Abstract
Completed genome sequences provide templates for the design of genome analysis tools in orphan species lacking sequence information. To demonstrate this principle, we designed 384 PCR primer pairs to conserved exonic regions flanking introns, using Sorghum/Pennisetum expressed sequence tag alignments to the Oryza genome. Conserved-intron scanning primers (CISPs) amplified single-copy loci at 37% to 80% success rates in taxa that sample much of the approximately 50-million years of Poaceae divergence. While the conserved nature of exons fostered cross-taxon amplification, the lesser evolutionary constraints on introns enhanced single-nucleotide polymorphism detection. For example, in eight rice (Oryza sativa) genotypes, polymorphism averaged 12.1 per kb in introns but only 3.6 per kb in exons. Curiously, among 124 CISPs evaluated across Oryza, Sorghum, Pennisetum, Cynodon, Eragrostis, Zea, Triticum, and Hordeum, 23 (18.5%) seemed to be subject to rigid intron size constraints that were independent of per-nucleotide DNA sequence variation. Furthermore, we identified 487 conserved-noncoding sequence motifs in 129 CISP loci. A large CISP set (6,062 primer pairs, amplifying introns from 1,676 genes) designed using an automated pipeline showed generally higher abundance in recombinogenic than in nonrecombinogenic regions of the rice genome, thus providing relatively even distribution along genetic maps. CISPs are an effective means to explore poorly characterized genomes for both DNA polymorphism and noncoding sequence conservation on a genome-wide or candidate gene basis, and also provide anchor points for comparative genomics across a diverse range of species.
The sequencing and detailed functional analysis of the genomes of a few select botanical models opens new doors into comparative biology of the angiosperms, with great potential benefits for improvement of many orphan crops that feed large populations but are understudied at the genomic level. Among 27 orphan crops collectively planted to 250-million ha/year and yielding $100 billion (US dollars)/year farm gate value in the developing world (Naylor et al., 2004), only four (barley [Hordeum vulgare], sorghum [Sorghum spp.], cassava [Manihot esculenta Crantz.], and sunflower [Helianthus annuus]) had appreciable numbers of sequences (>10,000) in GenBank as of February 11, 2005. Pearl millet (Pennisetum glaucum) and tef (Eragrostis tef) are prime examples of such orphan crops having utmost importance in feeding millions of people (Ketema, 1997; Qi et al., 2004), yet with limited resources in GenBank. Efficient methods to leverage genomic knowledge for botanical models in the study and improvement of orphan crops will play a central role in translation of genomic discovery research into improved human nutrition.
Similarity in the repertoire, sequence, and organization of genes has the consequence that genomic information for a sampling of members of a taxonomic family (such as Poaceae, the cereals) may be used to identify gene sequences likely to be shared by as-yet unstudied (or understudied) family members such as orphan crops. Using high-throughput single-nucleotide polymorphism (SNP)-based methods, direct analysis of many genes may come to replace the indirect analyses of diagnostic DNA markers that have been the focus of the past two decades of crop genomics (Bhattramakki and Rafalski, 2001; Gupta et al., 2001). Parallel or convergent evolution of many simple and complex phenotypes (Lin et al., 1995; Paterson et al., 1995; Gale and Devos, 1998; Bennetzen and Ma, 2003; Hu et al., 2003) suggests that the analysis of candidate genes from one taxon may accelerate identification of the genetic determinants of a trait in less-studied taxa, for example by using association approaches (Thornsberry et al., 2001).
Cross utilization of genomic tools to study genetic diversity requires resolution of a fundamental conflict between the need to identify genomic sequences that are conserved (largely or wholly) across many divergent taxa, and the need to identify DNA-level differences that reflect diversity at its most elemental level. The relatively high level of conservation of the locations (Quax-Jeuken et al., 1985), but not the sequences of introns, provides a potential resolution to this dilemma. The identification of vast numbers of probable gene and intron locations in the sequences of botanical models is becoming routine, and expressed sequence tag (EST) sequence representing diverse angiosperm nodes provides a means to assess relative degrees of conservation of exons. It should be noted that EST resources have been useful in the detection of simple sequence repeat (SSR) polymorphisms within exons, some of which function across taxa (Kantety et al., 2002).
Herein, we evaluate one approach to resolving this conflict. Conserved-intron scanning primers (CISP) within relatively conserved exons located near exon-intron boundaries, are used to scan introns for variation suitable for DNA-marker identification. The recent availability of full plant genomes (Arabidopsis Genome Initiative, 2000; Feng et al., 2002; Goff et al., 2002; Sasaki et al., 2002; Yu et al., 2002) makes it possible to greatly expand on earlier related concepts previously put forth in animal studies (Palumbi and Baker, 1994; Lyons et al., 1997), permitting systematic sampling of entire genomes for well-distributed markers, or targeted enrichment of particular regions containing a gene of interest. The close proximity of introns to exons makes them especially well suited for linkage disequilibrium studies that promise to add a powerful new dimension to the understanding and improvement of crop gene pools. The Poaceae family, which diverged from common ancestors about 50-million years ago and includes both well-studied models and many orphan crops of critical nutritional and economic importance, serves as an excellent test case in which to explore the strengths and limitations of the method.
