtFRO2 and AtFRO3), shoot and flowers (AtFRO5 and AtFRO6), as well as in cotyledons and trichomes (AtFRO7). Moreover, expression of AtFRO8 was found to be largely restricted in leaf veins (Wu et al., 2005). Therefore, it is likely that AtFRO5 through AtFRO8 function in the aerial portions of plants to reduce ferric iron.
Ferric-chelate reductase activity has been proposed in leaves such as LeFRO1 in tomato and PsFRO1 in pea, both of which are primarily expressed in leaves (Waters et al., 2002; Li et al., 2004). FROs in plant aerial portions appear to be regulated by a mechanism different from that of the root-specific activities. This notion is supported by the observation that FRO activity in leaves, but not in roots, is regulated by light (Brüggemann et al., 1993; de la Guardia and Alcantara, 1996; Gonzalez-Vallejo et al., 2000). For example, a light-dependent ferric-chelate reduction activity was reported in leaves of cowpea (Vigna unguiculata; Brüggemann et al., 1993) and sunflower (Helianthus annuus; de la Guardia and Alcantara, 1996), with an increase of 3- and 10-fold, respectively. Remarkably, a more than 35-fold increased light-dependent ferric-chelate reduction activity was detected in sugar beet (Beta vulgaris) protoplasts (Gonzalez-Vallejo et al., 2000).
Despite an increasingly accumulated body of knowledge on FROs in plants, thus far little is known about the mechanism of light-regulated activity of FROs in plant aerial tissues. In this report, expression of AtFRO6, a gene encoding a ferric-chelate reductase, is characterized in detail. The AtFRO6 gene is mainly expressed in green-aerial tissues in a light-dependent manner. Promoter deletion and site-directed mutation analyses defined multiple light-responsive elements (LREs) that are necessary for the light-dependent expression of AtFRO6. Moreover, an AT-1-like box is essential for the aerial green tissue-specific expression of AtFRO6. In undifferentiated calli derived from tissue-cultured explants or dedifferentiated calli of the korrigan1-2 (kor1-2) mutant, essentially no AtFRO6 expression was detected. These results suggest that the light-regulated expression of AtFRO6 is green tissue specific and cell differentiation specific.
RESULTS
AtFRO6 Specifically Expresses in Aerial Green Tissues
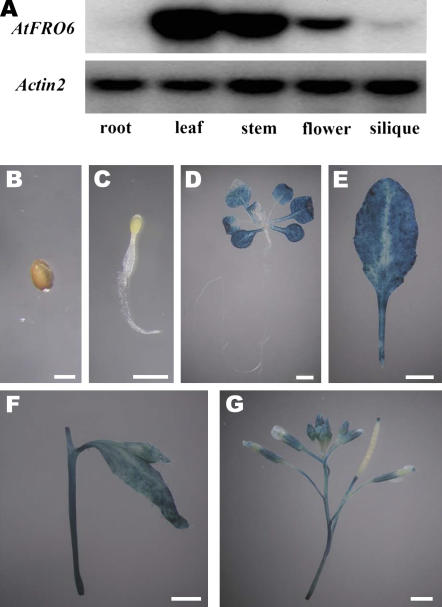
In a previous study, we reported the identification and characterization of eight putative AtFRO genes (Wu et al., 2005). When expressed in yeast cells, all tested AtFROs (AtFRO2 through AtFRO8) showed varying activities of FROs. Because AtFRO6 shows a low FRO activity in yeast cells (Wu et al., 2005), its precise biochemical nature remains to be verified. A reverse transcription (RT)-PCR analysis revealed that AtFRO6 (At5g49730) was expressed in shoots with no detectable expression in roots (Wu et al., 2005). In agreement with this result, a northern-blot analysis indicated that AtFRO6 predominantly expressed in leaves and stems, with a lower expression level in flowers and siliques (Fig. 1A). However, AtFRO6 expression was not detectable in roots.
Figure 1.
Expression patterns of AtFRO6. A, A northern-blot analysis of AtFRO6 expression. RNA was prepared from 3-week-old seedlings germinated and grown on Murashige and Skoog medium. Ten micrograms of RNA were used for northern-blot analysis using an AtFRO6 cDNA and an Actin2 cDNA as probes. B to G, Analysis of GUS expression in AtFRO6∷GUS transgenic plants by histochemical staining. Two-week-old seedlings were stained for 6 h, and all others were stained for 16 h. B, A germination seed. C, A 2-d-old seedling. D, A 2-week-old seedling. E, A rosette leaf collected from a soil-grown 4-week-old plant. F, A stem. G, Flowers and siliques. Bar = 2 mm.
To monitor the AtFRO6 expression in planta, we made an AtFRO6∷β-glucuronidase (GUS) reporter construct, which was stably transformed into Arabidopsis plants (Columbia-0 [Col-0]) by vacuum infiltration (Clough and Bent, 1998). The reporter construct, designated as pFul, contained a 1,093-bp Arabidopsis genomic sequence spanned from the 3′-untranslated region (UTR) of At5g49740 (in a head-to-tail configuration to At5g49730/AtFRO6) to sequences encoding for the first 11 amino acid residues of AtFRO6, in frame fused to the coding sequence of GUS. Therefore, this DNA fragment should encompass the entire AtFRO6 promoter sequence.
In germinating seeds, no GUS activity was detected (Fig. 1, B and C). However, consistent with the northern-blot analysis, the GUS reporter gene was strongly expressed in cotyledons, leaves, and stems, but not in roots and hypocotyls (Fig. 1, D–F). In flowers, the GUS activity appeared to be restricted to sepals (Fig. 1G). Weak GUS activity was also detected in siliques (Fig. 1G).
We also tested the AtFRO6 expression under different iron conditions. Under the conditions of iron starvation or externally supplied iron, no substantial alterations of the GUS expression were observed (data not shown), suggesting that the AtFRO6 expression is not regulated by iron availability.
