Abstract
Plants and animals share similar mechanisms in the heat shock (HS) response, such as synthesis of the conserved HS proteins (Hsps). However, because plants are confined to a growing environment, in general they require unique features to cope with heat stress. Here, we report on the analysis of the function of a novel Hsp, heat-stress-associated 32-kD protein (Hsa32), which is highly conserved in land plants but absent in most other organisms. The gene responds to HS at the transcriptional level in moss (Physcomitrella patens), Arabidopsis (Arabidopsis thaliana), and rice (Oryza sativa). Like other Hsps, Hsa32 protein accumulates greatly in Arabidopsis seedlings after HS treatment. Disruption of Hsa32 by T-DNA insertion does not affect growth and development under normal conditions. However, the acquired thermotolerance in the knockout line was compromised following a long recovery period (>24 h) after acclimation HS treatment, when a severe HS challenge killed the mutant but not the wild-type plants, but no significant difference was observed if they were challenged within a short recovery period. Quantitative hypocotyl elongation assay also revealed that thermotolerance decayed faster in the absence of Hsa32 after a long recovery. Similar results were obtained in Arabidopsis transgenic plants with Hsa32 expression suppressed by RNA interference. Microarray analysis of the knockout mutant indicates that only the expression of Hsa32 was significantly altered in HS response. Taken together, our results suggest that Hsa32 is required not for induction but rather maintenance of acquired thermotolerance, a feature that could be important to plants.
When cells are exposed to elevated temperature, their transcription and translation machineries are reprogrammed to activate multiple protection mechanisms, a phenomenon called heat shock (HS) response (HSR; Lindquist, 1986). In HSR, transient synthesis of a group of conserved HS proteins (Hsps) is universal and has been well characterized (Lindquist and Craig, 1988; Vierling, 1991). Previous studies have shown that many of these Hsps function as molecular chaperones in maintaining homeostasis of protein folding and are thought to be responsible for the acquisition of thermotolerance (Vierling, 1991; Parsell and Lindquist, 1993; Sung et al., 2003).
With the progress of genome-wide gene expression studies employing technologies such as microarray analysis, increasing data have shown that in addition to the well-characterized and conserved Hsps, many other genes are significantly up- or down-regulated with HS treatment (Gasch et al., 2000; Helmann et al., 2001; Shockley et al., 2003; Pysz et al., 2004; Rizhsky et al., 2004; Busch et al., 2005). In general, genes involved in protein folding and degradation, carbohydrate metabolism, reactive oxygen species scavenging, and signaling are up-regulated by HS. However, the functions of many HSR genes being unknown hinders a complete understanding of the HSR. Effort needs to be devoted to identifying the function of these genes in stress tolerance (Sung et al., 2003).
Land plants are sessile and constantly experiencing periodic temperature fluctuation. Thus, we hypothesize that plant-specific HS-responsive genes might be beneficial for plants to cope with heat stress. To identify and characterize possible plant-specific features in the HSR, we previously isolated a novel Hsp gene from a tomato (Lycopersicon esculentum) cDNA library generated by subtractive hybridization (Liu et al., 2006b). This putative protein does not share significant homology to any well-characterized Hsp. To avoid confusion with the known Hsp, we have named this gene Hsa32 for encoding a heat-stress-associated 32-kD protein. Homologous sequences of Hsa32 were found in the expressed sequence tag databases for various plant species but were absent in those for most other organisms. The gene exists as a single-copy gene in the Arabidopsis (Arabidopsis thaliana; At4g21320), rice (Oryza sativa; Os06g46900), and tomato genomes. Microarray study showed that Hsa32 is one of the top candidate targets of the key HS transcription factors, HsfA1a and/or HsfA1b, in Arabidopsis (Busch et al., 2005).
Currently, the locus of At4g21320 is annotated as phosphosulfolactate (PSL) synthase-related gene in The Arabidopsis Information Resource (TAIR; www.arabidopsis.org) on the basis of a weak similarity (about 34%) with the PSL synthase of Methanococcus jannaschii (Graham et al., 2002). PSL synthase is involved in the biosynthesis of PSL, the precursor of coenzyme M required for methane production in the hyperthermophilic euryarchaeon, and possibly sulfolactate, a major component (5% dry weight) of mature spores of Bacillus subtilis (Bonsen et al., 1969; Graham et al., 2002). Sequence comparison showed that, because of the three-dimensional structure of PSL synthase, most amino acid residues proposed to interact with the substrates (Wise et al., 2003) were not conserved in the plant Hsa32 orthologs (Liu et al., 2006b), which suggests that the plant protein does not have PSL synthase activity. As well, plants do not contain the remaining genes required for biosynthesis of coenzyme M (Graham et al., 2002). Hence, the function of Hsa32 was unknown.
Here, we report on the molecular biological and genetic analysis of Hsa32. We showed that Hsa32 was up-regulated by HS from lower to higher plants, which suggests an evolutionarily conserved role in HSR. Immunoblot analysis confirmed that the Arabidopsis gene encodes a protein significantly induced by HS treatment. Reverse genetic approach revealed that in Arabidopsis, Hsa32 is essential for tolerance against a severe heat challenge after a long recovery following acclimation treatment, which is apparently due to a fast decay of thermotolerance in the absence of Hsa32. This study provides direct evidence for plants requiring unique molecular features for thermotolerance not found in other organisms.