RESULTS
Pan-Poaceae PCR Amplification of Orthologs by CISPs
To design a pilot set of grass CISPs, we aligned sorghum (Sorghum bicolor and Sorghum propinquum) or buffelgrass (Pennisetum ciliare) EST sets to the Oryza (subsp. japonica) sequence. Sorghum and Pennisetum represent warm-season (C4) Panicoideae while Oryza represents cool-season (C3) Oryzoideae, thus these conserved regions have been maintained for about 42-million years of divergence. We hypothesized that the requirement of near-perfect conservation (0–1 mismatch) of an exon for CISP design (described in “Materials and Methods” and in Fig. 1) would increase the likelihood that these primers worked in additional grasses.
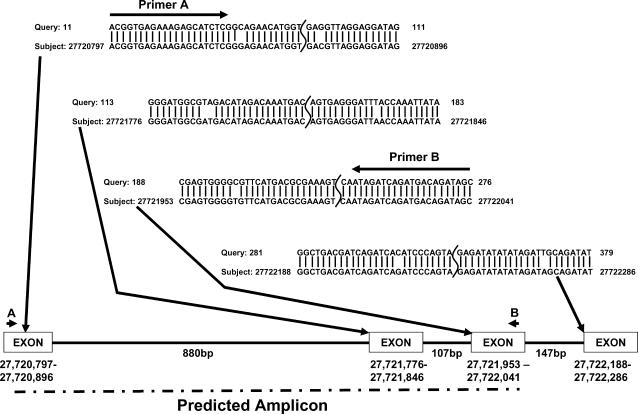
Figure 1.
CISP design process. An example alignment of a Pennisetum EST aligned to rice chromosome 4 is shown. The EST was split into four alignments by putative introns. Two primers are shown along with the predicted amplicon size.
A total of 384 CISP pairs (Supplemental Table I) were designed from 72 Oryza-Pennisetum and 312 Oryza-Sorghum alignments. The actual primer sequence was that of Oryza in all cases. On the source taxa, Sorghum and Oryza, similarly high percentages of 83.1% and 71.4% of primer sets amplified single bands, consistent with success rates for well-designed homologous primers. Using the Oryza-Sorghum-derived primer sets, a success rate of 55.4% for pearl millet (a panicoid but in the Paniceae group in contrast to the Andropogoneae for Sorghum) provided a first assessment of the degree to which the CISPs may work in orphan crops. A similar success rate of 57.3% for Cynodon, a member of the Chloridoideae, provided stronger support in suggesting that the CISPs would work outside of the taxonomic lineages used in primer design.
One application of these primer sets is the generation of anchor points between genomes, so it is important to verify that orthologous loci are amplified. At the primer design level, we applied filtering criteria that selected for single-copy rice (Oryza sativa) loci thereby reducing the chance of amplifying paralogous rice sequences. At the sequencing stage, we further reduced the possibility of sequencing-duplicated loci by selecting single-band PCR products. It is possible that paralogous loci of identical band size were occasionally amplified, but we do not see that as a major problem for two reasons. First, the sequences were trimmed for low-quality regions that would remove mixed sequence reads. In addition, polymorphisms were called with a high-quality criterion, so if high identity paralogs were in the same sequencing reaction, a false polymorphism would likely be removed due to a low-quality base call. Second, we BLAST aligned all the sequences to the rice genome and found which sequences derived from the same primer set hit the intended genomic position. Of the 215 loci that were successfully amplified and sequenced, only 11 (5.1%) hit an unexpected genomic region. Six of these unexpected hits were only seen in one species, so it is possible that these primer sets may amplify an orthologous yet unexpected region in other grasses. Furthermore, we BLAST aligned amplified rice and sorghum CISP sequences to 3,214,668 S. bicolor reads in the National Center for Biotechnology Information trace archive (approximately 3× coverage of the sorghum genome). Sixty-four percent of the rice and sorghum sequences derived from the same CISP primer set showed the same best BLAST hit. Therefore, we believe that a majority of the CISP sets will generate informative probes and genetic markers for comparative genome analysis.
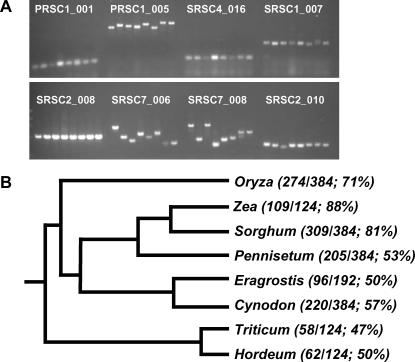
Out of 384 designed primer sets, 124 (32%) amplified successfully in all four test grasses (Oryza, Sorghum, Pennisetum, and Cynodon). These pan-grass primers (listed in Supplemental Table I) were tested for their ability to amplify across a wider evolutionary range of monocots (Fig. 2A), in particular sampling additional Chloridoid (Eragrostis) and Panicoid (Zea), as well as two Pooid grasses (Triticum and Hordeum). PCR success rates for Zea (88%) closely follow those for the other panicoids, while those for Eragrostis (50%) and the two pooids (47% and 50%) reinforce the utility of CISPs outside of the lineages used in primer design (Fig. 2B).