Light-Dependent Expression of AtFRO6 Regulated by Multiple LREs
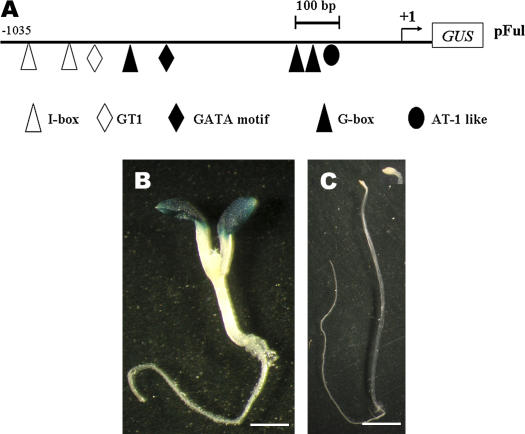
Sequence analysis of the AtFRO6 promoter revealed the presence of multiple putative LREs (Lescot et al., 2002), including G-box (Donald et al., 1990), GATA-motif (Lam and Chua, 1989), GT1-motif (Villain et al., 1996), and I-box (Giuliano et al., 1988; Figs. 2 and 3A). Moreover, data presented above indicated that AtFRO6 mainly expressed in aerial green tissues/organs. Therefore, we examined if the AtFRO6 expression is light dependent. Transgenic pFul seedlings germinated and grown in light or darkness were assayed for the GUS activity. Whereas strong GUS staining was observed in cotyledons of the light-grown seedlings (Fig. 3B), no GUS activity was detected in etiolated pFul seedlings at the same developmental stage (Fig. 3C). This result suggests that AtFRO6 expression is dependent on light in a tissue- or organ-specific manner.
Figure 2.
Nucleotide sequence of the AtFRO6 promoter region. The putative transcription initiation site (referred to as +1) is indicated by an arrow. Numbers at the left refer to the positions of nucleotides relative to the putative transcription initiation site. The putative LREs, the AT-1-like element, and restriction sites are shown in bold. The palindromic repeat is underlined (dots denote the space between the repeat).
Figure 3.
Light-dependent expression of AtFRO6. A, A diagram showing the AtFRO6∷GUS reporter construct (not in scale for GUS coding sequence). The putative transcription start site is indicated by an arrow (+1). Multiple putative LREs are shown. B, Seven-day-old pFul transgenic seedlings germinated and grown in the light were assayed for the GUS activity by histochemical staining (stained for 6 h). C, Seven-day-old pFul transgenic seedlings germinated and grown in the dark were assayed for GUS activity (stained for 16 h). The insert shows an enlarged view of the cotyledons. Bar = 2 mm.
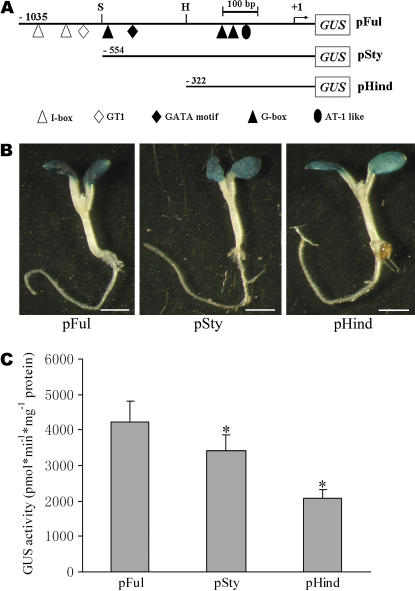
To characterize these putative LREs, we made two mutant constructs of AtFRO6∷GUS, which carried truncations from the distal end of the promoter (Fig. 4A). These constructs were stably transformed into wild-type Col-0 plants, and the GUS activity was analyzed in homozygous T2 or T3 plants. Note that, because the precise position of the transcription initiation site of AtFRO6 has not been determined, we deduced a transcription initiation site (referred to as +1) based on available cDNA sequences that contained the longest 5′-UTR of 58 bp (accession no. AY091140; see Fig. 2). Whereas the sequence upstream from −554 (StyI site) contained a putative GT1-box and several I-box-type cis-elements, the region between −554 and −322 (HindIII site) had a putative GATA-motif and a G-box (Fig. 4A). Deletion of these putative LREs caused substantially reduced GUS activities in pSty and pHind transgenic plants (80.9% and 49.7% relative to pFul, respectively; Fig. 4, B and C). These results suggest that these putative LREs may be involved in the maintenance of optimal promoter activity of AtFRO6. In particular, cis-elements located between −554 and −322 (covered by pSty and pHind), which contained a putative GATA-box and a putative G-box (Fig. 4A), contributed approximately 50% of the promoter activity (Fig. 4C).
Figure 4.
Deletion analysis of the AtFRO6 promoter. A, Schematic maps showing wild type (pFul) and truncation mutants (pSty and pHind) of the AtFRO6 promoter-GUS constructs (not in scale for GUS coding sequence). S and H denote StyI and HindIII restriction sites, respectively. B, Analysis of the GUS activity in transgenic plants by histochemical staining. Bar = 2 mm. C, Quantitative analysis of the GUS activity in transgenic plants by the fluorimetric assay. Data presented are mean values of three independent experiments. Bars, ses. *, Significant differences at P < 0.05 compared to the control.
The absence of AtFRO6 expression in roots under both the light and dark conditions may be caused by repressive or negative regulatory cis-elements in the AtFRO6 promoter. To test this possibility, we analyzed the GUS activity of pFul, pSty, and pHind transgenic plants germinated and grown in the dark. No detectable GUS activity was observed in these transgenic plants upon extensive staining (16–24 h; data not shown). This result rules out the possibility that the AtFRO6 expression is directly regulated by a light-repressive mechanism.
