Abstract
Messenger RNAs that do not contain a long open reading frame (ORF) or non-protein-coding RNAs (npcRNAs) are an emerging novel class of transcripts. Their functions may involve the RNA molecule itself and/or short ORF-encoded peptides. npcRNA genes are difficult to identify using standard gene prediction programs that rely on the presence of relatively long ORFs. Here, we used detailed bioinformatic analyses of expressed sequence tag/cDNA databases to detect a restricted set of npcRNAs in the Arabidopsis (Arabidopsis thaliana) genome and further characterized these transcripts using a combination of bioinformatic and molecular approaches. Compositional analyses revealed strong nucleotide strand asymmetries in the npcRNAs, as well as a biased GC content, suggesting the existence of functional constraints on these RNAs. Thirteen of these transcripts display tissue-specific expression patterns, and three are regulated in conditions affecting root architecture. The npcRNA 78 gene contains the miR162 sequence in an alternative intron and corresponds to the MIR162a locus. Although DICER-LIKE 1 (DCL1) mRNA is known to be regulated by miR162-guided cleavage, its level does not change in a mir162a mutant. Alternative splicing of npcRNA 78 leads to several transcript isoforms, which all accumulate in a dcl1 mutant. This suggests that npcRNA 78 is a genuine substrate of DCL1 and that splicing of this microRNA primary transcript and miR162 processing are competitive nuclear events. Our results provide new insights into Arabidopsis npcRNA biology and the potential roles of these genes.
In eukaryotes, several studies have revealed a new class of mRNAs containing only short open reading frames (sORFs), named either sORF-mRNAs, noncoding RNAs, or protein-lacking RNAs, but we will refer to them as non-protein-coding RNAs (npcRNAs). The lack of a long ORF indicates that npcRNA activity involves the RNA itself and/or sORF-encoded oligopeptides. These oligopeptides may act as signals in development (Lindsey et al., 2002). Both sORF translation and RNA structure can be involved in npcRNA function, as shown in several viruses (Erdmann et al., 2001) or in plants (Sousa et al., 2001). Alternatively, translation of sORFs present in npcRNAs may occur even though the main function of the gene lies in the RNA product, as shown for a five-amino acid peptide encoded by the Escherichia coli 23S ribosomal RNA (Tenson et al., 1996) or through immunological detection of a putative protein encoded by the H19 RNA (Leibovitch et al., 1991; Leighton et al., 1995). For certain npcRNAs, sequence conservation at the nucleotide but not at the amino acid level suggests that RNA can play an important role in their function (Erdmann et al., 2001; MacIntosh et al., 2001). However, evolutionary conservation cannot be used as a unique criterion because npcRNAs are generally much less conserved than protein-coding genes, as in the case of the Xist gene (Taylor et al., 2003).
Although npcRNAs are not easily detected by computational analyses due to the small sizes of the encoded sORFs (for review, see Eddy, 2002), the number of npcRNAs is expanding rapidly. A pioneer analysis of the Arabidopsis (Arabidopsis thaliana) genome identified 40 putative npcRNAs (MacIntosh et al., 2001). However, many of these transcripts could later be linked to protein-coding genes when more information on the mRNA molecules was available. The advent of genomic approaches (full-length cDNA analysis and genome tiling arrays [Numata et al., 2003; Yamada et al., 2003]) has revealed a large diversity of npcRNAs, including a surprising number of antisense RNA transcripts. Very recently, an analysis of Arabidopsis expressed sequence tag (EST) data identified 591 orphan transcripts not previously assigned to genomic loci, including putative noncoding and peptide-coding RNAs (Riano-Pachon et al., 2005). Additionally, a novel class of small npcRNAs has been characterized in several species as playing regulatory roles in a diversity of developmental processes (He and Hannon, 2004). These 20- to 24-nucleotide (nt) RNAs, called microRNAs (miRNAs) or trans-acting small interfering RNAs (tasiRNAs; Vazquez et al., 2004b), are produced through the action of DICER, a specific type III ribonuclease. In Arabidopsis, DICER-LIKE 1 (DCL1) is the main enzyme responsible for the production of regulatory miRNAs. mi/tasiRNAs are transcribed as part of longer primary transcripts (called pri-miRNAs in the case of miRNAs) that are processed to release the small RNAs in their mature forms. miRNA and tasiRNA primary transcripts appear to be longer npcRNAs, although few have been fully characterized (Kurihara and Watanabe, 2004; Vazquez et al., 2004b; Baker et al., 2005). Recently, transcription start sites consistent with an RNA polymerase II mechanism of transcription were mapped for 52 Arabidopsis MIRNA loci (Xie et al., 2005).
npcRNAs playing regulatory roles, the so-called riboregulators, have been shown to participate in diverse processes such as organization of the embryo cytoplasm, mRNA translation or stability, and protein secretion or silencing (Erdmann et al., 2001). These functional RNAs associate with RNA-binding proteins to form ribonucleoprotein (RNP) particles, which may have various cellular roles (Dreyfuss et al., 2002; Joyce, 2002; He and Hannon, 2004), as shown for the intron-encoded small nucleolar RNAs (Kiss, 2002), the miRNAs (He and Hannon, 2004), or the BC1 transcript in mammals (Zalfa et al., 2003). The RNA molecule seems to determine the functional specificity of the complex. Much remains to be known about the molecular mechanisms of action of npcRNAs in eukaryotes (Erdmann et al., 2001). In plants, npcRNAs of the AtIPS1/At4 family are highly induced during phosphate starvation in Arabidopsis and Medicago roots (Franco-Zorrilla et al., 2002), whereas the npcRNA enod40 is involved in the formation of symbiotic nitrogen-fixing nodules in legumes (Campalans et al., 2004). Interestingly, these npcRNAs are regulated in conditions or developmental processes affecting root tissues. RNA-mediated regulation may thus be particularly relevant for processes requiring rapid developmental plasticity, like the adaptation of root architecture to environmental conditions (Franco-Zorrilla et al., 2002).