RESULTS
Hsa32 Is a Conserved HS-Inducible Gene in Land Plants
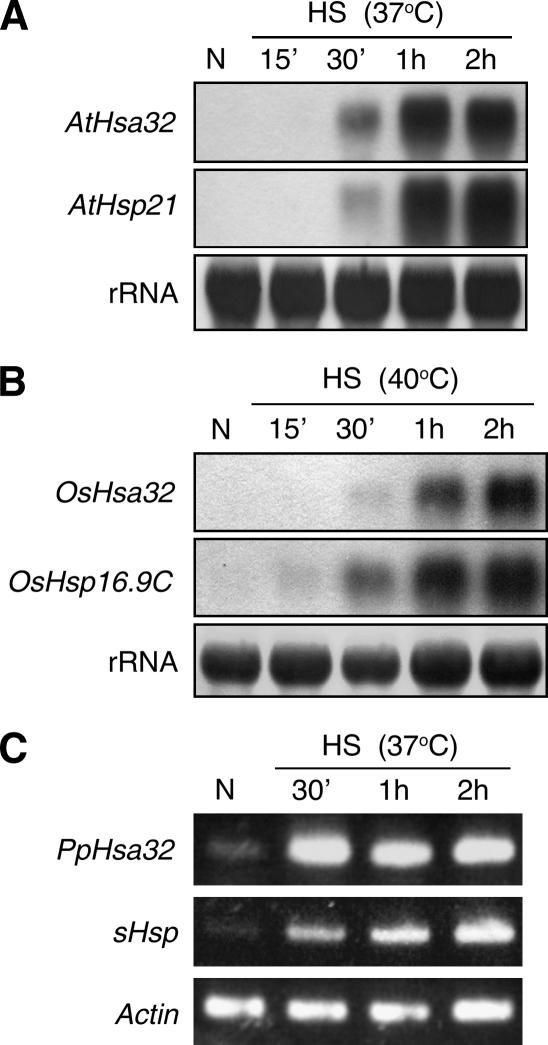
We have previously identified Hsa32 as a novel HS-responsive gene from tomato, and its orthologs are mainly found in land plants (Liu et al., 2006b). We wanted to know whether Hsa32 is also responsive to HS in a wide range of plant species. We examined the expression of Hsa32 after HS treatment in Physcomitrella patens, Arabidopsis, and rice, three commonly used model plants. Northern-blot or reverse transcription (RT)-PCR analysis showed that Hsa32 was significantly induced by HS treatment in all plants, as were the other Hsp controls (Fig. 1). These results indicate that the regulation of Hsa32 by HS is evolutionarily conserved in plant species, which suggests a common role of the gene in HSR.
Figure 1.
Hsa32 was responsive to HS from lower to higher plants. Hsa32 transcript levels in Arabidopsis (A) and rice (B) seedlings were revealed by northern blot and in P. patens (C) by RT-PCR. Plants were either without HS treatment (N) or treated at the indicated temperatures for various times. The transcript levels of Arabidopsis Hsp21, rice OsHsp16.9C, and P. patens sHsp were shown as positive controls. Ribosomal RNA (A and B) or actin (C) was shown to ensure equal loading of samples.
Hsa32 Protein Is Synthesized in Response to HS
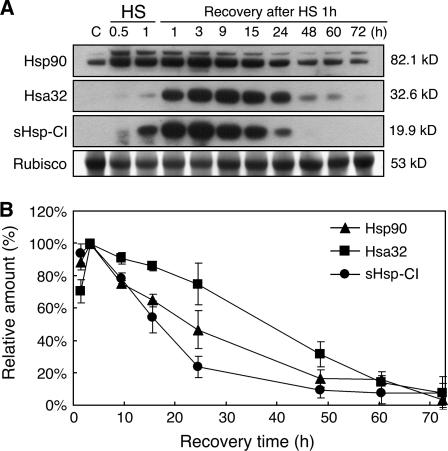
To examine the level of Hsa32, a specific antibody was raised against the Arabidopsis recombinant Hsa32 for immunoblot analysis. Arabidopsis seedlings without HS treatment showed no, or a very low level of, Hsa32, whereas a nonlethal HS treatment (37°C for 1 h) resulted in a protein band of approximately 32 kD (Fig. 2A), which is very close to the predicted size (Mr 32,638) of Arabidopsis Hsa32. However, the level was relatively low as compared with Hsp90 and class I small Hsp (sHsp-CI) during the first hour of HS treatment (Fig. 2A). After the HS treatment, the levels of Hsa32, Hsp90, and sHsp-CI all peaked at approximately 3 h, then slowly declined during recovery at room temperature, but Hsa32 declined at a relatively slower pace than that of the other two Hsps (Fig. 2B). After 72 h of recovery, a low level of Hsa32 was still detectable if the x-ray film was exposed to the chemiluminescence signals for a longer time, whereas sHsp-CI was not detected (data not shown). Calculation of the apparent half-life of the steady state for each of these Hsps resulted in variable numbers from time to time, probably because of the limitation of chemiluminescence detection method. Nevertheless, in all similar experiments, Hsa32 always decayed slower than the other two Hsps. The calculated maximal amount of Hsa32 under the tested conditions reached about 0.01% of total protein. These results confirm that Arabidopsis Hsa32 indeed responds to HS both at the transcription and translation levels. The strong induction of the protein by heat suggests that Hsa32 is involved in acquired thermotolerance. We have tried to detect the Hsa32 protein in tomato or rice samples following HS treatment with the same antibody. However, no cross-reaction was observed, probably because of low cross-reactivity of the antibody against Hsa32 from other sources.
Figure 2.
Hsa32 protein accumulates in response to HS and declines during recovery. A, Immunoblotting analysis of Hsa32, sHsp-CI, and Hsp90 in Arabidopsis 3-d-old seedlings during and after HS treatment at 37°C for indicated time. In each lane, 50 μg of protein was loaded. Rubisco large subunit stained by Amido black was shown to ensure equal loading. Calculated molecular mass of each protein based on mobility was indicated. B, Semiquantification of Hsa32, sHsp-CI, and Hsp90 levels of immunoblots according to conditions described in A. The relative amount of each protein was expressed as percentage of the highest signal detected after normalized by the level of Rubisco. The data were means of two biological repeats. Error bars represent sd.
Defect in Acquired Thermotolerance Was Manifested after a Long But Not Short Recovery in the Hsa32 Knockout Mutant
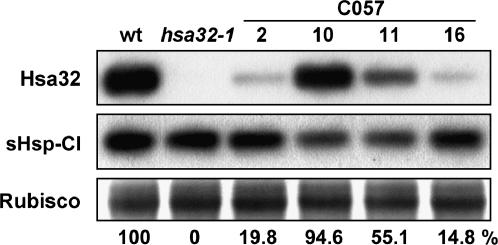
To elucidate the biological function of Hsa32, we have characterized a knockout mutant, hsa32-1, with a T-DNA insertion at the third exon of the gene. The homozygous hsa32-1 is a null mutant that did not synthesize Hsa32 mRNA (data not shown) or protein (Fig. 3) after HS treatment, which confirms that the antibody recognized the authentic Hsa32. Similarly, suppression of Hsa32-1 expression by RNA interference (RNAi) also led to a decreased level of Hsa32 protein induced by HS, whereas the induction of sHsp-CI was not affected (Fig. 3). When grown under normal conditions, hsa32-1 plants exhibited no obvious phenotypic difference in terms of germination time and rate, growth rate, time to flowering, and seed yield as compared to the wild type, which suggests that Hsa32 is not essential for normal growth and development.