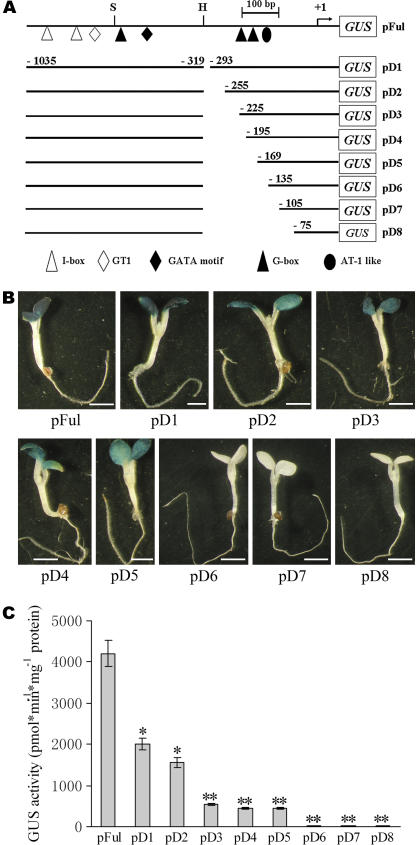
Compared to pFul, pSty and pHind showed a substantially reduced promoter activity. However, the light-dependent and tissue-specific expression pattern was not altered in pSty and pHind (Fig. 4, B and C), suggesting that the sequence between −322 and +1 contained additional regulatory elements sufficient for maintaining the tissue- or organ-specific expression pattern of AtFRO6. To identify possible cis-acting elements in this region, we made a series of internal deletion mutants based on pFul (Fig. 5A). These mutant AtFRO6 promoter-GUS constructs, designated as pD1 through pD8, were introduced into wild-type Arabidopsis plants. Multiple independent T2 or T3 lines homozygous for single T-DNA insertions were identified and used for subsequent experiments. Deletions up to −169 (pD5) had no apparent effect on the aerial tissue-specific expression pattern of the reporter gene (Fig. 5B). As expected, pD1 through pD4, which had shorter deletions in this region, displayed an expression pattern similar to that of pD5 (Fig. 5B). Deletions of additional sequences toward the proximal end completely abolished the expression of the reporter gene (pD6, pD7, and pD8; Fig. 5B).
Figure 5.
Mapping of cis-regulatory elements in the AtFRO6 promoter by internal deletions. A, Schematic maps of wild type (pFul) and internal deletion mutants (pD1–pD8) of the AtFRO6 promoter-GUS constructs (not in scale for GUS coding sequence). B, Analysis of the GUS activity in transgenic seedlings by histochemical staining. pFul seedlings were stained for 6 h and all others were stained for 16 h. Bar = 2 mm. C, Quantitative analysis of the GUS activity in transgenic plants. Data presented are mean values of three independent experiments. Bars, ses. *, Significant differences at P < 0.05 compared to the control. **, Significant differences at P < 0.01 compared to the control.
Similar to that of pSty and pHind, although the aerial tissue-specific expression pattern was maintained in pD1 through pD5, the GUS activity was remarkably reduced. Quantitative fluorometric GUS assay indicated that pD1 and pD2 maintained approximately 50% activity, whereas pD3, pD4, and pD5 had approximately 10% activity relative to that of wild-type promoter pFul (Fig. 5C). The region covered by pD3 through pD5 (−225 to approximately −169) contains two putative G-box cis-elements (Fig. 5A). Deletion of these elements resulted in a 90% loss of the GUS activity (Fig. 5C), suggesting that these elements are crucial for the promoter activity. Consistent with this observation, a longer staining time was required for pD3, pD4, and pD5 transgenic plants compared to that of wild-type pFul transgenic plants (16–24 versus 4–6 h).
Taken together, data presented above suggest that multiple LREs in the AtFRO6 promoter are involved in the control of the promoter activity.
An AT-1-Like Box Is Essential for Aerial Green Tissue-Specific Expression of AtFRO6
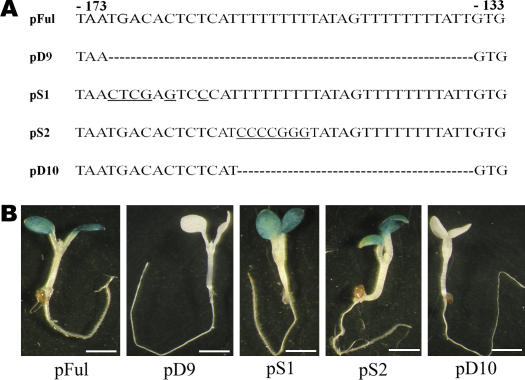
Data presented above suggest that sequences between −195 and −135 (covered by pD4 through pD6) are required for light-inducible expression of AtFRO6 in the aerial green tissues. The loss of promoter activity in pD6 (deletion of −318 to −136) may be caused by nonspecific alterations of the promoter structure because of a relatively large deletion in this construct (183 bp). Alternatively, the aerial green tissue-specific expression of AtFRO6 is regulated by a cis-element located in this region. To distinguish these two possibilities, we made an additional mutant pD9, which contained a 35-bp deletion between −170 and −136 (Figs. 2 and 6A). As shown in Figure 6B, pD9 did not show any detectable GUS activity, suggesting that this 35-bp sequence is essential for the promoter activity.
Figure 6.
Definition of an AT-1-like box essential for tissue-specific and light-dependent AtFRO6-GUS expression. A, The AtFRO6 promote sequences of wild-type (pFul) and mutant (pD9, pS1, pS2, and pD10) constructs. Deleted sequences are represented by dashed lines, and substituted nucleotides are underlined. B, Analysis of the GUS activity of the transgenic plants by histochemical staining. Bar = 2 mm.
In this 35-bp region, we noticed two putative cis-elements that might be responsible for the regulation of the promoter. Whereas a perfect palindromic repeat was present at the distal end (AATGACACTCTCATT), two T-repeats were found at the proximal end (Figs. 2 and 6A). In particular, the proximal T-repeat (TATAGTTTTTTTTATT) is structurally similar to the previously characterized AT-1-box (AATATTTTTATT) found in the pea RbcS-3A promoter (Datta and Cashmore, 1989). To determine which element(s) is required for the regulation, we made three additional mutant promoter-GUS constructs that were tested in stably transformed transgenic plants. We first introduced substitution mutations at multiple positions to disrupt the palindromic repeat (pS1) or the distal T-repeat (pS2; see Fig. 6A). Both mutants displayed a phenotype similar to that of wild-type pFul (Fig. 6B). This result suggests that none of these two elements was required for the promoter activity. However, deletion of a 23-bp sequence between −158 and −136 (pD10; Fig. 6A), which included both T-repeats, resulted in a complete loss of the promoter activity (Fig. 6B). Because disruption of the distal T-repeat (pS2) did not cause any altered promoter activity, we concluded that the proximal T-repeat or the AT-1-like box is essential for a fully functional AtFRO6 promoter.