Figure 2.
Effectiveness of CISPs across the grass family. A, Representative 1.5% agarose gel of eight CISP-PCR products. From left to right: Oryza, Sorghum, Zea, Pennisetum, Cynodon, Eragrostis, Triticum, and Hordeum. B, Approximate dendrogram illustrating the evolutionary relationship of monocots used in this study. The numbers in parentheses are successful PCR reactions of the total tested CISP sets.
DNA Polymorphism Detection
The number of loci that were sequenced from at least two genotypes and could therefore be scanned for polymorphisms (i.e. scannable loci) were 114, 167, 110, and 59 for Oryza, Sorghum, Pennisetum, and Cynodon, respectively (Table I). DNA polymorphisms (SNP and insertion-deletion [INDEL]) were detected from ClustalW-derived forced alignments and phred-derived quality scores. Inclusive of all genotypes tested, 73.7%, 58.1%, 35.5%, and 23.7% of scannable loci were polymorphic for Oryza, Sorghum, Pennisetum, and Cynodon, respectively (Table I). The higher number of polymorphic loci in Oryza and Sorghum is probably due to the larger number of genotypes compared (eight and three, respectively). The reduced success rate in Cynodon is probably due to one genotype being an autotetraploid (Cynodon dactylon). Since an autotetraploid may contain up to four alleles at a locus, a frame shift in any one allele would result in gibberish sequence and would reduce the number of loci for which both reads are interpretable from two genotypes. The breakdown of the polymorphism types for each species is shown in Supplemental Figure 1.
Table I.
PCR and sequencing success rates
| Trait
|
PCR Success
|
Genotypes Sequenced per Locus
|
Scannable Loci
|
||||
|---|---|---|---|---|---|---|---|
| 0a | 1b | 2+c | MONO | POLY | Percent Polymorphic | ||
| Oryza | 274/384 | 12 | 8 | 114 | 30 | 84 | 73.7% |
| Sorghum | 309/384 | 61 | 34 | 167 | 70 | 97 | 58.1% |
| Pennisetum | 205/384 | 58 | 29 | 110 | 71 | 39 | 35.5% |
| Cynodon | 220/384 | 83 | 41 | 59 | 45 | 14 | 23.7% |
Zero quality reads.
One genotype with a quality read.
Two or more genotypes with quality reads (i.e. scannable locus).
A breakdown of the polymorphic versus monomorphic loci on a per-genotype basis is shown in Supplemental Table II. The overall polymorphism rates are shown in Supplemental Figure 2. The exact polymorphisms and flanking nucleotide sequence for each genotype can be found at http://www.plantgenome.uga.edu/CISP/. In addition to SNP and INDEL polymorphisms, we searched all sequenced loci for SSR signatures (2–6 bp repeats), even if there was only a single read for the locus. We found that 9/122 Oryza, 19/201 Sorghum, 15/139 Pennisetum, and 2/100 Cynodon loci contained putative SSRs (Supplemental Table III). This demonstrates that intron scanning with CISPs is able to detect various marker classes.
We also compared the efficacy of detecting polymorphisms in forced sequence alignments with that of widely used Polybayes (Marth et al., 1999). Polybayes looks for polymorphisms in multiple sequence alignments in single-nucleotide slices, then uses a Bayesian model to distinguish paralogs, sequencing errors, and inferred SNPs. Polybayes is only able to detect single-base polymorphisms as opposed to our method that is capable of detecting extended polymorphisms. While our method does not employ a paralog/sequencing error detection mechanism by virtue of the primer design strategy, virtually all reads tested here should be from a single orthologous locus within a species (as confirmed above for rice) and we used a high-quality cutoff (Q ≥ 20) that should decrease false-positive SNPs. Our method found many more polymorphisms than Polybayes (Supplemental Fig. 3), mainly due to the fact that Polybayes does not identify extended polymorphisms and cannot use forced alignments as input.
Intron Size Stability and Conserved Noncoding Motifs at Multiple CISP Loci
Across eight taxa, intron length was constant for a remarkably high number; 23 (19%) of the 118 loci studied (see Fig. 2A for examples and Supplemental Table I). Interestingly, nucleotide diversity does not correlate with implied intron size, and the average polymorphism rate was not statistically different between introns that showed static length (S-loci) and those that showed different lengths across taxa (P = 0.67). Furthermore, although short (8–20 bp) conserved sequences occur between genera in the S-loci (data not shown), there are clear differences in sequence as would be expected in noncoding DNA. Therefore, it appears that in about 19% of cases, individual nucleotides are free to evolve in S-loci, yet the intron size is constrained.