Figure 3.
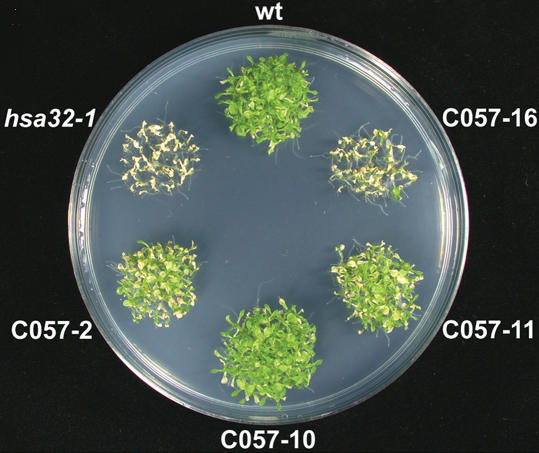
Suppression of HS induction of Hsa32 by T-DNA insertion and RNAi. The Arabidopsis wild type (wt), Hsa32 T-DNA knockout mutant (hsa32-1), and RNAi lines (C057-2, -10, -11, and -16) seedlings were treated at 37°C for 1 h and recovered for 3 h, then harvested for protein extraction. Hsa32 and sHsp-CI levels were revealed by immunoblotting. In each lane, 50 μg of protein was loaded. Rubisco large subunit stained by Amido black was shown to ensure equal loading. The numbers below indicate the relative amount of Hsa32.
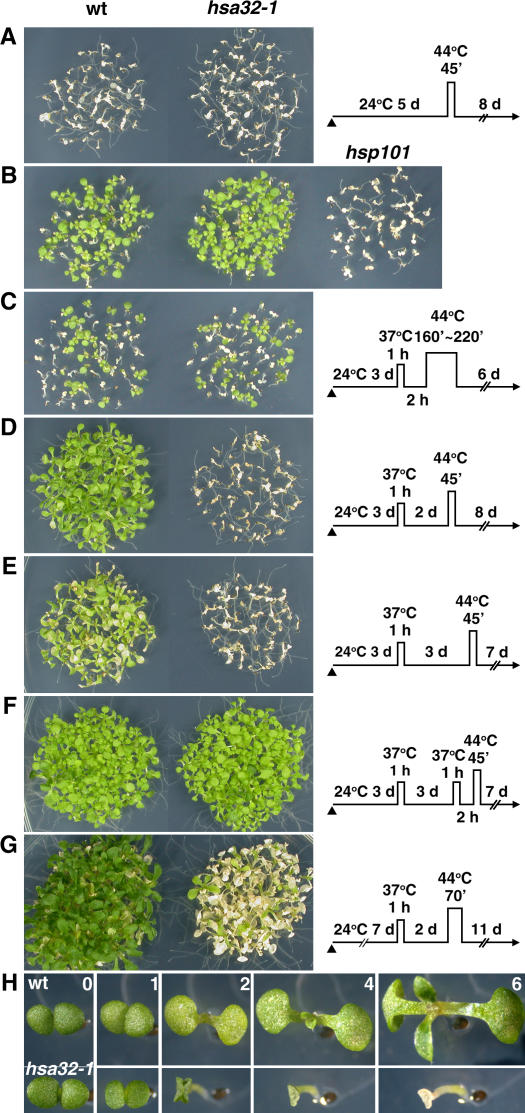
Since Hsa32 was not present in 3-d-old seedlings but was strongly induced by heat (Fig. 2A), we thus compared the development of acquired thermotolerance of the mutant and wild type at this stage. Seedlings first underwent acclimation treatment with HS at 37°C for 1 h, recovery for 2 h at 22°C, then severe HS challenge at 44°C for various times. The treatment at 44°C for 45 min killed both the wild-type and hsa32-1mutant 3- to 6-d-old seedlings without acclimation (Fig. 4A; only the results of 5-d-old seedlings are shown). The acclimation treatment led to enhanced tolerance of a similar level in the wild type and hsa32-1 against the HS challenge at 44°C up to 220 min (Fig. 4, B and C), but both lines were killed at 44°C for 250 min (data not shown). In contrast, the Hsp101 knockout line completely lost the acquired thermotolerance after treatment at 44°C for 160 min (Fig. 4B), which agrees well with previous reports that lack or mutations of Hsp101 significantly compromise thermotolerance within a short (2-h) recovery period (Hong and Vierling, 2000; Queitsch et al., 2000).
Figure 4.
Disruption of Hsa32 by T-DNA insertion leads to heat-sensitive phenotype after a long recovery. The phenotypes of the wild-type (wt) and hsa32-1 seedlings were revealed after treatment by different HS regimes schematically shown on the right of each section except B. The times of HS treatment at 44°C in B and C were 160 and 220 min, respectively. The arrowheads indicate the end of seed imbibitions. In B, phenotype of Hsp101 (At1g74310) T-DNA knockout plants (hsp101) was shown as a comparison. The survival rates of wild-type and hsa32-1 lines in C were 43% and 44%, respectively. In G, older seedlings were employed for the thermotolerance test. The plants in A to G were photographed 6 to 11 d after HS treatment. Seedlings of each section were grown on the same plate and reorganized for presentation. H shows the progression of phenotypes of representative wild-type (top) and hsa32-1 (bottom) seedlings 0 to 6 d after treatment by the same HS regime shown in D. The bar represents 1 mm.
Because of the slow degradation rate of Hsa32 during recovery (Fig. 2B), we thought that this protein might play some role in the duration of acquired thermotolerance. To test this possibility, seedlings underwent various recovery times at room temperature between the acclimation and severe HS treatments to examine the duration of tolerance to heat. In the wild type, the thermotolerance acquired by acclimation treatment lasted up to 72 h but decayed gradually, with a more pale-green color occurring with longer recovery before the challenge (Fig. 4, D and E). Again, no significant difference was observed between the wild-type and hsa32-1 seedlings if the severe HS challenge was applied after 24 h of recovery (data not shown). However, a great loss of thermotolerance was observed in the mutant if the challenge was applied after 48 h (Fig. 4D) or 72 h of recovery (Fig. 4E). This observation suggests that the apparent duration of thermotolerance of hsa32-1 was shortened under the tested conditions. The decreased thermotolerance after long recovery in hsa32-1, however, was reversible. A second acclimation treatment 2 h before the severe HS challenge after 48 h (data not shown) or 72 h (Fig. 4F) of recovery protected the hsa32-1 plants from being killed, which suggests that the acquisition of a subsequent thermotolerance does not require Hsa32 and seems to overcome the thermotolerance defect of the mutant. The death of hsa32-1 caused by HS was progressive. Immediately after the severe HS challenge following long recovery, the hsa32-1 plants appeared to be identical to the wild type. However, the cotyledons of the mutant plants started to fold up or curl after 2 d and subsequently became bleached and dry, whereas the wild-type plants continued to grow (Fig. 4H). The acquired thermotolerance of older seedlings of hsa32-1was also significantly compromised after long recovery (Fig. 4G), which suggests that the mutant phenotype did not occur only in one stage.