Cell Differentiation-Specific Expression of AtFRO6
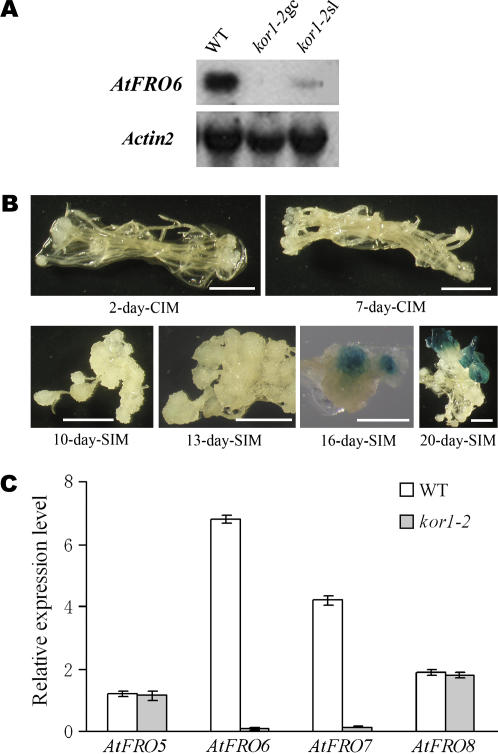
A previous study suggested that AT-1-box-containing promoters display the photosynthetic cell-specific expression pattern (Datta and Cashmore, 1989). Consistent with these findings, AtFRO6 expression is restricted to some light-grown green tissues/organs, suggesting that light and/or photosynthesis are necessary for the AtFRO6 expression. However, it is not known if light and/or photosynthesis are sufficient for the AtFRO6 expression. To address this question, we examined the AtFRO6 expression pattern in the kor1-2 mutant. The kor1-2 mutation causes all adult organs to become transformed into green calli (Zuo et al., 2000). Thus, kor1-2 green calli should represent a group of cells that are photosynthetically active but become dedifferentiated. Figure 7A shows that no AtFRO6 expression was detected in kor1-2 green callus. This result suggests that photosynthesis may not be sufficient for AtFRO6 expression or the kor1-2 mutation may somehow affect AtFRO6 expression. To distinguish these two possibilities, we performed the following experiments. We noted that shoot-like structures can be occasionally formed from the kor1-2 green calli upon longer culture (8–10 weeks). If KOR1 indeed plays a role in regulating AtFRO6, no AtFRO6 expression should be seen in the kor1-2 shoot-like structures. A northern-blot analysis revealed weak AtFRO6 expression in the kor1-2 shoot-like structures (Fig. 7A), thereby ruling out the possibility that KOR1 plays a direct regulatory role in AtFRO6 expression.
Figure 7.
Cell differentiation-specific expression of AtFRO6. A, A northern-blot analysis of AtFRO6 expression in wild-type (C24) and kor1-2 mutant plants. Ten micrograms of RNA were used for northern-blot analysis using an AtFRO6 cDNA and an Actin2 cDNA as probes. WT, Three-week-old wild-type plants; kor1-2gc, 8-week-old green calli of the mutant; and kor1-2sl, shoot-like structures derived from 8-week-old kor1-2 calli. All materials were grown or cultured on Murashige and Skoog medium. B, Analysis of the GUS activity in calli derived from pFul transgenic plants. Root explants derived from pFul transgenic plants were cultured on callus-induction medium (CIM; see “Materials and Methods”) and then transferred onto SIM (see “Materials and Methods”). Samples were collected at different time points and stained for the GUS activity. Bar = 1 mm. C, Real-time quantitative RT-PCR analysis of AtFRO5, AtFRO6, AtFRO7, and AtFRO8 expression in wild-type (C24) and kor1-2 mutant plants. Relative expression level of AtFRO5 through AtFRO8 was shown as percentage of the expression level of Actin8 that serves as an internal control. Mean values obtained from two independent experiments were shown in the histogram. Bars, ses. WT, Three-week-old wild-type plants; kor1-2, 8-week-old green calli of the mutant.
To further test if cell differentiation is a perquisite for the AtFRO6 expression, we examined the AtFRO6 expression during in vitro shoot regeneration. Root explants derived from pFul were cultured in the callus induction medium for 2 d and then transferred onto shoot induction medium (SIM; Koncz et al., 1989; see “Materials and Methods” for technical details). During this course, explants are presumed to first acquire competence to shoot induction signals and then become committed to shoot development (Sugiyama, 1999; Che et al., 2002). As shown in Figure 7B, whereas specific GUS expression was detected in green calli and shoots derived from these green calli, no GUS activity was found in root explants or undifferentiated calli. This result suggests that AtFRO6 expression may mark cell differentiation or tissue specification in plants. We conclude from the above results that, whereas greening or photosynthesis is not sufficient for the expression of AtFRO6, cell or organ differentiation is necessary for the AtFRO6 expression.
Expression of AtFRO5, AtFRO7, and AtFRO8 in kor1-2 Green Calli
Our previous studies showed that AtFRO5, AtFRO7, and AtFRO8 had an expression pattern similar to that of AtFRO6. These four genes have substantial expression in shoots, but with a significantly reduced expression level (AtFRO5 and AtFRO8) or no detectable expression (AtFRO6 and AtFRO7) in roots. Moreover, expression of AtFRO5 and AtFRO8 appears to be inducible by iron deficiency (Wu et al., 2005). To reveal if expression of the three genes is also regulated by cell or tissue differentiation, we analyzed their expression patterns in the kor1-2 mutant by quantitative real-time PCR. Both AtFRO5 and AtFRO8 had a similar expression level in wild-type and kor1-2 plants. However, similar to that of AtFRO6, expression of AtFRO7 was remarkably reduced in the kor1-2 mutant (Fig. 7C). This result suggests expression of AtFRO7 is also regulated by a mechanism similar to that of AtFRO6, in a cell differentiation- or tissue differentiation-dependent manner.