In this article, we searched for Arabidopsis npcRNAs with the aim of obtaining a restricted and experimentally verified collection of npcRNA genes, rather than a large set of computer-derived candidates. We thus used a combination of bioinformatic and molecular approaches to characterize 43 npcRNA genes. Thirteen of these npcRNAs display diverse tissue-specific expression patterns and three of them are regulated in response to phosphate starvation or cytokinin treatment of roots. One npcRNA corresponds to the differentially spliced miR162a primary transcript that targets DCL1. This npcRNA contains the miRNA sequence within an alternative intron. Deregulation of all npcRNA isoforms in a dcl1 mutant allows us to propose that splicing of this primary transcript and miR162 processing are competitive nuclear events.
RESULTS
In Silico Identification of 43 npcRNAs
In an effort to uncover putative regulatory or peptide-encoding RNAs for subsequent experimental testing, we mined public EST and cDNA libraries for transcripts lacking significant ORFs. Alignment of EST and cDNA sequences was used to localize candidate npcRNA genes on the Arabidopsis genome. The selection process (detailed in “Materials and Methods”) allowed us to identify 46 putative npcRNAs positioned on intergenic regions of the five chromosomes. Among the 46 npcRNAs, we identified three previously well-described putative peptide-coding RNAs, namely, DEVIL20 (DVL20), POLARIS (PLS), and RPL41F, encoding, respectively, 57-amino acid (Wen et al., 2004) and 36-amino acid (Casson et al., 2002) peptides and a 25-amino acid ribosomal peptide (Barakat et al., 2001). These genes were not further analyzed. Table I features relevant data on gene structure, homologies, and putative function for the 43 remaining npcRNA genes. In particular, based on the size of the largest ORF, its coding capacity, its proportion relative to the full transcript size, the presence of putative signal peptides, as well as EST/mRNA homologies, npcRNAs were tentatively classified as putative functional RNAs (pfRNAs) or peptide-encoding RNAs (sORF-RNAs). The presence of ORFs more conserved at the amino acid than the nucleotide level was a major criterion to define sORF-RNAs (Table I, homologies column). We thus obtained a set of 28 pfRNAs and 15 sORF-RNAs.
Table I.
npcRNA gene features
Columns contain, respectively, name; genomic localization (i.e. chromosome number, positions of gene start and strand [w, Watson; c, Crick]); number of exons; gene and mRNA lengths; TAIR or At_oRNA equivalent (*, additional atorphan gene, At_oRNA_572, corresponds to this npcRNA); the longest ORF start position on the transcript; ORF length in nucleotides; number of ATGs before the putative coding ORF; probability of coding of this ORF [+, P (coding) in the frame of the ORF >0.5; −, P (noncoding, independent of the frame) >0.5; ±, others]; presence of homologous ORFs in ESTs from other species (number of species; Bn, Brassica napus) or nucleotide homologies independent of encoded peptides (+); and final status of the gene (pf, pfRNA; sORF, putative peptide-encoding gene [see “Materials and Methods”]). npcRNA genes were predicted from full-length cDNAs (npcRNAs 2–86) or from clustered ESTs (npcRNAs 111–431).