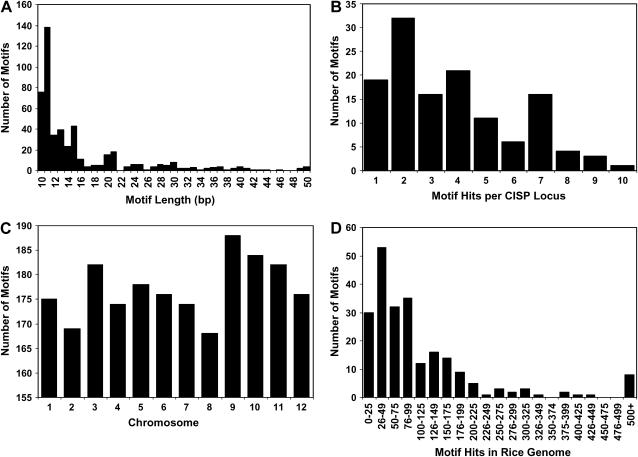
In addition to the intron size constraints, we were able to detect conserved-noncoding sequence (CNS) motifs that contain two or more sequence reads in at least two species. First, sequences were collapsed into a consensus sequence for each genus from which at least one read was available. Next, these sequences were then masked of all known grass ESTs (see “Materials and Methods”) to remove transcriptionally active DNA. Then, motifs were discovered in 129 CISP loci using the motif elicitation program, MEME (Bailey and Elkan, 1995). A total of 487 CNS motifs in 129 loci were identified that were 10 to 50 bp long (Fig. 3). Most of the motifs were between 10 to 15 bp in length (Fig. 3A), and there were between one and 10 motif hits per locus (Fig. 3B). These motifs were then searched for in the rice genome using the motif search program, MAST (Bailey and Gribskov, 1998). The number of hits per chromosome is shown in Figure 3C, and there was a significant difference in the number of hits between chromosomes. Motif hit frequency ranged from 4.1 hits/megabase (chromosome 1) to 8.9 hits/megabase (chromosome 9). Most of the motifs had a genome hit frequency of 1 to 100 (65.8%), while 22.4% hit the genome 101 to 200 times (Fig. 3D). The position-specific scoring matrices for these motifs are available as supplemental data.
Figure 3.
CNS intronic motifs. A total of 487 grass CNS motifs were identified in 129 CISP using the MEME/MAST system. A, Motif length distribution is shown for motifs (>10 bp). B, The frequency of motif occurrence is shown in the 129 CISP loci. C, CNS motif hits per rice chromosome. D, Number of motifs that hit the rice genome in given ranges.
Genomic Distribution of CISP Loci
To evaluate the probable genome coverage of CISPs, we assessed their physical distributions in rice. Much of the genome is sampled by our relatively small number of tested CISP sets (Fig. 4). To expand genomic coverage, we used EC_oligos (Liu et al., 2004) and in-house software to design all possible CISP sets between a 17,714 sorghum unigene set and 2,074 annotated rice bacterial artificial chromosome (BAC) sequences representing 61% of the rice genome. This resulted in 6,062 CISP pairs that should amplify 1,676 (9.5%) unique Sorghum unigenes, and would permit one to evaluate either single or multiple sites within these genes. Assuming a Sorghum gene space of 30,000 genes, this technique could provide approximately 5.6% Sorghum transcriptome coverage. The total CISP number will increase with further characterization of the sorghum gene space (expansion of the unigene set), and inclusion of more annotated rice BACs. It should be noted that these primer sets were not tested at the bench, but they were designed with the same criteria as the manually designed CISPs for which empirical testing is provided (88% PCR success rate in Sorghum).
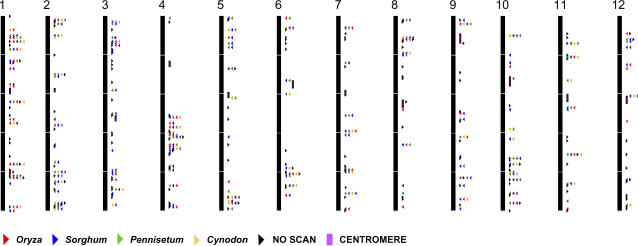
Figure 4.
Distribution of CISP loci within the rice genome. Primer sets indicate where a locus was successfully scanned for polymorphisms in at least two genotypes from an individual species (red triangle, Oryza; blue triangle, Sorghum; green triangle, Pennisetum; orange triangle, Cynodon; black triangle, unscannable locus; and purple box, centromere).
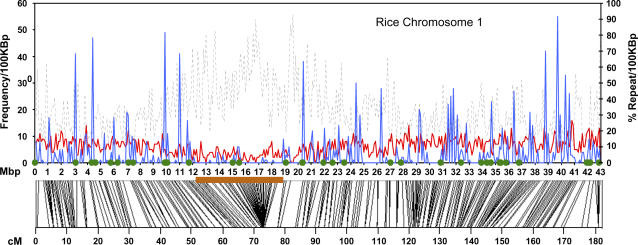
The distribution of the 6,062 primer sets across the rice genome showed enrichment in recombinogenic regions, with chronic gaps near the centromeres. An example can be seen in rice chromosome 1 in which the CISP frequency drops off around the centromere, which is rich in repetitive DNA (Feltus et al., 2004) and where Rice Genome Project genetic marker (http://rgp.dna.affrc.go.jp/publicdata/geneticmap2000/index.html) recombination is clearly inhibited (Fig. 5, region from 13–18 Mb). As a formal test of the association of CISPs with recombinogenicity, we estimated the portions of the rice genome assembly that are heterochromatic or euchromatic (respectively) based upon the cytological studies of Cheng et al. (2001). The average CISP frequency was 1.06 CISPs/kb in euchromatin versus 0.32 CISPs/kb in heterochromatin, a statistically significant difference (P = 1.4 × 10−20). These results suggest that CISPs are more frequently found in recombinogenic, euchromatic regions.