DISCUSSION
In strategy I plants, a critical step for iron acquisition from soil is to reduce ferric to ferrous iron catalyzed by FROs that are encoded by evolutionarily conserved gene families FRO in various plant species. Expression of FRO genes shows distinctive patterns during plant growth and development and is regulated by various environmental factors (Robinson et al., 1999; Wu et al., 2005). Although light-dependent Fe(III)-chelate reduction has been observed in several plant species (Brüggemann et al., 1993; de la Guardia and Alcantara, 1996; Gonzalez-Vallejo et al., 2000), the molecular mechanism for this regulation remains largely unknown. In this study, we have characterized the expression of the Arabidopsis AtFRO6 gene, which encodes a putative FRO (Wu et al., 2005). We found that expression of AtFRO6 was regulated by a novel mechanism involved in both light and cell differentiation.
The AtFRO6 expression was found to restrict to aerial green tissues but not in roots, suggestive of the involvement of a possible light regulatory mechanism. This view is supported by the observation that AtFRO6∷GUS is expressed in light-grown but not in etiolated seedlings. Consistent with this light-dependent expression pattern, multiple putative LREs were found in the AtFRO6 promoter. By analyzing GUS expression driven by a series of mutant AtFRO6 promoters in transgenic Arabidopsis plants, we have been able to define several LREs that are involved in the light-dependent expression of the gene. All these LREs appear to contribute to the promoter activity at different degrees. Whereas deletion of three LREs upstream from −320 results in an approximate 50% reduction of the promoter activity, two G-box-type cis-elements upstream from −169 account for 90% activity of the promoter activity. Despite the importance of these LREs as highlighted above, however, deletion of the AT-1-like box upstream from −189 causes a complete loss of the GUS activity. Note that no detectable reporter activity in this mutant may attribute to detect limitation under the assay conditions. Nevertheless, mutations in any LREs cause substantially reduced promoter activities. One explanation for these results is that the AT-1-like box may be essential for basal transcription of the promoter. However, considering the position of this cis-element in various promoters, including in pea rbcS (Datta and Cashmore, 1989) and AtFRO6, and the nature of a tobacco nuclear protein that binds to an AT-1-like box (Tjaden and Coruzzi, 1994), it appears to be unlikely that the AT-1-like box is a core element of a minimal promoter. Alternatively, the AT-1-like box and other upstream LREs may act synergistically to control the promoter activity. This notion is supported by the fact that none of the LREs identified thus far alone is able to confer light responsiveness to a minimal promoter; instead, distinctive combinations of LREs are both necessary and sufficient for the light inducibility of a promoter (Terzaghi and Cashmore, 1995; Puente et al., 1996).
In addition to light responsiveness, AtFRO6 appears to be cell differentiation-specific regulated. In most, if not all, cases, expression of a light-regulated gene is restricted to photosynthetic active cells and displays a tissue- or development-specific expression mode (Terzaghi and Cashmore, 1995). The mutagenesis analysis of a series of promoter-GUS reporter genes suggests that AtFRO6 may have a similar regulatory mechanism. However, AtFRO6 does not have detectable expression in dedifferentiated green calli of kor1-2 but has weak expression in the shoot-like structures derived from the mutant calli. These observations suggest that cell or tissue differentiation is necessary for the AtFRO6 promoter activity, whereas light or photosynthetic activity is necessary but not sufficient for AtFRO6 expression. A similar observation was made in tobacco (Nicotiana tabacum) TID748, a partial cDNA clone identified by a subtractive hybridization from genetic tumors derived from the interspecific hybrid between Nicotiana glauca and Nicotiana langsdorffii. TID748 contains a partial open reading frame encoding a polypeptide of 184 amino acid residues (accession no. BAA05478), which shares considerable homology with carboxyl termini of FRO proteins (51% identical to AtFRO2, AtFRO4, and AtFRO5; 27% identical to AtFRO6; Fujita et al., 1994). Expression of TID748 appears to mark cell differentiation in the genetic tumors at the onset of organogenesis, although no substantial expression was detected in aerial green tissues of both parents (Fujita et al., 1994). Moreover, a 9-fold increase in ferric reductase activity was observed during the monocyte to macrophage differentiation, suggesting that cell differentiation is able to stimulate ferric reductase activity (Partridge et al., 1998), albeit it is not known whether the regulation occurs at the transcriptional or posttranscriptional level. Notably, expression of rbcS and CAB2, two typical light-responsive genes, is substantially down-regulated in kor1-2 green calli (H. Feng and J. Zuo, unpublished data), suggesting that this cell differentiation-specific regulatory mechanism may also be employed by other light-regulated genes.
Among the identified AtFROs, AtFRO6 shows the lowest FRO activity in yeast cells (Wu et al., 2005). However, AtFRO7, another member of the family that shares the highest homolog with AtFRO6 (92.54% identity of the protein sequences), displays considerable FRO activity (more than 4-fold higher than the control; Wu et al., 2005). The strong homology between these two proteins suggests that AtFRO6 may encode a functional FRO, although it remains unclear why AtFRO6 has a lower FRO activity in yeast cells. Consistent with their high degree of sequence similarity, AtFRO6 and AtFRO7 display a similar expression pattern. Expression of both genes appears to be restricted in shoots or green aerial tissues without detectable expression in roots (Wu et al., 2005 and this study). Moreover, expression of both genes is remarkably reduced in kor1-2 green calli. A reduced expression level of AtFRO6 and AtFRO7 in kor1-2 green calli appears to be specific, because neither AtFRO5 nor AtFRO8 had an apparently altered expression level in the mutant. These observations suggest that expression of AtFRO6 and AtFRO7 is likely regulated by a similar, if not identical, mechanism.
Iron plays a great deal of roles in chloroplast development and function, including respiration, chlorophyll biosynthesis, and photosynthetic electron transfer (Leonhardt and Straus, 1994; Robinson et al., 1999; Cody et al., 2000; Waters et al., 2002). In addition, iron homeostasis is also critical for cells against the oxidative stress of reactive oxygen species (ROS) generated by PSI and PSII (Michel and Pistorius, 2004). This is not only because many ROS-detoxifying enzymes, such as catalase, peroxidase, and some superoxide dismutases, require iron as cofactors (Isaac and Dawson, 1999; Matsunaga and Shiro, 2004), but also due to an increased ROS level generated by PSI and PSII under the iron limiting conditions (Aro et al., 1993; Ghassemian et al., 1994). Therefore, an increased requirement of iron during light-induced development or cell differentiation is obviously responded by an elevated ferric reductase activity, which appears to be regulated at both transcriptional and posttranscriptional levels.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Transformation of Plants
Arabidopsis (Arabidopsis thaliana) Col-0 and C24 ecotypes were used in this study. The kor1-2 mutant is in the C24 background (Zuo et al., 2000). Whereas C24 wild-type plants were used in experiments related to kor1-2, Col-0 plants were used in all other experiments. Unless indicated otherwise, plants were grown in continuous white light as described previously (Sun et al., 2003). Transformation of Arabidopsis was carried out by vacuum infiltration (Clough and Bent, 1998).