According to an antibiotics resistance test, the hsa32-1 mutant contains two or more T-DNA insertional events. The great loss of acquired thermotolerance of hsa32-1 shown in Figure 4, D and E, should be caused by disruption of Hsa32 instead of other T-DNA insertional events or a secondary mutation in the genome of the mutant line because suppression of Hsa32 expression by RNAi led to similar results (Fig. 5). However, some of the RNAi lines (057-2 and -11) exhibited a relatively less severe phenotype as compared with hsa32-1 and 057-16, probably because of a relatively higher level of Hsa32 remaining in these lines (Fig. 3). Nevertheless, the protein level of Hsa32 seemed to correlate well with thermotolerance in these transgenic lines. Genetic analysis showed that the hsa32-1 allele is recessive since the Hsa32/hsa32-1 heterozygote exhibited a wild-type phenotype, and about one-quarter of the offspring derived from self-pollination of the heterozygous line showed the mutant phenotype, which is consistent with the classic Mendelian ratio.
Figure 5.
Heat sensitivity of Hsa32 RNAi lines after a long recovery. The 3-d-old seedlings of the wild type (wt), hsa32-1, and RNAi (C057-2, -10, -11, and -16) lines were subjected to acclimation at 37°C for 1 h and recovered for 2 d, challenged at 44°C for 60 min, then recovered at 22°C for 8 d before the photograph was taken.
We further checked the basal thermotolerance of the wild-type and hsa32-1 seedlings by heating them at various temperatures (from 45°C–50°C in 1-degree increments) for 5 min, with no significant difference in survival rate between the wild-type and hsa32-1 plants. Both were killed at or above 48°C but survived at or below 47°C, which suggests that Hsa32 is not required for basal thermotolerance and agrees with the absence of the protein in the seedlings without HS treatment (Fig. 2). No significant difference in growth between the wild-type and hsa32-1 plants was observed when they were grown at 35°C for up to 17 d. Growth under this temperature was severely retarded for both plants.
Acquired Thermotolerance Decays Faster in the Absence of Hsa32
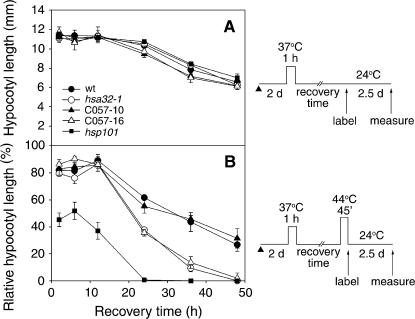
To further confirm our findings and determine the decay rate of acquired thermotolerance during recovery, we adopted the quantitative hypocotyl elongation assay (Hong and Vierling, 2000), which is sensitive in measuring the thermotolerance level. Etiolated 2-d-old vertically grown seedlings underwent acclimation treatment at 37°C for 1 h and then severe HS challenge at 44°C for 45 min. The hypocotyl length was measured after further growth for 2.5 d at 22°C in the dark. Here, various lengths of recovery time were introduced between the acclimation and the severe HS treatments to reveal the kinetics of thermotolerance decay. Hypocotyls without the acclimation treatment could not elongate if they were directly treated with the severe HS. With the seedlings growing older, hypocotyl elongation was retarded gradually after acclimation treatment, but no significant difference was observed among the wild-type, hsa32-1, and RNAi lines without the severe HS treatment (Fig. 6A). Figure 6B shows the relative hypocotyl elongation after severe HS treatment following different recovery times. The longer the recovery period, the less the hypocotyls could elongate, which indicates the decay of acquired thermotolerance during recovery. For all samples, the tolerance level started to decline after 10 h of recovery, and the decay was much faster in the hsa32-1 and RNAi (C057-16) lines than in the wild type and the other RNAi line, C057-10, which had about the same Hsa32 level as the wild type (Fig. 3). Severe HS treatment after 48 h of recovery inhibited hypocotyl elongation of hsa32-1 and C057-16 by almost 100%, whereas the wild-type and C057-10 seedlings retained about 30% elongation capacity (Fig. 6B). In contrast, a small or no difference was observed for the severe HS challenge applied within 10 h after the acclimation treatment. As a positive control, the Hsp101 knockout mutant showed a severe defect in acquired thermotolerance during short and long recovery (Fig. 6B). The results of the hypocotyl elongation assay were consistent with those of the survival rate data shown in Figure 4.
Figure 6.
Acquired thermotolerance decays faster in the absence of Hsa32. The wild type, hsa32-1, RNAi lines C057-10 and -16, and hsp101 seedlings were first conditioned at 37°C for 1 h then without (A) or subject to (B) severe HS treatment after recovery for the indicated times as shown schematically at the right. Then, the elongation of hypocotyl during 2.5 d of recovery was measured as indicated. The relative hypocotyl length in B was expressed as percentage of the numbers in A. Error bars represent sd and are based on data in four separate duplicates of five seedlings of each line. The arrowheads indicate end of seed imbibitions.