| Name | Chromosome | Start Position | Strand | No. Exons | Gene Length | mRNA Length | TAIR Accession or At_oRNA Identifier | ORF Start | ORF Length | No. ATGs | P (Coding) | Homologies | Status |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2 | 1 | 749046 | c | 1 | 945 | 945 | AT1G03106 | 408 | 195 | 8 | − | pf | |
| 4 | 1 | 1862196 | w | 1 | 548 | 548 | AT1G06135 | 111 | 207 | 0 | + | sORF | |
| 14 | 1 | 17298005 | w | 1 | 735 | 735 | 636 | 90 | 7 | − | pf | ||
| 15 | 1 | 18141841 | w | 1 | 833 | 833 | 143 | 171 | 2 | − | pf | ||
| 17 | 1 | 19687168 | c | 1 | 555 | 555 | AT1G52855 | 158 | 204 | 0 | + | 10 | sORF |
| 21 | 1 | 26183015 | c | 3 | 3,700 | 767 | 565 | 123 | 13 | − | pf | ||
| 26 | 2 | 2330796 | w | 1 | 698 | 698 | 474 | 63 | 3 | − | pf | ||
| 29 | 2 | 6652614 | c | 1 | 1,253 | 1,253 | 390 | 189 | 2 | − | pf | ||
| 30 | 2 | 8129244 | w | 1 | 912 | 912 | 352 | 147 | 6 | − | pf | ||
| 33 | 2 | 9410332 | w | 1 | 557 | 557 | AT2G22122 | 111 | 180 | 0 | + | Bn | sORF |
| 34 | 2 | 14603700 | w | 1 | 921 | 921 | AT2G34655 | 216 | 66 | 4 | − | pf | |
| 40 | 3 | 5200640 | w | 4 | 882 | 592 | AT3G15395 | 206 | 177 | 0 | + | 12 | sORF |
| 41 | 3 | 5861497 | w | 2 | 948 | 556 | AT3G17185, TAS3 | 58 | 150 | 0 | + | + | sORF |
| 43 | 3 | 6956996 | w | 1 | 692 | 692 | 266 | 87 | 2 | + | pf | ||
| 48 | 3 | 11511953 | w | 1 | 983 | 983 | 741 | 120 | 9 | − | pf | ||
| 51 | 3 | 17719894 | c | 2 | 1,004 | 525 | AT3G47965 | 201 | 132 | 4 | − | pf | |
| 52 | 3 | 17783023 | c | 1 | 863 | 863 | 705 | 120 | 9 | − | pf | ||
| 58 | 4 | 2095257 | w | 2 | 1,266 | 585 | 107 | 195 | 0 | − | sORF | ||
| 60 | 4 | 7494232 | c | 1 | 475 | 475 | AT4G12735 | 51 | 180 | 0 | + | sORF | |
| 62 | 4 | 9244083 | w | 1 | 589 | 589 | At_oRNA_394 | 153 | 129 | 1 | − | sORF | |
| 72 | 4 | 16617625 | w | 1 | 887 | 887 | 273 | 177 | 4 | − | pf | ||
| 75 | 4 | 18152553 | c | 1 | 804 | 804 | 228 | 75 | 3 | − | pf | ||
| 78 | 5 | 2635438 | c | 4 | 1,368 | 635 | AT5G08185, MIR162a | 128 | 135 | 2 | − | pf | |
| 79 | 5 | 3396474 | w | 3 | 1,591 | 1,393 | AT5G10745 | 81 | 153 | 0 | + | 5 | sORF |
| 82 | 5 | 8151927 | w | 2 | 944 | 576 | AT5G24105 | 174 | 189 | 0 | + | 9 | sORF |
| 83 | 5 | 15909210 | c | 1 | 706 | 706 | 94 | 132 | 0 | − | 5 | sORF | |
| 86 | 5 | 22989737 | c | 2 | 603 | 341 | 71 | 84 | 0 | ± | pf | ||
| 111 | 1 | 17256342 | c | 1 | 936 | 936 | 222 | 147 | 3 | − | pf | ||
| 113 | 1 | 20126539 | c | 1 | 520 | 520 | 164 | 93 | 2 | + | pf | ||
| 131 | 2 | 13453123 | w | 1 | 684 | 684 | At_oRNA_588* | 61 | 120 | 0 | − | sORF | |
| 149 | 3 | 9103055 | w | 1 | 782 | 782 | AT3G24927 | 498 | 195 | 8 | + | pf | |
| 150 | 3 | 9666799 | w | 1 | 788 | 788 | At_oRNA_581 | 461 | 207 | 5 | − | pf | |
| 155 | 3 | 22001256 | w | 1 | 509 | 509 | At_oRNA_506 | 89 | 135 | 0 | + | Bn | sORF |
| 156 | 4 | 2376362 | c | 2 | 790 | 654 | AT4G04692 | 62 | 138 | 2 | − | pf | |
| 157 | 4 | 4866645 | w | 3 | 1,695 | 848 | At_oRNA_589 | 311 | 171 | 5 | − | + | pf |
| 311 | 1 | 3545472 | c | 1 | 488 | 488 | 319 | 96 | 5 | − | + | pf | |
| 325 | 1 | 11453600 | w | 2 | 415 | 344 | At_oRNA_416 | 87 | 156 | 0 | − | sORF | |
| 326 | 1 | 12582099 | w | 2 | 609 | 517 | 278 | 99 | 4 | + | pf | ||
| 351 | 1 | 25061142 | w | 1 | 782 | 782 | At_oRNA_576 | 497 | 99 | 4 | ± | pf | |
| 370 | 2 | 3300379 | c | 1 | 522 | 522 | 118 | 90 | 3 | − | pf | ||
| 375 | 2 | 3438412 | c | 1 | 832 | 832 | 418 | 138 | 6 | − | + | pf | |
| 415 | 3 | 2808273 | c | 1 | 679 | 679 | AT3G09162 | 99 | 198 | 0 | + | sORF | |
| 431 | 3 | 11155388 | w | 1 | 340 | 340 | At_oRNA_485 | 264 | 30 | 0 | − | + | pf |
Although most of the npcRNA genes were predicted from full-length cDNAs (npcRNAs 2–86), 16 genes were predicted from clusters of two or more ESTs (npcRNAs 111–431). As ESTs are often partial cDNAs, the predicted sORFs deduced to propose a npcRNA might be the result of an EST cluster representing a long 5′- or 3′-untranslated region (UTR). In particular, a longer 5′ region may introduce new ATGs upstream from the predicted longest sORF start codon and define a longer ORF. Hence, RNA ligase-mediated (RLM) 5′-RACE was performed for a subset of the npcRNAs predicted from EST clusters (npcRNAs 113, 155, 156, 311, 351, and 375) to map the 5′ ends of these transcripts. The 5′-RACE products were cloned and the transcription start site was deduced from the most abundant 5′ position represented among five to 10 clones randomly selected for sequencing. In each of these cases, the transcription initiation site either matched the one predicted by the EST clusters (npcRNAs 155 and 156) or fell within a few nucleotides of this position (113: +10; 311: +5; 351: −13; 375: +14). These results indicate that the 5′ ends of these npcRNAs based on EST clusters are accurate and that the ESTs do not correspond to long 3′-UTRs of larger protein-coding transcripts.