In vitro regeneration of shoots from root explants was carried out as previously described (Sun et al., 2003). Briefly, root explants were cultured on the callus-induction medium (1× B5 salts, 2% Glc, 0.5 g/L MES, 0.5 mg/L 2,4-dichlorophenoxyacetic acid, 0.05 mg/L kinetin, and 0.2% Phytagel, pH 5.7) for 2 to 3 d and then transferred onto the SIM (1× Murashige and Skoog salts, 1% Suc, 0.5 g/L MES, 1 mg/L N6-Δ2-isopentenyladenine, 0.15 mg/L indole-3-acetic acid, and 0.2% Phytagel, pH 5.7) for varying times. Samples collected at different time points were assayed for the GUS activity by histochemical staining (see below). The samples were photographed under a light microscope before and after GUS staining.
In Silico Analysis of the AtFRO6 Promoter
Search for putative cis-acting regulatory elements in the AtFRO6 promoter was performed using PlantCARE as described (Lescot et al., 2002).
Construction of the AtFRO6∷GUS Reporter Genes
Plasmid construction was performed by standard methods (Sambrook and Russell, 2001). All AtFRO6∷GUS reporter genes contained 5′-UTR and various promoter sequences of the AtFRO6 gene. In all these constructs, sequences encoding the first 11 amino acid residues (including the translation start codon) of AtFRO6 were included, which were in-frame fused to the GUS coding sequence.
The AtFRO6 (At5g49730) promoter sequence was obtained by PCR amplification of a 1.1-kb DNA fragment, which included the entire promoter (extended to the 3′-UTR of At5g49740), 5′-UTR, and a part of the coding sequence of AtFRO6. The PCR fragment was cloned into a pGEM-T vector (Promega) to yield pT-Ful. There are two StyI sites in this region, located at the −554 (Fig. 3A) and 31 bp downstream from the putative translation start codon, respectively. To construct pFul, a 1.1-kb DNA fragment released from pT-Ful by SalI and StyI (partially digested with StyI and then filled in by the Klenow enzyme; the StyI site is located between codons 10 and 11 of the AtFRO6 coding sequence) was ligated to SalI- and SmaI-digested pBI101-1 (CLONTECH). Therefore, the first 11 residues of AtFRO6 were in-frame fused to GUS. To make pT-Sty, pT-Ful was digested with SalI/StyI (partial digestion), blunted with Klenow enzyme, and then religated. After the religation, the StyI site was eliminated and the SalI site was maintained. pSty was made by a similar approach as for pFul. To make pHind, pFul was digested with SalI and HindIII, filled in by Klenow, and then religated. All other mutant constructs (internal deletions and substitutions) were first made in a pGEM-T vector by PCR using appropriate primers, and the mutant DNA fragments were then cloned into pFul using SalI and SnaBI sites (SnaBI is in the GUS coding region). All constructs were confirmed by DNA sequencing.
Primer pairs used in PCR (all sequences are from 5′-end to 3′-end) were as follows: D0F (GGTCGACGATGCTCTCAAGGCCAAAGA) and ATGStyB1 (GAGTCCTTGGACAAAAGAGGGGTT; StyI site is underlined) for pFul; and D0F and Hind3B1 (GGGAATTCGCTTACATTACCTGAATCTGAACA; EcoRI site is underlined) for the distal fragments of pD1 to pD8. The proximal fragments of pD1 to pD8, which contained partial sequences of the AtFRO6 promoter and the GUS coding region, were amplified by using pFul plasmid DNA as a PCR template and the following forward primers: D1F, ATAATAACCATCTTATCTATACT; D2F, CTCAGTCTCTCACCTTACCCT; D3F, TTGAAATCAACCTAACGTGACAC; D4F, CAAGTACGTTACTCTCACGTGT; D5F, GACACTCTCATTTTTTTTTATAGTT; D6F, GTGTACTAGGATTGTTGTCCGA; D7F, TTCATAAATGAGAATGATAGAAACC; and D8F, GGTCCAGAAAATTTTACTTGACTC. GUSB2 (GGCGGGATAGTCTGCCAGTTCA; located 3′-end of SnaBI site) was used as a backward primer in all these reactions. These two sets of fragments were ligated using EcoRI sites, located in the backward primer Hind3B1 of the distal fragment (see above) and the pGEM-T vector immediately flanking the proximal fragments, respectively. To make pS1, the distal fragment was amplified by D0F and S1B (ACTCGAGTTAACACGTGAGAGTAA; XhoI site is underlined), and the proximal fragment was amplified by S1F (ACTCGAGTCCCATTTTTTTTTATAGTTTTTTTTATTGTG; XhoI site is underlined) and GUSB2. These two fragments were ligated using XhoI sites. To make pS2, the distal fragment was amplified by D0F and S2B (CCCCGGGTATAGTGTTTTTTATTGTGTAC; SmaI site is underlined), and the proximal fragment was amplified by S2F (ACCCGGGGATGAGAGTGTCATTAACA; SmaI site is underlined) and GUSB2. These two fragments were ligated using SmaI sites. For pD9 and pD10, S1B and S2B were used, combined with D0F, for the amplification of the distal fragments, respectively, which were then ligated to SalI- and SpeI-digested pD6 (SpeI site was derived from pGEM-T vector).
Analysis of GUS Activity
GUS activity was assayed by histochemical and fluorimetric analyses as described (Jefferson et al., 1987).
RNA Isolation and Gel-Blot Analysis
RNA northern-blotting analyses were carried out as described previously (Sun et al., 2003) using an AtFRO6 cDNA and an Actin2 cDNA (AT3G18780) as probes.