Except for Hsa32, HSR in hsa32-1 Is Normal at the Transcriptional Level
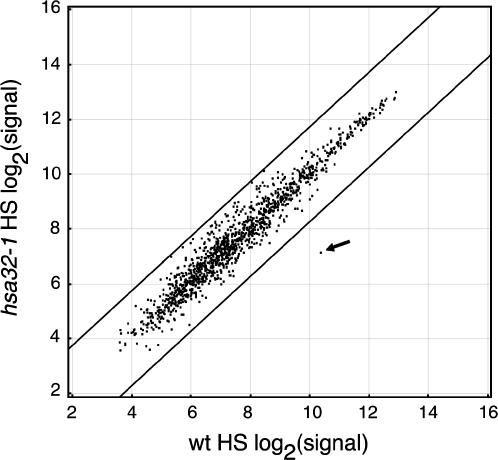
To test whether disruption of Hsa32 leads to an alteration in HSR, we have compared the expression profiles of the wild-type and hsa32-1 plants by employing the Affymetrix ATH1 genome array, which contains 22,500 probe sets representing approximately 24,000 genes. Normalized signals derived from the arrays representing expression levels were directly compared by scatterplot to view changes. Under the tested conditions, no significant difference was observed between the expression profiles of the wild-type and mutant plants under control conditions (data not shown), whereas HS treatment significantly up-regulated the expression of a number of genes both in the wild type and the mutant. When the hybridization signals of the HS-responsive genes of the HS-treated wild type were plotted against those of the HS-treated hsa32-1, only the signal of Hsa32 was significantly changed in the mutant, whereas the signals of the other genes retained less than a 3-fold difference between the wild type and the mutant (Fig. 7). A similar result was seen for signals of all expressed genes (data not shown). In addition, immunoblot analysis of Hsp90 and sHsp-CI levels in the wild-type and mutant plants after HS treatment showed no significant difference (Supplemental Fig. 1). These data suggest that the defect of thermotolerance observed in hsa32-1 is primarily due to the disruption of Hsa32, not because of a pleiotropic effect on HSR per se.
Figure 7.
Comparison of HSR in the wild-type and hsa32-1 plants by microarray analysis. Relative changes in Arabidopsis gene expression were studied in the hsa32-1 knockout mutant after HS treatment (37°C 2 h) by employing Arabidopsis ATH1 genome array. Expression signals (based on log2) of 1,306 HS-responsive genes (see “Materials and Methods” for definition) in the HS-treated wild-type plants were plotted against those of the HS-treated hsa32-1 mutant as illustrated by the scatterplot. The signal of Hsa32 was indicated by an arrow. Lines of 3-fold difference in expression between the wild-type and mutant plants were shown to assist reading. The data were the means of two biological replicates for the wild type and mutant.
DISCUSSION
This work was started by asking what special features plants may need to cope with heat stress in the environment, and Hsa32, which is mainly found in land plants, serves as a good candidate. The absence of a Hsa32 homolog in most other organisms, such as animals, suggests that Hsa32 may function as a unique feature specifically required for plants. This suggestion agrees with the view from an evolutionary perspective that the HSR in plants differs qualitatively from that observed for most other organisms (Waters, 2003). The observation that Hsa32 was up-regulated by HS treatment from lower to higher plants (Fig. 1) suggests that the gene plays a conserved role in HSR during plant evolution.
Similar to the well-characterized sHsp-CI, Hsa32 protein is also transiently synthesized during and after HS treatment (Fig. 2), thus confirming its status as a novel Hsp. Like Hsp101 (Queitsch et al., 2000), Has32 also is not required for the normal growth and development of Arabidopsis. Transcriptional and translational regulation of Hsa32 by heat stress suggested that it is required for thermotolerance of plants. To reveal the biological role of an unknown function gene, a common approach is knocking out the target gene and looking for a phenotype, which, however, is difficult because of redundancy of the gene or signaling networks (Cutler and McCourt, 2005). For a stress-response gene, finding the right assay condition is critical to revealing a mutant phenotype because plants without the gene of interest often grow like the wild type under normal or even the stress condition that induces it. In this study, we were not able to show the mutant phenotype by adopting a routine procedure, challenging the heat-acclimated plants after a short recovery time (Fig. 4, B and C). Thus, we turned to consider the importance of the duration of acquired thermotolerance, which has often been neglected in the past. Indeed, how long a previously acquired thermotolerance may be sustained apparently is important for the immobile plants that frequently face unpredictable fluctuation of temperature and other stresses, such as drought. Experimental design based on this idea has enabled us to discover the phenotype of hsa32-1 (Fig. 4, D and E), which would be difficult to identify otherwise.
It is intriguing that no obvious thermotolerance defect was observed for the hsa32-1 mutant within a short period of time after the acclimation HS treatment, when Hsa32 was accumulated to a high level in the wild type (Fig. 2). Our explanation is that Hsa32 is not essential for thermotolerance when other Hsps are also present at a sufficient level that could compensate for the lack of Hsa32. It was shown that overexpression of Hsp70 could partially compensate for the thermotolerance defect of lack of Hsp104 in Saccharomyces cerevisiae (Sanchez et al., 1993). Constitutive expression of Hsp101 alone in Arabidopsis confers tolerance against sudden shifts in extreme temperatures (Queitsch et al., 2000). Therefore, the function of Hsa32 being replaced by that of other abundant Hsps is not unlikely. However, when the levels of other Hsps are too low to confer such compensation after a longer recovery, the mutant thus becomes more susceptible than the wild type to the severe HS challenge. Alternatively, Hsa32 may become effective during the long recovery period by protein modification or by altered distribution in cells. Nevertheless, a second acclimation treatment during the long recovery period overcoming the lack of Hsa32 (Fig. 4F) favors our compensation hypothesis.
In addition to the survival rate results, similar conclusions were drawn by employing the quantitative hypocotyl elongation assay developed by Hong and Vierling for screening hot (sensitive to hot temperatures) mutants (Hong and Vierling, 2000). In the wild-type plants, after severe HS challenge, the hypocotyl elongation of acclimated seedlings was inhibited increasingly with longer recovery time (Fig. 6B), which can be regarded as a facet of thermotolerance decay. It is important to note that the difference between the hypocotyl elongation rate for the wild-type and mutant plants was manifested only after 10 h of recovery (Fig. 6B). This observation may explain why defect in Hsa32 (on chromosome 4) was not identified in a previous genetic screening of the hot mutants (hot1, hot2, and hot3 located on chromosome 1 and hot4 on chromosome 5) with hypocotyl elongation assay (Hong et al., 2003), in which conditioned seedlings were challenged by severe HS after 2 h of recovery. This assay is sensitive and easy to use to compare the relative contribution of Hsps to the development of acquired thermotolerance, as demonstrated by the distinct profiles of hsa32-1 and hsp101 mutants (Fig. 6B). The results from both assays suggest that Hsa32 might be a negative regulator of decay or a positive regulator of the maintenance of acquired thermotolerance, thus leading to rapid decay of thermotolerance in its absence. Hsa32 might directly or indirectly affect the profiles of HS-induced proteins or metabolites (Kaplan et al., 2004). Although we did not observe a significant alteration in transcriptomic profile (Fig. 7) and levels of Hsp90 and sHsp-CI (Supplemental Fig. 1), except Hsa32 in the hsa32-1 mutant, we cannot exclude the possibility that alterations in the transcript or protein level of other Hsps during or after the severe HS challenge after long recovery might affect the level of acquired thermotolerance. Detailed and systematic study of the transcriptomic, proteomic, or metabolomic profiles of the mutant and the wild type may provide insight into the action of Hsa32.