Figure 5.
CISP density correlates with recombogenicity. Rice chromosome 1 is shown. Top section, The left y axis shows the frequency of EST-verified genes (solid red line) and CISPs (solid blue line) per 100-kb interval. The right y axis shows the percent repetitive DNA (dotted black line). Green dots show the location of CISP primer sets tested in this study. The x axis shows the base pair location on the chromosome in Megabases. Bottom section, Rice Genome Project genetic marker locations are shown as lines to their chromosomal position (top x axis). The brown box indicates the estimated heterochromatic region. The bottom x axis shows the centiMorgan location on rice linkage group one.
DISCUSSION
CISPs can provide large numbers of pan-grass tools suitable for linking genomics research in many orphan crops of critical nutritional and economic importance but that lack appreciable sequence information, to burgeoning knowledge in botanical models and better-studied crops. About one-half of CISPs worked in individual taxa for which DNA sequence information was either lacking (Chloridoids) or was not considered in primer design (Pooids), and one-third (124/384) worked in all Panicoid, Chloridoid, and Oryzoid grasses tested.
CISP loci make excellent anchor points for comparative genomics in grasses and other crops, balancing the need for conservation across taxa with the need to detect diversity within a taxon. Prior knowledge of colinearity/synteny among well-studied crops provides an initial framework from which to make educated guesses about regions of fully sequenced genomes from which to seek CISPs likely to be informative in Panicoid, Chloridoid, Oryzoid, or Pooid orphan crops. Initial small samples of CISPs will provide de novo information, either confirming predicted relationships or revealing taxon-specific rearrangements. Once comparative relationships are validated, then targeted enrichments of specific genomic regions become feasible: by loosening the strict selection criteria (<2 nucleotide difference) or using alternative design criteria, one might design CISP-like primers for a much larger fraction of the gene space including specific functional candidates.
Utilization of introns as the primary amplicon yielded high levels of DNA polymorphism and increased the scannable genome space. Introns have less evolutionary constraint than exons and should therefore be more likely to identify polymorphism. We verified this in our Oryza set (Supplemental Table IV): On a per-locus basis intron polymorphisms averaged 12.1/kb while exon polymorphisms averaged 3.6/kb (a difference that was statistically significant at P = 0.0003). In addition, exons can be quite small, so implied intron size can be used as a design criterion to a degree appropriate for experimental goals. For example, in initial mapping of closely related genotypes, fewer longer introns might be preferred to maximize the number of loci that yield polymorphisms. By contrast, evaluation of high-priority candidate genes in targeted regions might warrant scanning of all introns to increase information for association or linkage disequilibrium studies.
In some taxa, recent gene or genome duplications complicate SNP detection; for example, many CISP loci that amplified in Cynodon were uninformative due to its autopolyploidy. Whether or not the CISP locus has been recently duplicated in another grass must be assessed, although we tried to filter recently duplicated loci in rice from consideration. Gene duplications that trace back to the approximately 70-million years ago event that affected all Poaceae genomes (Paterson et al., 2003, 2004) usually are sufficiently diverged that locus-specific primers can be designed.
Sequence orthology plus improvements in polymorphism detection increased overall efficiency. Targeting of primers to conserved low-copy exons assures that virtually all PCR products from different genotypes within a genus were orthologous and permitted us to force alignments. This is a different paradigm from contig building and subsequent polymorphism detection in large sequence samples such as ESTs, which must be assumed to contain paralogs. The extended detection capabilities of our method significantly increase the number of polymorphisms detected relative to Polybayes (Marth et al., 1999).
The CISPs validated herein represent only a small sample of the numbers available, using an automated framework for design and testing. We demonstrated this with the 6,062 Oryza-Sorghum unigene sets derived from 17,714 Sorghum unigenes and 2,074 annotated Oryza BACS. While the Panicoid, Oryzoid, and Pooid grasses enjoy a wealth of EST resources that help to guide CISP selection and design, valuable additions to round out our knowledge of Poaceae transcriptomes would be substantial EST collections for members of the Arundinoid, Bambusoid, and Chloridoid grasses. While CISPs tend to be localized to recombinogenic regions of the genome, this is where most linkage and association information is derived anyway, so we do not see this as a large impediment.
Further study of many CISPs in diverse taxa may shed new light on intron evolution. For 19% of tested loci, amplicon (essentially intron) size remained fixed across all taxa tested while per-nucleotide polymorphism rates were indistinguishable from those found among variably sized introns. Growing evidence suggests that recombination rates have a strong influence on intron length, with longer introns in regions of low recombination (Carvalho and Clark, 1999; Comeron and Kreitman, 2000; Lynch, 2002). However, we found no tendency for the S-loci to be enriched in low recombinogenic regions relative to the different-length loci on a gross scale (Supplemental Fig. 2). Further, this would not explain why introns would be fixed in length, despite considerable sequence polymorphism, across 50 million years of taxonomic divergence. It is unclear why some introns remain fixed in length and the answer will require additional gene structure data from divergent species and wet-lab testing.