Analysis of Gene Expression by Real-Time PCR
Expression of AtFRO5 through AtFRO8 was analyzed by real-time quantitative RT-PCR as described (Li et al., 2005). Primer pairs used in the RT-PCR analyses were (all sequences are from 5′-end to 3′-end) as follows: AtFRO5F, TGATGTCTCGAGGCACGATTCT and AtFRO5R, TCACGGAAATGAAGGGTGTAACT; AtFRO6F, AATCCGAGCCTCGCTTGGA and AtFRO6R, TGGTCCGTGGTAGCTTGACAGAA; AtFRO7F, TCGCTGTTTTACCCGGAGTTATCA and AtFRO7R, TCACGGTGTGAAGCGATGTGA; AtFRO8F, GGGACTCTCGATGTTCCGGTTACT and AtFRO8R, CGGTCCCGAACCACACATGA; and Actin8F, TTGCAGACCGTATGAGCAAAGAGA and Actin8R, TGGTGCCACGACCTTAATCTTCA.
Acknowledgments
We thank the Arabidopsis Biological Resources Center for cDNA clones. We are grateful to Dr. Nam-Hai Chua of Rockefeller University for critically reading the manuscript. J.Z. acknowledges that this work was initiated in the lab of Dr. Nam-Hai Chua at Rockefeller University.
This work was supported by the National Natural Science Foundation (grant nos. 30125025 and 30221002 to J.Z.; grant nos. 30225029 and 30530460 to H.-Q.L.), by the Chinese Academy of Sciences (grant no. KSCX2–SW–308 to J.Z.), and by HarvestPlus Program-China (to H.-Q.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Jianru Zuo (jrzuo@genetics.ac.cn).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074138.
References
- Aro EM, Virgin I, Andersson B (1993) Photoinhibition of Photosystem II. Inactivation, protein damage and turnover. Biochim Biophys Acta 1143: 113–134 [DOI] [PubMed] [Google Scholar]
- Brüggemann W, Maas-Kantel K, Moog P (1993) Iron uptake by leaf mesophyll cells: the role of the plasma membrane-bound ferric-chelate reductase. Planta 190: 151–155 [Google Scholar]
- Che P, Gingerich DJ, Lall S, Howell SH (2002) Global and hormone-induced gene expression changes during shoot development in Arabidopsis. Plant Cell 14: 2771–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cody GD, Boctor NZ, Filley TR, Hazen RM, Scott JH, Sharma A, Yoder HS Jr (2000) Primordial carbonylated iron-sulfur compounds and the synthesis of pyruvate. Science 289: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Colangelo EP, Guerinot ML (2004) The essential basic helix-loop-helix protein FIT1 is required for the iron deficiency response. Plant Cell 16: 3400–3412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly EL, Fett JP, Guerinot ML (2002) Expression of the IRT1 metal transporter is controlled by metals at the levels of transcript and protein accumulation. Plant Cell 14: 1347–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curie C, Alonso JM, Le Jean M, Ecker JR, Briat JF (2000) Involvement of NRAMP1 from Arabidopsis thaliana in iron transport. Biochem J 347: 749–755 [PMC free article] [PubMed] [Google Scholar]
- Dancis A, Klausner RD, Hinnebusch AG, Barriocanal JG (1990) Genetic evidence that ferric reductase is required for iron uptake in Saccharomyces cerevisiae. Mol Cell Biol 10: 2294–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N, Cashmore AR (1989) Binding of a pea nuclear protein to promoters of certain photoregulated genes is modulated by phosphorylation. Plant Cell 1: 1069–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Guardia MD, Alcantara E (1996) Ferric chelate reduction by sunflower (Helianthus annuus L.) leaves: influence of light, oxygen, iron deficiency and leaf age. J Exp Bot 47: 669–675 [Google Scholar]
- Donald RG, Schindler U, Batschauer A, Cashmore AR (1990) The plant G box promoter sequence activates transcription in Saccharomyces cerevisiae and is bound in vitro by a yeast activity similar to GBF, the plant G box binding factor. EMBO J 9: 1727–1735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide D, Broderius M, Fett J, Guerinot ML (1996) A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc Natl Acad Sci USA 93: 5624–5628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming MD, Trenor CC III, Su MA, Foernzler D, Beier DR, Dietrich WF, Andrews NC (1997) Microcytic anaemia mice have a mutation in Nramp2, a candidate iron transporter gene. Nat Genet 16: 383–386 [DOI] [PubMed] [Google Scholar]
- Fujita T, Kouchi H, Ichikawa T, Syono K (1994) Cloning of cDNAs for genes that are specifically or preferentially expressed during the development of tobacco genetic tumors. Plant J 5: 645–654 [DOI] [PubMed] [Google Scholar]
- Ghassemian M, Wong B, Ferreira F, Markley JL, Straus NA (1994) Cloning, sequencing and transcriptional studies of the genes for cytochrome c-553 and plastocyanin from Anabaena sp. PCC 7120. Microbiology 140: 1151–1159 [DOI] [PubMed] [Google Scholar]
- Giuliano G, Pichersky E, Malik VS, Timko MP, Scolnik PA, Cashmore AR (1988) An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc Natl Acad Sci USA 85: 7089–7093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vallejo EB, Morales F, Cistue L, Abadia A, Abadia J (2000) Iron deficiency decreases the Fe(III)-chelate reducing activity of leaf protoplasts. Plant Physiol 122: 337–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerinot ML, Yi Y (1994) Iron: nutritious, noxious, and not readily available. Plant Physiol 104: 815–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunshin H, Mackenzie B, Berger UV, Gunshin Y, Romero MF, Boron WF, Nussberger S, Gollan JL, Hediger MA (1997) Cloning and characterization of a mammalian proton-coupled metal-ion transporter. Nature 388: 482–488 [DOI] [PubMed] [Google Scholar]
- Hell R, Stephan UW (2003) Iron uptake, trafficking and homeostasis in plants. Planta 216: 541–551 [DOI] [PubMed] [Google Scholar]
- Henriques R, Jasik J, Klein M, Martinoia E, Feller U, Schell J, Pais MS, Koncz C (2002) Knock-out of Arabidopsis metal transporter gene IRT1 results in iron deficiency accompanied by cell differentiation defects. Plant Mol Biol 50: 587–597 [DOI] [PubMed] [Google Scholar]