Unlike the well-characterized Hsps, which belong to multigene families (Agarwal et al., 2001; Hill and Hemmingsen, 2001; Krishna and Gloor, 2001; Lin et al., 2001; Miernyk, 2001; Scharf et al., 2001), Hsa32 is a single-copy gene in Arabidopsis. Given the status of Hsa32 as a single gene in a complex context of HSR involving multiple-member Hsps, how Hsa32 participates in cytoprotection and why it seems to be plant specific are interesting. We thought that Hsa32 plays a direct role in protecting the cells from heat damage instead of being a component in HS signaling because the hsa32-1 mutant exhibits a normal HSR according to the microarray data (Fig. 7) and a second acclimation treatment restored thermotolerance to the knockout mutant (Fig. 4F). The possible molecular function of Hsa32 has been mentioned previously. From sequence homology, Hsa32 was suggested to have PSL synthase activity and be involved in sulfolipid biosynthesis (Graham et al., 2002). However, we do not think this hypothesis is likely because of the weak similarity between the 2 sequences and the substrate binding sites being not conserved (Liu et al., 2006b). Moreover, the hsa32-1 mutant and wild-type plants show no significant difference in sulfolipid level under normal conditions or after HS treatment (Liu et al., 2006a). The Sqd1 and Sqd2 knockout mutants, which are devoid of sulfolipid (Yu et al., 2002), did not show the same defect in acquired thermotolerance as did the Hsa32 null mutant, which suggests that the sulfolipid level is not related to the HS-sensitive phenotype of hsa32-1. Further study of the biochemical properties of the Hsa32 protein is needed to demonstrate the mechanism of its action. Nevertheless, obtaining a knockout mutant with a significant phenotype first could greatly facilitate later biochemical study.
The requirement of Hsa32 for thermotolerance during long recovery implies that the protein is an important feature for plants to achieve better protection under the circumstances that plants most likely face. For example, the requirement reinforces the thermotolerance of plants that experience a sudden shift to extreme temperature after a previously acquired thermotolerance gradually decayed to a suboptimal range. We speculated that this scenario is probably common in nature because of the presence of Hsa32 in many land plants and because of the assay condition for revealing the hsa32-1 phenotype. To our knowledge, the importance of acquired thermotolerance after long recovery has not been emphasized in prior studies and the assay condition reported here has not been applied previously for characterizing any other Hsp or for isolation of mutants (Burke, 2001). Severe HS challenge during a long recovery after acclimation could efficiently reveal a significant phenotype for other Hsp mutants. Thus, we expect that our assay condition might be applicable for forward and reverse genetic study of HSR components involving Arabidopsis. To further explore the possible network of plant HSR genes, we are currently phenotyping the Arabidopsis T-DNA knockout mutants of other HS-responsive genes with this protocol. Our preliminary data showed that at least one more knockout mutant of a well-known Hsp exhibited a thermotolerance phenotype similar to that of hsa32-1. Taken together, our results demonstrate the feasibility of identifying the biological function of a novel Hsp by a straightforward approach of adopting a new assay condition, which might be useful in characterizing other Hsps.
MATERIALS AND METHODS
Growth of Plants and RNA Analysis
Arabidopsis (Arabidopsis thaliana) seeds were sterilized and sown on solid medium (25 mL of 0.5×Murashige and Skoog containing 0.8% agar and 1% Suc) in a petri dish (90 × 15 mm) and incubated for 3 d at 4°C for synchronized germination. Rice (Oryza sativa cv Tainung no. 67) seeds were also sterilized and grown on the same medium in a Magenta box. The seeds were grown at 24°C with 16 h light (130 μmol m−2 s−1) before HS treatment. Physcomitrella patens plants were grown on solid Knop medium as described previously (Strepp et al., 1998). After HS treatment, the plants were harvested and quickly frozen with liquid nitrogen with TRIZOL reagent (Invitrogen) for total RNA isolation. Gene-specific primers were designed for RT-PCR analysis or cloning of Hsa32 cDNA from Arabidopsis (AK118775), rice (AY623907), and P. patens (AY772008). The cloned cDNAs were used as templates for producing digoxigenin-labeled antisense RNA probes for RNA gel-blot analysis as described previously (Li et al., 2003). Transcript levels of Arabidopsis Hsp21 (X54102), rice OsHsp16.9C (U81385), P. patens sHsp (BQ827513), and actin (AW698983) were determined as positive or loading controls. The sequences of the gene-specific primers used in this study are provided in Supplemental Table I.
Arabidopsis T-DNA Knockout and Transgenic Plants
The Arabidopsis (Columbia) Hsa32 T-DNA insertion line 268A08, named hsa32-1 here, was generated in the context of the GABI-Kat program (Rosso et al., 2003) and provided by Bernd Weisshaar (Max-Planck-Institute for Plant Breeding Research, Cologne, Germany). The Hsp101 (At1g74310) T-DNA insertion line SALK_066374 (Alonso et al., 2003) was obtained from the Arabidopsis Biological Resource Center. Location of the T-DNA insertion was confirmed by PCR and sequencing. Homozygous, hemizygous, and azygous lines of the mutant allele were identified by PCR analysis. To suppress the expression of Hsa32 by the RNAi method, a 442-bp fragment from +1 to +442 of the AtHsa32 coding region was introduced into pHELLSGATE2 (CSIRO Plant Industry) in forward and reverse directions by an in vitro recombinase system (Wesley et al., 2001). The RNAi construct, pYC057, was transferred into Agrobacterium tumefaciens LBA4404 strain for Arabidopsis (Columbia) transformation by the floral dip method (Clough and Bent, 1998). Transformants were selected on Murashige and Skoog medium plates containing 50 μg/mL kanamycin. Homozygous transgenic lines (C057-2, -10, -11, and -16) with a single T-DNA insertion event were obtained through Southern-blot analysis and kanamycin-resistant tests. For all experiments, T3 seeds of the RNAi lines were used.