Since the onset of this work, 15 npcRNAs from our dataset have been annotated as expressed or unknown proteins (see Table I for The Arabidopsis Information Resource [TAIR] annotations). It is worth noting that five npcRNAs are located within highly dense genomic environments in the current annotated genome (see Supplemental Fig. 1). Aside from npcRNA 78, which contains the miR162a precursor sequence, no miRNA precursors referenced in the miRBase database (http://microrna.sanger.ac.uk/sequences) were identified among the npcRNAs. npcRNA 41 has recently been shown to be a tasiRNA precursor (TAS3; Allen et al., 2005). Additionally, the sequence of npcRNA 86 is almost identical to that of the metallothionein MT1b (ATU11254), a gene predicted to encode a 45-amino acid protein (Zhou and Goldsbrough, 1994). However, this npcRNA contains a frameshift mutation, suggesting that the npcRNA 86 gene is an expressed pseudogene. Very recently, eight other npcRNAs have been independently retrieved in a screen for transcripts not previously assigned to genomic loci and described as orphan transcripts or At_oRNAs by Riano-Pachon et al. (2005). The corresponding identifiers in the Arabidopsis orphan RNA database (http://atornadb.bio.uni-potsdam.de) are also indicated in Table I. Finally, npcRNAs 52 and 149 contain two short ESTs (R87017 and T13664, representing 21% and 27% of these genes, respectively) previously proposed to be noncoding or peptide-coding genes (MacIntosh et al., 2001). The remaining 20 npcRNAs are currently not annotated in the updated Arabidopsis genome database. The entire set of npcRNAs has been included in the publicly available FlagDB++ database (http://urgv.evry.inra.fr/projects/FLAGdb++/HTML/index.shtml).
npcRNA Sequence Features
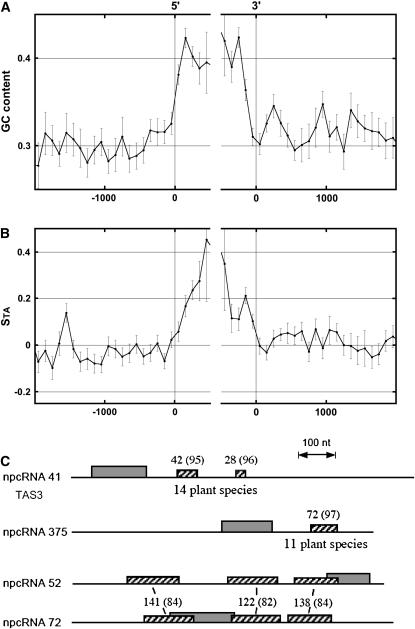
To examine whether the candidate npcRNA genes identified here present particular nucleotide composition properties that could be related to functional sequence elements, we examined their GC content and nucleotide compositional strand asymmetries (Fig. 1, A and B). These asymmetries (expressed as compositional skews; see Fig. 1B) correspond to the deviation from equality of the relative proportions of A and T and of G and C nucleotides calculated on one strand. In Arabidopsis, only small values of strand asymmetries could be associated with the transcription process by comparing the central regions of introns of protein-coding genes and intergenic regions. Conversely, the intronic borders revealed strong biases, spanning more than 500 nt, likely due to numerous T- and G-rich targets involved in the splicing process (see “Discussion”; Touchon et al., 2004). Here, comparison of all the Arabidopsis npcRNAs with their neighboring 5′ and 3′ intergenic sequences showed large values of the STA bias in the transcripts (Fig. 1B; the SGC bias did not deviate significantly from zero values). In addition, the GC content of these regions was significantly higher than that of the neighboring 5′ and 3′ intergenic sequences (Fig. 1A). This suggests that the npcRNAs display skews more similar to the functionally relevant intronic borders than to the central regions (neutral) of introns.
Figure 1.
Bioinformatic analysis of compositional biases and nucleotide conservation of npcRNA genes. A, GC content in the regions surrounding the 5′- and 3′-npcRNA gene extremities calculated in adjacent windows starting from each gene extremity in both directions. In the abscissa, the distance (in nucleotides) of each 100-bp window to the indicated gene extremity is presented, zero values of the abscissa corresponding to 5′ (left) or to 3′ (right) gene extremities. In the ordinate, the mean values, for all npcRNAs, of the GC content are calculated at the corresponding abscissa in the corresponding windows; vertical bars indicate se. B, Strand asymmetries STA = (T − A)/(T + A), where A and T are the numbers of the corresponding nucleotides in the sequence window; abscissa is as in A. C, Conserved nucleotide regions in npcRNAs. Regions conserved in ESTs from other species are indicated for npcRNAs 41 (TAS3) and 375. For npcRNA 41 and npcRNA 375, homologies are present in 14 (e.g. Oryza sativa, Populus tremula, Glycine max, and Picea glauca) and 11 (e.g. Oryza sativa, Citrus sinensis, Antirrhinum majus, and Hordeum vulgare) species, respectively. npcRNA 52 and 72 genes are two paralogous candidate genes. Hatched boxes, Conserved nucleotide fragments; gray boxes, longest ORF. Numbers above boxes indicate the size of the matching regions in nucleotides (in Arabidopsis); the percentage of identity is indicated in parentheses.