Multiple reports of CNSs have appeared for plants and animals (Inada et al., 2003; Shin et al., 2005; Woolfe et al., 2005). Herein, we identify 487 conserved motifs of unknown function. Due to the presence of these sequences within the same intron, we speculate that these motifs are involved with the gene in which they are found, possibly in a regulatory or other role. However, it cannot be ruled out that these sequences are affecting nearby linked genes. The discovery of CNSs and fixed introns demonstrates a separate utility for CISP primers described here, beyond their utility in polymorphism discovery.
MATERIALS AND METHODS
Plant Materials
Twenty-two genotypes from eight genera were tested: rice (Oryza sativa IR20, IR52561, IR58821, IR62266, IR64, Nootripathu, CT9993, and Azucena), sorghum (Sorghum propinquum, Sorghum Bicolor BTx623, and IS18551), maize (Zea mays CML268), pearl millet (Pennisetum glaucum 841B and 863B), bermuda grass (Cynodon dactylon T89 [4X] and Cynodon transvaalensis T574 [2X]), tef (Eragrostis tef Kaua murri and Eragrostis pilosa), wheat (Triticum aestivum M6 and Opata), and barley (Hordeum vulgare Steptoe and Morex).
Manual CISP Design
Primers were designed to span introns (CISPs) and be located within highly conserved (0–1 nucleotide mismatch) exons. ESTs from buffelgrass (Pennisetum ciliare), pearl millet, and sorghum (S. bicolor/S. propinquum) were aligned to targeted segments of rice chromosomes 1 to 12 (The Institute for Genomic Research [TIGR] v1.0, www.tigr.org) using BLASTN (E ≤ 1 × 10−10). Redundant hits were removed and PCR primers were designed by hand inspection from highly conserved (0–2 mismatches) alignments between Oryza-Pennisetum or Oryza-Sorghum considering intergeneric sequence conservation (Fig. 1). Primer design criteria included implied intron size (200–1,500 bp), oligonucleotide melting temperature (58°C–62°C), size range (18–22 bp), gas chromatography content (50%), and primer-dimer formation potential (minimized). In particular, design criteria were focused on compatibility with common sets of PCR conditions (specifically, target annealing temperatures of 55°C/60°C). A total of 384 intron-scanning primer sets were designed, synthesized, and tested (Supplemental Table I).
Automated CISP Design
EC_oligos (Liu et al., 2004) software was used to design all possible intron-spanning conserved PCR primer sets between a sorghum unigene set (Feltus et al., 2004) and annotated rice BAC sequences. All 17,714 sorghum unigenes were BLAST aligned to the rice genome (E < 1 × e−10; TIGRv2 [ftp://ftp.tigr.org/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_2.0/]). Unigenes with at least two exons that hit only a single chromosome (15,403 high-scoring segment pairs/4,768 unigenes) were identified and placed into FASTA input file for EC_oligos. The second input file for EC_oligos was 2,074 coding sequence-annotated japonica rice BACs (423 MB) downloaded from GenBank (http://www.ncbi.nlm.nih.gov/) on August 2, 2004 (Entrez query string: txid39947[Organism:noexp] 50000:200000[SLEN] AND cds NOT chloroplast NOT mitochondria). CISPs were then discovered with EC_oligos with the command line: EC_oligos -f A.genbank −2 B.fasta -v -s -p 200 2000 -d 2 -t 58 62 -a 1024. An in-house Perl script parsed 271,848 CISP sets from the output file. These CISP sets were BLAST aligned (E ≤ 0.002) to the rice genome (TIGRv2) and the results loaded into an ACCESS (Microsoft) database. Of 193,961 that were verified to span introns, 188,672 matched rice at only a single locus, and 6,062 were unique and nonoverlapping. All CISP sets can be found at http://www.plantgenome.uga.edu/CISP/.
PCR Product Sequencing
PCR buffer conditions were the same for all primers. Reaction mixtures included 1 ng/μL genomic DNA, 0.2 mm dNTPs (Amersham), 1.25 units of Taq (Promega), 0.0626 units cloned Pfu (Stratagene), 3.0 μm degenerate primer, 4 μm of MgSO4, and 3 μL 10× Cloned Pfu buffer (Stratagene) in a total reaction volume of 30 μL. PCR (MJ Research PTC-100) cycling parameters were similar in all cases. Cycling parameters were repeated for 35 cycles: 94°C for 5 min, 94°C for 30 s, 55°C or 60°C for 45 s, 72°C for 60 s, and a final extension 72°C for 10 min. PCR products were visualized on a 1.5% agarose gel stained with ethidium bromide. Loci were classified (0–3) according to whether they produced no product (0), a single band (1), two bands (2), or three or more bands (3). Relative differences in product size were noted between species.