Immunoblotting
To produce the antibody against AtHsa32, the full-length recombinant protein with a C-terminal tag of six His residues (rHsa32-His6) was produced by use of the pET24d+ (Novagen) and Escherichia coli BL21(DE3) system as described previously (Charng et al., 2001). The recombinant protein was purified from the inclusion body by use of the His⋅Bind column (Novagen). The purity of the recombinant protein was more than 90% as estimated by SDS-PAGE and Coomassie Blue staining. The identity of the purified protein was confirmed by N-terminal peptide sequencing performed by the Core Facilities for Proteomics Research of Academia Sinica. The purified protein was then used as antigen to immunize rabbit. Immunization and serum collection were performed by LTK Biotech. The antibody was further purified by an affinity purification method (Smith and Fisher, 1984) to avoid nonspecific bands shown on immunoblotting. The polyclonal antibody against rice sHsp-CI was kindly provided by Dr. Chu-Yung Lin (National Taiwan University). Cross-reactivity of the rice antibody against Arabidopsis sHsp-CI has been demonstrated previously (Jinn et al., 1993). Monoclonal antibody against Hsp90 was purchased from Sigma. The total protein of plant samples was extracted with Tris-HCl buffer (60 mm, pH 8.5, containing 2% SDS, 2.5% glycerol, 0.13 mm EDTA, and 1% protease inhibitor cocktail). The protein amount was measured by use of DC Protein Assay reagents (Bio-Rad) with bovine serum albumin as a standard. For immunoblot analysis, protein was separated by SDS-PAGE with a precast minigel assembly (NuPAGE 4%–12% BisTris gel + MOPS SDS running buffer; Invitrogen) and transferred onto a nitrocellulose membrane for antibody probing. The amount of antigen was detected by use of the Super Signal West Dura Extended Duration Substrate system (Pierce). Following the chemiluminescence detection, the membrane was stained with 0.1% (w/v) Amido black to ensure equal loading of protein. The quantity of Hsa32 in the plant crude extract was estimated by scanning and analyzing the chemiluminescent signals with use of Image Gauge V3.12 (Fujifilm), with the known amount of purified rHsa32-His6 used as a standard.
Thermotolerance Test
For the acquired thermotolerance test, Arabidopsis seedlings were grown in a petri dish (90 × 15 mm) with 25 mL of solid medium and under the growth conditions described above. Heat treatment usually began at 10 am; the plate containing 3-d-old seedlings was sealed with plastic electric tape and submerged in a water bath at a temperature indicated in the figures. Treatment at different times of the day, such as noon or afternoon, from 2 to 4 pm, led to a similar phenotype in hsa32-1. The rate of temperature increase and decrease in a sealed petri dish was monitored with use of a compact temperature data logger (model SK500; Dickson) or an inserted lab thermometer. The medium and air were heated to 44°C exponentially from room temperature at about 9 and 15 min, respectively, and cooled to 30°C and 25°C linearly from 44°C at 20 min, respectively. During recovery from each HS treatment, the plate was removed from the water bath and kept at the previous growth condition under the same light/dark cycles. For basal thermotolerance test, seedlings were grown and treated similarly except in a plate containing 10 mL of solid medium for fast heating and cooling. The plates were heated at various temperatures in water bath for 5 min and then immediately cooled by fanning. For these tests, the thermotolerance was evaluated by survival rate. The quantitative hypocotyl elongation assay was performed as described previously (Hong and Vierling, 2000) except that 2-d-old etiolated seedlings were used and the agar plate was heated in a water bath as above. The recovery periods applied in Figure 6 were 2, 6, 12, 24, 36, and 48 h.
Microarray Analysis
For expression profiling of Arabidopsis genes, total RNA was isolated from the shoots of 15-d-old wild-type and hsa32-1 seedlings (a pool of about 100 plants per treatment in duplicates) harvested immediately after treatment at 37°C (HS) or 22°C (control) for 2 h (began at about 10 am). The plants were grown and treated in petri dish with 25 mL of solid medium as described above. Examination of the RNA quality and processing of ATH1 GeneChip arrays (Affymetrix) were performed by Vita Genomics according to the manufacturer's protocol. Scanning was performed with use of the GeneArray 2500 scanner (Affymetrix). In this experiment, eight chips were used, one each for two biological replicates of the control and HS-treated samples for the wild-type and hsa32-1 plants. To compare the profile derived from each chip, the data were scaled by use of the Global Scaling (all probe sets) method to a target intensity of 500 and then normalized by use of the Robust Multichip Average program (Irizarry et al., 2003). The log-transformed data were analyzed and visualized by use of the Spotfire DecisionSite 8.0 package. To show the effect of disruption of Hsa32 on general HSR, the averaged signals of two biological replicates of HSR genes of the HS-treated wild-type and hsa32-1 plants were compared by scatterplot (Fig. 7). The HS-responsive genes mentioned here were defined as those responding to the HS treatment with 2-fold up- or down-regulation in the wild-type plants. The microarray data reported here were deposited in the Gene Expression Omnibus at the National Center for Biotechnology Information (accession no. GSE4062).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AK118775, AY623907, and AY772008.
Supplementary Material
Acknowledgments
We thank Chai-Fen Lee, Chiu-Chung Wang, Shu-Hua Wu, and Wan-Jen Hsieh for technical support. We also thank Drs. Jong-Ching Su, Kuo-Chen Yeh, and Tzyy-Jen Chiou for critically reading and editing the manuscript. We are grateful to Drs. Wolfgang Frank and Ralf Reski for providing Physcomitrella plants and growth protocols.
This work was supported by the National Science Council (grant nos. 91–3112–P–001–036–Y and 94–2311–B–001–058) and by Academia Sinica, Taiwan, Republic of China.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yee-yung Charng (yycharng@gate.sinica.edu.tw).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074898.