- Isaac IS, Dawson JH (1999) Haem iron-containing peroxidases. Essays Biochem 34: 51–69 [DOI] [PubMed] [Google Scholar]
- Jakoby M, Wang HY, Reidt W, Weisshaar B, Bauer P (2004) FRU (BHLH029) is required for induction of iron mobilization genes in Arabidopsis thaliana. FEBS Lett 577: 528–534 [DOI] [PubMed] [Google Scholar]
- Jefferson R, Kavanagh T, Bevan MW (1987) GUS fusion: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz C, Martini N, Mayerhofer R, Koncz-Kalman Z, Korber H, Redei GP, Schell J (1989) High-frequency T-DNA-mediated gene tagging in plants. Proc Natl Acad Sci USA 86: 8467–8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam E, Chua NH (1989) ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1: 1147–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonhardt K, Straus NA (1994) Photosystem II genes isiA, psbDI and psbC in Anabaena sp. PCC 7120: cloning, sequencing and the transcriptional regulation in iron-stressed and iron-repleted cells. Plant Mol Biol 24: 63–73 [DOI] [PubMed] [Google Scholar]
- Lescot M, Dehais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouze P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Cheng X, Ling HQ (2004) Isolation and characterization of Fe(III)-chelate reductase gene LeFRO1 in tomato. Plant Mol Biol 54: 125–136 [DOI] [PubMed] [Google Scholar]
- Li X-B, Fan X-P, Wang X-L, Cai L, Yang W-C (2005) The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell 17: 859–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Bauer P, Bereczky Z, Keller B, Ganal M (2002) The tomato fer gene encoding a bHLH protein controls iron-uptake responses in roots. Proc Natl Acad Sci USA 99: 13938–13943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling HQ, Koch G, Baumlein H, Ganal MW (1999) Map-based cloning of chloronerva, a gene involved in iron uptake of higher plants encoding nicotianamine synthase. Proc Natl Acad Sci USA 96: 7098–7103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, San Diego
- Matsunaga I, Shiro Y (2004) Peroxide-utilizing biocatalysts: structural and functional diversity of heme-containing enzymes. Curr Opin Chem Biol 8: 127–132 [DOI] [PubMed] [Google Scholar]
- Michel KP, Pistorius EK (2004) Adaptation of the photosynthetic electron transport chain in cyanobacteria to iron deficiency: the function of IdiA and IsiA. Physiol Plant 120: 36–50 [DOI] [PubMed] [Google Scholar]
- Partridge J, Wallace DF, Raja KB, Dooley JS, Walker AP (1998) Monocyte-macrophage ferric reductase activity is inhibited by iron and stimulated by cellular differentiation. Biochem J 336: 541–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente P, Wei N, Deng XW (1996) Combinatorial interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J 15: 3732–3743 [PMC free article] [PubMed] [Google Scholar]
- Robinson NJ, Procter CM, Connolly EL, Guerinot ML (1999) A ferric-chelate reductase for iron uptake from soils. Nature 397: 694–697 [DOI] [PubMed] [Google Scholar]
- Roman DG, Dancis A, Anderson GJ, Klausner RD (1993) The fission yeast ferric reductase gene frp1+ is required for ferric iron uptake and is homologous to the gp91-phox subunit of the human NADPH phagocyte oxidoreductase. Mol Cell Biol 13: 4342–4350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römheld V, Marschner H (1986) Evidence for a specific uptake system for iron phytosiderophores in roots of grasses. Plant Physiol 80: 175–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sugiyama M (1999) Organogenesis in vitro. Curr Opin Plant Biol 2: 61–64 [DOI] [PubMed] [Google Scholar]
- Sun J, Niu QW, Tarkowski P, Zheng B, Tarkowska D, Sandberg G, Chua NH, Zuo J (2003) The Arabidopsis AtIPT8/PGA22 gene encodes an isopentenyl transferase that is involved in de novo cytokinin biosynthesis. Plant Physiol 131: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terzaghi WB, Cashmore AR (1995) Photomorphogenesis. Seeing the light in plant development. Curr Biol 5: 466–468 [DOI] [PubMed] [Google Scholar]
- Thomine S, Wang R, Ward JM, Crawford NM, Schroeder JI (2000) Cadmium and iron transport by members of a plant metal transporter family in Arabidopsis with homology to Nramp genes. Proc Natl Acad Sci USA 97: 4991–4996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjaden G, Coruzzi GM (1994) A novel AT-rich DNA binding protein that combines an HMG I-like DNA binding domain with a putative transcription domain. Plant Cell 6: 107–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto C, Maiwald D, Pesaresi P, Jahns P, Salamini F, Leister D (2002) The metal ion transporter IRT1 is necessary for iron homeostasis and efficient photosynthesis in Arabidopsis thaliana. Plant J 31: 589–599 [DOI] [PubMed] [Google Scholar]
- Vert G, Grotz N, Dedaldechamp F, Gaymard F, Guerinot ML, Briat JF, Curie C (2002) IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14: 1223–1233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villain P, Mache R, Zhou DX (1996) The mechanism of GT element-mediated cell type-specific transcriptional control. J Biol Chem 271: 32593–32598 [DOI] [PubMed] [Google Scholar]
- Waters BM, Blevins DG, Eide DJ (2002) Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol 129: 85–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Li L, Du J, Yuan Y, Cheng X, Ling HQ (2005) Molecular and biochemical characterization of the Fe(III) chelate reductase gene family in Arabidopsis thaliana. Plant Cell Physiol 46: 1505–1514 [DOI] [PubMed] [Google Scholar]
- Yuan YX, Zhang J, Wang DW, Ling HQ (2005) AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res 15: 613–621 [DOI] [PubMed] [Google Scholar]
- Zuo J, Niu QW, Nishizawa N, Wu Y, Kost B, Chua NH (2000) KORRIGAN, an Arabidopsis endo-1,4-β-glucanase, localizes to the cell plate by polarized targeting and is essential for cytokinesis. Plant Cell 12: 1137–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]