The npcRNA genes present only small nucleotide regions conserved in orthologous npcRNAs, as revealed by BLASTn and BLASTp analyses (http://www.ncbi.nlm.nih.gov/BLAST) on EST databases and the rice (Oryza sativa) genome. Conserved sORFs were identified for several npcRNAs in different species as indicated in Table I (homologies column) and mainly corresponded to predicted sORF-RNAs. Intergenomic analyses identified short, conserved segments with high nucleotide identity levels in several plant species. For five candidates, nucleotide homologies were found outside the proposed sORF regions (Table I, homologies column “+”). For npcRNA 41 and 375, highly homologous nucleotide boxes were detected in 14 and 11 species, respectively, including rice (Fig. 1C). The region conserved in the former npcRNA spans the tasiRNA-generating regions (Allen et al., 2005). In addition, the npcRNA 52 and 72 genes seem to be paralogous and share three nucleotide regions not corresponding to their predicted sORFs.
Functional RNA domains often involve secondary structures. Detection of such structures by genomic sequence analysis is not an easy task without the help of phylogenetic data. The comparison of ΔG values between functional and nonrelevant structures cannot be used directly because these values strongly depend both on the size and the GC content of the folded regions. To test for an exceptional stability of a candidate RNA structure, the corresponding ΔG value has been compared to the distribution of ΔG values computed for a large number of sequences presenting the same size and composition as the sequence of interest. This comparison, monitored as a Z score (Z = d/σ, where d is the distance separating the mean ΔG value of the distribution from the actual ΔG value and σ is the sd of the distribution), has already been used successfully to predict functional RNA structures (Crespi et al., 1994; Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Wang et al., 2004). As shown in Table II, 11 npcRNA transcripts contain regions that display significant Z-score values (potential secondary structures are proposed in Supplemental Fig. 2). These structures span 8% of the total length of all the candidates. As a comparison, the same analysis was performed on coding sequences from protein-coding genes of Arabidopsis with a GC content similar to that of the npcRNAs (yielding 1,488 sequences; see “Materials and Methods”). In this case, statistically significant structures span only 2% of the total length, supporting the hypothesis of functional roles for npcRNA secondary structures. Interestingly, eight of the npcRNAs showing significant Z scores were classified as pfRNAs.
Table II.
npcRNAs may contain highly stable structures
Sequence fragments within each npcRNA associated with a statistically significant stable RNA secondary structure were selected (Z scores > 5). The three columns contain, respectively, the name of the npcRNA, the ends of the selected fragment, and the values of the Z scores computed for this fragment.
| npcRNA | Fragment | Z Score |
|---|---|---|
| 2 | 31–270 | 5.7 |
| 41 | 361–430 | 6.4 |
| 48 | 31–290 | 5.6 |
| 75 | 31–220 | 5.3 |
| 78 | 251–380 | 5.2 |
| 82 | 11–530 | 9.0 |
| 83 | 71–430 | 16.7 |
| 150 | 231–380 | 5.6 |
| 311 | 221–300 | 6.4 |
| 351 | 21–310 | 7.2 |
| 375 | 571–700 | 5.1 |
Expression Patterns of the npcRNAs
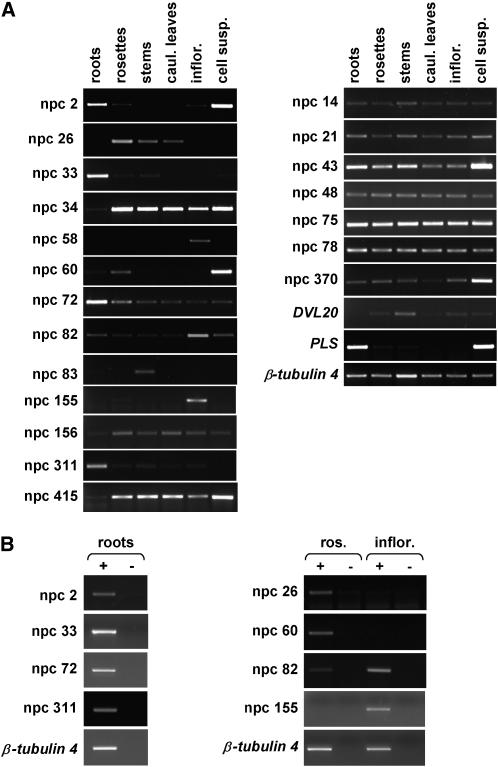
Expression of the npcRNAs was investigated in different plant tissues (Fig. 2). We were able to detect 43 of 46 npcRNAs (including DVL20, PLS, and RPL41F) for which gene-specific primers could be designed using reverse transcription (RT)-PCR, although several RNAs accumulated to very low levels in all tissues analyzed (no specific primer pairs could be designed for npcRNAs 17, 51, and 86).
Figure 2.