Prior to sequencing, PCR product cleanup involved enzymatic digestion with Exonuclease I/shrimp alkaline phosphatase, adding 5 μL of a mixture of 1% Exonuclease I and 10% shrimp alkaline phosphatase to 25 μL of PCR product, followed by a brief centrifugation then incubation at 37°C for 15 min (to react) and 80°C for 15 min (to terminate reaction). Cleaned high-quality PCR products were amplified using the ABI Big Dye 3.1 cycle sequencing kit (Applied Biosystems) and standard protocols. Finished cycle sequencing reaction products were treated with a dilute (2.2%) SDS solution then passed through homemade Sephadex filter plates into Perkin-Elmer MicroAmp Optical 96-well reaction plates. The filtered sequence reaction products were analyzed on an ABI Prism 3700 or 3730 automated DNA sequencer (Applied Biosystems).
Polymorphism Detection
Trace files for each locus were divided into separate projects in a genus-specific manner using the phred (www.phrap.org; Ewing et al., 1998) directory structure. Reads were base called, sequence quality determined, and end trimmed with phred (trim). Phred-processed sequences and quality scores were used as input to DNA polymorphism detection.
Sequences were aligned by ClustalW (Chenna et al., 2003) in a locus- and species-specific manner. The GDE-format alignment and corresponding quality files for each locus were adjusted to match, and loci with at least one read from two genotypes were scanned for high-quality polymorphisms (Q ≥ 20) using in-house Python scripts (S.R. Schulze and A.H. Paterson, unpublished data). Three basic polymorphism classifications were used: SNPs, polymorphisms that extend for more than one base (EXTENDED), and insertion/deletions (INDEL).
We also searched for single-base INDELs and SNPs with Polybayes (Marth et al., 1999). We used Phrap (www.phrap.org) aligned sequences from each locus project folder as input to Polybayes (priorPoly 0.001) with paralog filtering off.
CNS Detection
DNA sequences from each amplified loci were aligned in a species-dependent manner. Aligned contigs (and singletons) were converted into consensus sequences using CONS (EMBOSS; Olson et al., 2002). The best consensus sequence for each taxa was placed into a single FASTA file and masked for transcribed sequences using Cross_match (www.phrap.org; minmatch 10 and minscore 20). The mask file contained 2,212,811 mRNA sequences (as defined by GenBank) for the grass family Poaceae (txid4479). CNSs were then detected in the transcription-masked sequences with the motif elicitation program, MEME (mod oops, nmotifs 20, minw 10, maxw 50, and revcomp; Bailey and Elkan, 1995). The best scoring (LLR ≥ 14) position-specific scoring matrix was extracted from the MEME output using custom Perl scripts and used to probe the rice genome (TIGRv2; http://www.tigr.org) with MAST (mt 1 × e−7; Bailey and Gribskov, 1998). MAST output was parsed with custom Perl scripts.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers CW883294 to CW884906.
Supplementary Material
Acknowledgments
We thank A. Reddy, R.C. Babu, and S. McCouch for rice DNA, and M. Sorrells for tef, barley, and wheat DNA.
This work was supported by grants from the Rockefeller Foundation and the U.S. Agency for International Development Cereals Comparative Genomics Initiative (to A.H.P., F.A.F., and H.P.S.); the International Society for Plant Molecular Biology (to T.D.S.); and the Biotechnology Overseas Associateship of the Department of Biotechnology, Ministry of Science and Technology, Government of India program (to H.C.L.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: A.H. Paterson (paterson@uga.edu).
The online version of this article contains Web-only data.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Elkan CP (1995) Unsupervised learning of multiple motifs in biopolymers using expectation maximization. Machine Learning 21: 51–83 [Google Scholar]
- Bailey TL, Gribskov M (1998) Methods and statistics for combining motif match scores. J Comput Biol 5: 211–221 [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Ma J (2003) The genetic colinearity of rice and other cereals on the basis of genomic sequence analysis. Curr Opin Plant Biol 6: 128–133 [DOI] [PubMed] [Google Scholar]
- Bhattramakki D, Rafalski A (2001) Discovery and application of single nucleotide polymorphism markers in plants. In RJ Henry, ed, Plant Genotyping: The DNA Fingerprinting of Plants. CABI Publishing, Wallingford, UK
- Carvalho AB, Clark AG (1999) Intron size and natural selection. Nature 401: 344. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11: 2133–2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron JM, Kreitman M (2000) The correlation between intron length and recombination in drosophila: dynamic equilibrium between mutational and selective forces. Genetics 156: 1175–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing B, Hillier L, Wendl MC, Green P, Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, et al (1998) Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res 8: 175–185 [DOI] [PubMed] [Google Scholar]
- Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH (2004) An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res 14: 1812–1819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Zhang Y, Hao P, Wang S, Fu G, Huang Y, Li Y, Zhu J, Liu Y, Hu X, et al (2002) Sequence and analysis of rice chromosome 4. Nature 420: 316–320 [DOI] [PubMed] [Google Scholar]
- Gale MD, Devos KM (1998) Comparative genetics in the grasses. Proc Natl Acad Sci USA 95: 1971–1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296: 92–100 [DOI] [PubMed] [Google Scholar]
- Gupta G, Roy JK, Prasad M (2001) Single nucleotide polymorphisms: a new paradigm for molecular marker technology and DNA polymorphism detection with emphasis on their use in plants. Curr Sci 80: 524–535 [Google Scholar]
- Hu FY, Tao DY, Sacks E, Fu BY, Xu P, Li J, Yang Y, McNally K, Khush GS, Paterson AH, et al (2003) Convergent evolution of perenniality in rice and sorghum. Proc Natl Acad Sci USA 100: 4050–4054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inada DC, Bashir A, Lee C, Thomas BC, Ko C, Goff SA, Freeling M (2003) Conserved noncoding sequences in the grasses. Genome Res 13: 2030–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48: 501–510 [DOI] [PubMed] [Google Scholar]
- Ketema S (1997) Promoting the Conservation and Use of Underutilized and Neglected Crops. International Plant Genetic Resources Institute, Rome
- Lin YR, Schertz KF, Paterson AH (1995) Comparative analysis of QTLs affecting plant height and maturity across the Poaceae, in reference to an interspecific sorghum population. Genetics 141: 391–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Tinker NA, Molnar SJ, Mather DE (2004) EC_oligos: automated and whole-genome primer design for exons within one or between two genomes. Bioinformatics 20: 3668–3669 [DOI] [PubMed] [Google Scholar]
- Lynch M (2002) Intron evolution as a population-genetic process. Proc Natl Acad Sci USA 99: 6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons LA, Laughlin TF, Copeland NG, Jenkins NA, Womack JE, O'Brien SJ (1997) Comparative anchor tagged sequences (CATS) for integrative mapping of mammalian genomes. Nat Genet 15: 47–56 [DOI] [PubMed] [Google Scholar]
- Marth GT, Korf I, Yandell MD, Yeh RT, Gu Z, Zakeri H, Stitziel NO, Hillier L, Kwok PY, Gish WR, et al (1999) A general approach to single-nucleotide polymorphism discovery. Nat Genet 23: 452–456 [DOI] [PubMed] [Google Scholar]
- Naylor RL, Falcon WP, Goodman RM, Jahn MM, Sengooba T, Tefera H, Nelson RJ (2004) Biotechnology in the developing world: a case for increased investments in orphan crops. Food Policy 29: 15–44 [Google Scholar]
- Olson SA, Bailey TL, Gribskov M, Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2002) EMBOSS opens up sequence analysis: European molecular biology open software suite. Brief Bioinform 3: 87–91 [DOI] [PubMed] [Google Scholar]
- Palumbi SR, Baker CS (1994) Contrasting population structure from nuclear intron sequences and mtDNA of humpback whales. Mol Biol Evol 11: 426–435 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Chapman BA (2004) Ancient polyploidization predating divergence of the cereals, and its consequences for comparative genomics. Proc Natl Acad Sci USA 101: 9903–9908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Peterson DG, Estill JC, Chapman BA (2003) Structure and evolution of cereal genomes. Curr Opin Genet Dev 13: 644–650 [DOI] [PubMed] [Google Scholar]
- Paterson AH, Lin YR, Li Z, Schertz KF, Doebley JF, Pinson SRM, Liu S-C, Stansel JW, Irvine JE (1995) Convergent domestication of cereal crops by independent mutations at corresponding genetic loci. Science 269: 1714–1718 [DOI] [PubMed] [Google Scholar]
- Qi X, Pittaway TS, Lindup S, Liu H, Waterman E, Padi FK, Hash CT, Zhu J, Gale MD, Devos KM (2004) An integrated genetic map and a new set of simple sequence repeat markers for pearl millet, Pennisetum glaucum. Theor Appl Genet 109: 1485–1493 [DOI] [PubMed] [Google Scholar]
- Quax-Jeuken Y, Quax W, van Rens G, Khan PM, Bloemendal H (1985) Complete structure of the alpha B-crystallin gene: conservation of the exon-intron distribution in the two nonlinked alpha-crystallin genes. Proc Natl Acad Sci USA 82: 5819–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Matsumoto T, Yamamoto K, Sakata K, Baba T, Katayose Y, Wu J, Niimura Y, Cheng Z, Nagamura Y, et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420: 312–316 [DOI] [PubMed] [Google Scholar]
- Shin JT, Priest JR, Ovcharenko I, Ronco A, Moore RK, Burns CG, MacRae CA (2005) Human-zebrafish non-coding conserved elements act in vivo to regulate transcription. Nucleic Acids Res 33: 5437–5445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornsberry JM, Goodman MM, Doebley J, Kresovich S, Nielsen D, Buckler ES (2001) Dwarf8 polymorphisms associate with variation in flowering time. Nat Genet 28: 286–289 [DOI] [PubMed] [Google Scholar]
- Woolfe A, Goodson M, Goode DK, Snell P, McEwen GK, Vavouri T, Smith SF, North P, Callaway H, Kelly K, et al (2005) Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol 3: e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Hu S, Wang J, Wong GK, Li S, Liu B, Deng Y, Dai L, Zhou Y, Zhang X, et al (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. indica). Science 296: 79–92 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.