References
- Agarwal M, Katiyar-Agarwal S, Sahi C, Gallie DR, Grover A (2001) Arabidopsis thaliana Hsp100 proteins: kith and kin. Cell Stress Chaperones 6: 219–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Bonsen PP, Spudich JA, Nelson DL, Kornberg A (1969) Biochemical studies of bacterial sporulation and germination. XII. A sulfonic acid as a major sulfur compound of Bacillus subtilis spores. J Bacteriol 98: 62–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke JJ (2001) Identification of genetic diversity and mutations in higher plant acquired thermotolerance. Physiol Plant 112: 167–170 [Google Scholar]
- Busch W, Wunderlich M, Schoffl F (2005) Identification of novel heat shock factor-dependent genes and biochemical pathways in Arabidopsis thaliana. Plant J 41: 1–14 [DOI] [PubMed] [Google Scholar]
- Charng YY, Chou SJ, Jiaang WT, Chen ST, Yang SF (2001) The catalytic mechanism of 1-aminocyclopropane-1-carboxylic acid oxidase. Arch Biochem Biophys 385: 179–185 [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cutler S, McCourt P (2005) Dude, where's my phenotype? Dealing with redundancy in signaling networks. Plant Physiol 138: 558–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasch AP, Spellman PT, Kao CM, Carmel-Harel O, Eisen MB, Storz G, Botstein D, Brown PO (2000) Genomic expression programs in the response of yeast cells to environmental changes. Mol Biol Cell 11: 4241–4257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham DE, Xu H, White RH (2002) Identification of coenzyme M biosynthetic phosphosulfolactate synthase: a new family of sulfonate-biosynthesizing enzymes. J Biol Chem 277: 13421–13429 [DOI] [PubMed] [Google Scholar]
- Helmann JD, Wu MF, Kobel PA, Gamo FJ, Wilson M, Morshedi MM, Navre M, Paddon C (2001) Global transcriptional response of Bacillus subtilis to heat shock. J Bacteriol 183: 7318–7328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JE, Hemmingsen SM (2001) Arabidopsis thaliana type I and II chaperonins. Cell Stress Chaperones 6: 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-W, Lee U, Vierling E (2003) Arabidopsis hot mutants define multiple functions required for acclimation to high temperatures. Plant Physiol 132: 757–767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SW, Vierling E (2000) Mutants of Arabidopsis thaliana defective in the acquisition of tolerance to high temperature stress. Proc Natl Acad Sci USA 97: 4392–4397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP (2003) Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics 4: 249–264 [DOI] [PubMed] [Google Scholar]
- Jinn T, Wu SH, Yeh CH, Hsieh MH, Yeh YC, Chen YM, Lin CY (1993) Immunological kinship of class I low molecular weight heat shock proteins and thermostabilization of soluble proteins in vitro among plants. Plant Cell Physiol 34: 1055–1062 [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL (2004) Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol 136: 4159–4168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishna P, Gloor G (2001) The Hsp90 family of proteins in Arabidopsis thaliana. Cell Stress Chaperones 6: 238–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li HY, Chang CS, Lu LS, Liu CA, Chan MT, Charng YY (2003) Over-expression of Arabidopsis thaliana heat shock factor gene (AtHsfA1b) enhances chilling tolerance in transgenic tomato. Bot Bull Acad Sinica (Taiwan) 44: 129–140 [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist S (1986) The heat-shock response. Annu Rev Biochem 55: 1151–1191 [DOI] [PubMed] [Google Scholar]
- Lindquist S, Craig EA (1988) The heat-shock proteins. Annu Rev Genet 22: 631–677 [DOI] [PubMed] [Google Scholar]
- Liu NY, Hsieh WJ, Liu HC, Charng YY (2006. a) Hsa32, a phosphosulfolactate synthase-related heat-shock protein, does not involve in sulfolipid biosynthesis in Arabidopsis. Botanical Studies (in press)
- Liu NY, Ko SS, Yeh KC, Charng YY (2006. b) Isolation and characterization of tomato Hsa32 encoding a novel heat-shock protein. Plant Sci 170: 976–985 [Google Scholar]
- Miernyk JA (2001) The J-domain proteins of Arabidopsis thaliana: an unexpectedly large and diverse family of chaperones. Cell Stress Chaperones 6: 209–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsell DA, Lindquist S (1993) The function of heat-shock proteins in stress tolerance: degradation and reactivation of damaged proteins. Annu Rev Genet 27: 437–496 [DOI] [PubMed] [Google Scholar]
- Pysz MA, Ward DE, Shockley KR, Montero CI, Conners SB, Johnson MR, Kelly RM (2004) Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8: 209–217 [DOI] [PubMed] [Google Scholar]
- Queitsch C, Hong SW, Vierling E, Lindquist S (2000) Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell 12: 479–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizhsky L, Liang H, Shuman J, Shulaev V, Davletova S, Mittler R (2004) When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 134: 1683–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B (2003) An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol Biol 53: 247–259 [DOI] [PubMed] [Google Scholar]
- Sanchez Y, Parsell DA, Taulien J, Vogel JL, Craig EA, Lindquist S (1993) Genetic evidence for a functional relationship between Hsp104 and Hsp70. J Bacteriol 175: 6484–6491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharf KD, Siddique M, Vierling E (2001) The expanding family of Arabidopsis thaliana small heat stress proteins and a new family of proteins containing alpha-crystallin domains (Acd proteins). Cell Stress Chaperones 6: 225–237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shockley KR, Ward DE, Chhabra SR, Conners SB, Montero CI, Kelly RM (2003) Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl Environ Microbiol 69: 2365–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DE, Fisher PA (1984) Identification, developmental regulation, and response to heat shock of two antigenically related forms of a major nuclear envelope protein in Drosophila embryos: application of an improved method for affinity purification of antibodies using polypeptides immobilized on nitrocellulose blots. J Cell Biol 99: 20–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95: 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung D-Y, Kaplan F, Lee K-J, Guy CL (2003) Acquired tolerance to temperature extremes. Trends Plant Sci 8: 179–187 [DOI] [PubMed] [Google Scholar]
- Vierling E (1991) The roles of heat shock proteins in plants. Annu Rev Plant Physiol Plant Mol Biol 42: 579–620 [Google Scholar]
- Waters ER (2003) Molecular adaptation and the origin of land plants. Mol Phylogenet Evol 29: 456–463 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang M, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Wise EL, Graham DE, White RH, Rayment I (2003) The structural determination of phosphosulfolactate synthase from Methanococcus jannaschii at 1.7-A resolution: an enolase that is not an enolase. J Biol Chem 278: 45858–45863 [DOI] [PubMed] [Google Scholar]
- Yu B, Xu C, Benning C (2002) Arabidopsis disrupted in SQD2 encoding sulfolipid synthase is impaired in phosphate-limited growth. Proc Natl Acad Sci USA 99: 5732–5737 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.