Expression profiling of npcRNAs. Total RNA from roots, rosettes, stems, cauline leaves, inflorescences, and cell suspensions was assayed by semiquantitative RT-PCR. Fourteen npcRNAs (including DVL20) display specific developmental expression patterns. Other npcRNAs are broadly expressed in all tissues analyzed. PLS (POLARIS) mRNA, which accumulates preferentially in roots, was used as a tissue-specific control. β-Tubulin 4 mRNA served as a constitutively expressed control. B, DNA contamination controls for several npcRNAs with highly specific expression patterns. ±, RT-PCR performed with or without reverse transcriptase.
Several npcRNAs displayed highly specific expression patterns. These include npcRNAs 2, 33, 72, and 311, which accumulated preferentially in roots (npcRNA 2 also accumulated in cell suspensions). npcRNA 26 was detected only in leaves and stems, whereas npcRNA 60 accumulated specifically in rosette leaves and cell suspensions. A weak signal for npcRNAs 83 and DVL20 was detected mainly in stems, whereas npcRNAs 58 and 155 were detected only in inflorescences and npcRNA 82 levels were severalfold higher in inflorescences compared to other tissues. Three other npcRNAs (34, 156, and 415) were more broadly expressed, albeit displaying aerial organ-specific expression. Finally, npcRNAs 14, 21, 43, 48, 75, 78, and 370 are examples of RNAs that could be detected at comparable levels in all tissues examined using this semiquantitative approach. The 21 other RNAs analyzed were also broadly expressed in the different tissues analyzed (see Supplemental Fig. 3). The previously described PLS gene served as a control for our expression-profiling experiments. The corresponding transcript is expressed predominantly in embryonic and seedling roots (Casson et al., 2002), as confirmed in our RT-PCR analysis.
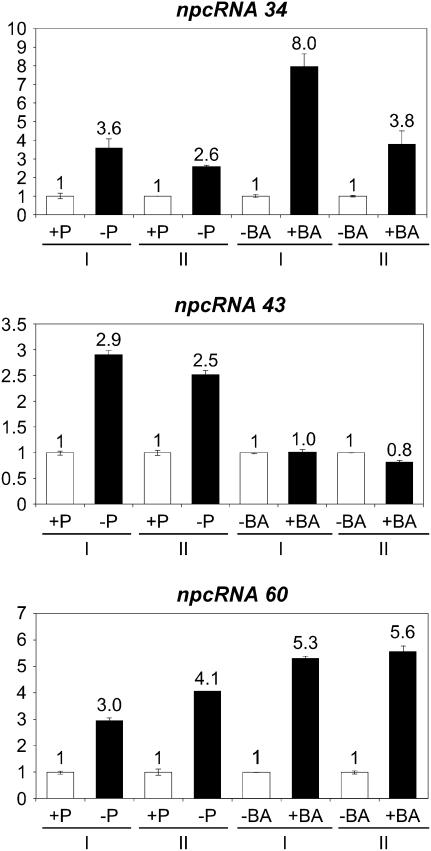
Based on the results showing the involvement of several npcRNAs in root differentiation processes, we speculated that some npcRNAs from our set may be regulated in conditions affecting root development and architecture. The entire set of npcRNAs was surveyed for regulation by salt stress, phosphate starvation, or cytokinin treatment in roots, using semiquantitative RT-PCR (data not shown). No salt-regulated RNAs were identified for the considered time point. Three npcRNAs regulated by phosphate starvation and/or cytokinin treatment were identified, and their accumulation in phosphate-starved/cytokinin-treated roots was further validated by real-time RT-PCR (Fig. 3). Expression levels of npcRNAs 34 and 60 increased severalfold in phosphate-starved and in 6-benzylaminopurine (BA)-treated roots; npcRNA 43 levels also increased over 2-fold in phosphate-starved roots but were unchanged in cytokinin-treated roots.
Figure 3.
Regulation of npcRNAs under conditions modifying root architecture. Expression of npcRNAs in roots of plants grown in low (−P) or high (+P) phosphate conditions and in the presence of cytokinin (BA) was examined. Real-time RT-PCR was performed for npcRNAs 34, 43, and 60 and data were normalized with ACTIN2. Values for roots grown in high-phosphate conditions or not treated with 0.1 μm BA are arbitrarily fixed to 1. For each cDNA synthesis, quantifications were made in triplicate and two biological replicates (I and II) were analyzed. Values are means ± sds.
Identification of a Putative miR162 pri-miRNA: An Alternatively Spliced Transcript
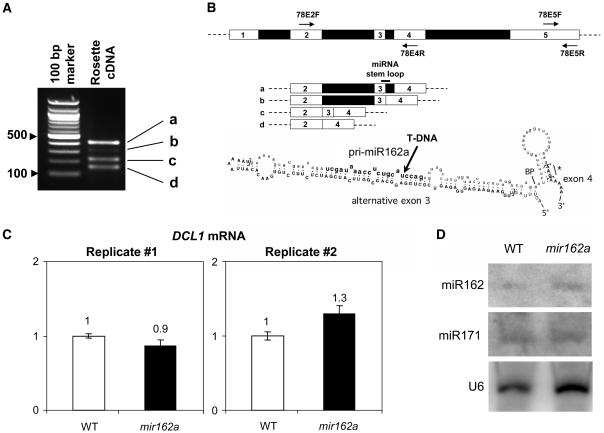
Among the npcRNA set, one candidate particularly drew our attention because it contained the miR162 sequence (Reinhart et al., 2002). This candidate, npcRNA 78, contains the miR162a sequence and may be the miR162a primary transcript (pri-miR162a). This gene will henceforth be referred to as MIR162a. To examine the expression of this pri-miRNA, we performed RT-PCR using primers in the predicted exons flanking the miRNA-containing region. The MIR162a locus produces at least four transcripts (Fig. 4A) in all tissues analyzed (see Supplemental Fig. 4). These splicing variants (a, b, c, and d) are generated by intron retention and skipping of exon 3, as determined by cloning and sequencing of the PCR products. The predicted miR162a hairpin contains both the entire exon 3 in its left arm and a large part of intron 3 in its right arm, including the miR162 sequence (Fig. 4B). Strikingly, the most likely putative branch point within intron 3 lies just downstream of the 3′ extremity of the predicted hairpin structure (Tolstrup et al., 1997). Additionally, two weak alternative 3′ acceptor splice sites (AAG) were identified (Fig. 4B, star). Such a situation has already been described in primates, where a weak acceptor site (GAG) is redundant and leads to two isoforms derived from the constitutive use of both acceptor sites (Manrow and Berger, 1993).
Figure 4.
Analysis of the npcRNA 78/MIR162a transcripts and molecular characterization of the mir162a mutant. A, RT-PCR detection of different npcRNA 78/MIR162a transcripts. RT-PCR was performed using npcRNA 78-specific primers located in exons 2 and 4 (78E2F/78E4R). B, Diagrammatic representation of the differentially spliced transcripts of npcRNA 78 as deduced from A and annotated structure of the miR162a predicted hairpin region. Exonic sequences are in uppercase bold letters, intronic sequences are in lowercase letters, and the miR162 sequence is in bold lowercase letters. Square brackets, 5′ and 3′ splice sites; BP, likely branch point; star, alternative codon. The arrow indicates the position of the SALK_107598 T-DNA insertion. C, Uncleaved DCL1 mRNA levels are similar in mir162a T-DNA insertion mutant and wild-type inflorescences. Real-time RT-PCR was performed using primers flanking the miR162-directed cleavage site in the DCL1 mRNA. Quantifications were normalized with ACTIN2. Values in wild-type inflorescences were arbitrarily fixed to 1. Quantifications were made in triplicate (error bars represent sds). Results for two biological replicates are presented. D, miR162 levels are comparable in wild-type and mir162a plants. Blots of RNA extracted from inflorescences of wild-type and mir162a plants were successively hybridized with miR162 and miR171 LNA-modified probes. Hybridization to a U6 probe served as a loading control.
npcRNA 78/MIR162a Transcripts Accumulate in a dcl1 Mutant
miR162 targets the DCL1 mRNA, which encodes the enzyme responsible for miRNA production (Xie et al., 2003). To analyze the function of the MIR162a npcRNA, a T-DNA insertion mutant (SALK_107598) in the miR162 sequence of this gene was identified (Fig. 4B). There were no detectable MIR162a transcripts in rosette leaves or inflorescences of the mutant (data not shown). No significant phenotype was observed in mir162a aerial organs or roots, although the initial insertion line displayed an altered floral phenotype that did not segregate with the T-DNA insertion in MIR162a (data not shown). Real-time RT-PCR experiments showed that the steady-state level of uncleaved DCL1 mRNA was unchanged in inflorescences of homozygote mir162a plants compared to their wild-type siblings (Fig. 4C). Moreover, miR162 was detected in mir162a plants at levels comparable to those of wild-type plants (Fig. 4D).
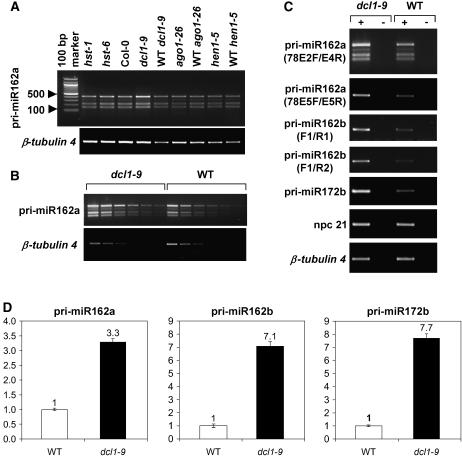
The accumulation of MIR162a transcripts was then assayed in several silencing-related mutants (hst, ago1, hen1, and dcl1; Fig. 5, A and B). All MIR162a transcripts accumulated in the partial loss-of-function dcl1-9 mutant, as did the well-characterized miR172b primary transcript (Aukerman and Sakai, 2003), suggesting that the MIR162a npcRNA is a genuine substrate of DCL1 and processed in vivo to yield miR162. This miRNA can be derived from two distinct loci: indeed, the Arabidopsis genome contains a second predicted miR162 locus, MIR162b (Reinhart et al., 2002). Because expression data concerning the MIR162b gene were lacking, we designed primers to detect the miR162b primary transcript. Semiquantitaive and quantitative RT-PCR experiments indicated very low abundance of this transcript in wild-type plants, whereas it accumulated in dcl1-9 plants (Fig. 5, C and D). The two sets of primer pairs used in these experiments (pri-miR162b-F1/R1 and pri-miR162b-F1/R2; see Supplemental Table II) did not reveal any alternative splicing of the miR162b primary transcript, suggesting that miR162 does not derive from an alternative intron of this transcript. Moreover, based on these RT-PCR experiments, there appear to be at least two alternative 3′ ends for the MIR162b primary transcript because we were able to amplify a PCR product using a reverse primer located 258 nt downstream from the 3′ end of this transcript proposed by Xie et al. (2005) based on 3′-RACE experiments.
Figure 5.
npcRNA 78/MIR162a transcripts are stabilized in the dcl1-9 mutant. A, RT-PCR was performed using npcRNA 78-specific primers located in exons 2 and 4 (78E2F/78E4R) on RNA from rosettes of RNAi-related mutants. B, Dilution series of cDNAs from dcl1-9 and wild-type inflorescences shows consistently higher amplification by RT-PCR in the mutant when using the above-mentioned primers. C, Accumulation of miR162a and b primary transcripts in the dcl1-9 mutant. Specific primer pairs used for npcRNA 78/MIR162a amplification were located either in exons 2 and 4 (78E2F/78E4R) or in exon 5 (78E5F/78E5R). Primers pairs used for amplification of pri-miR162b (pri-miR162b-F1/R1 and pri-miR162b-F1/R2) did not reveal any alternative splicing for this transcript. The miR172b (EAT) primary transcript is a known pri-miRNA. npcRNA 21 is not affected by the dcl1 mutation. ±, RT-PCR performed with or without reverse transcriptase. D, Accumulation of miR162a and b primary transcripts in the dcl1-9 mutant measured by real-time RT-PCR. Primers used for npcRNA 78/MIR162a amplification were located in exon 5 (78E5F/78E5R). Primers pri-miR162b-F1 and R1 were used for amplification of pri-miR162b. Data were normalized with ACTIN2. Values in wild-type inflorescences were arbitrarily fixed to 1. Three technical replicates were performed for wild-type and dcl1-9 inflorescences. Values are means ± sds. For all these RT-PCR experiments, analyses were performed on homozygous mutants and their wild-type siblings.
Taken together, these results suggest that the MIR162b gene may compensate for the loss of function of MIR162a and that miR162 is probably released from the MIR162a primary transcript.
DISCUSSION
npcRNAs, an Intriguing Portion of the Arabidopsis Transcriptome
In our initial computational screen, over 1,000 candidates could be sorted, but novel intergenic npcRNA transcripts were selected using strict criteria. Although we probably lost a number of bona fide npcRNAs, the 43 npcRNAs we did retain constitute a restricted set of very reliable npcRNAs. It must be noted that, in the absence of information on the size of these RNAs, some predicted npcRNA genes may turn out to be protein coding upon further analysis. Nevertheless, this new, highly curated and experimentally supported collection of npcRNAs expands the list of transcripts previously suggested to be Arabidopsis npcRNAs (e.g. AtGUT15, AtCR20-1, At4, AtIPS1, and JAW; see the database of plant noncoding RNAs [http://www.prl.msu.edu/PLANTncRNAs]) and broadens our view of potential peptide-coding and functional RNAs in plants. In a recent computational search for orphan transcripts not previously assigned to genomic loci, a set of approximately 560 putative noncoding or peptide-coding RNAs was identified (Riano-Pachon et al., 2005), but only nine of these transcripts were found in our dataset, among which two correspond to the same npcRNA. Sixty of these orphan transcripts encode ORFs longer than 70 amino acids (data not shown) and many others are based on single ESTs. In this work, we favored the detection of bona fide npcRNAs for future biological studies by selecting those containing very short ORFs and detected by at least two ESTs.
Using massively parallel signature sequencing, Meyers et al. (2004) detected 4,698 antisense signatures with unique genomic positions and 3,455 signatures mapping to unique positions in intergenic regions of the Arabidopsis genome. In another study, antisense transcripts for over 7,500 annotated genes were detected using whole-genome tiling microarrays and transcriptional activity was detected in 2,000 intergenic regions (Yamada et al., 2003). These large numbers do not exclusively reflect the number of npcRNAs in the Arabidopsis genome because a portion of these sequences may contain long ORFs and encode proteins, constitute UTRs of incompletely annotated genes, and, in the case of sequences detected on tiling arrays, may result from cross-hybridization. Such whole-genome approaches tend to yield large sets of candidate npcRNAs but cannot be fully exploited to sort true npcRNAs without detailed analysis of genomic contexts. The number of npcRNAs in the genome of Arabidopsis could consequently be anywhere between two extremes: our strict underestimated set and these sets that likely contain a significant portion of non-npcRNA sequences. Taken together, our work and these different studies demonstrate that npcRNAs undoubtedly constitute a substantial and so far overlooked portion of the transcriptome.
Under no strand bias conditions, the frequencies of A and T and of G and C should be equal in each DNA strand (Lobry, 1995). Deviations from these intrastrand equimolarities have been extensively studied in prokaryotic, organelle, and viral genomes (Grigoriev, 1998; Mrazek and Karlin, 1998; Frank and Lobry, 1999). Recently, deviations from these rules have also been established in several eukaryotic genomes, a property that has been linked to asymmetries in transcription (Touchon et al., 2004). However, in contrast to other genomes, the analysis of a large number of protein-coding genes from Arabidopsis showed that transcribed regions containing no selected sequence element present very small values of the TA and GC biases. Conversely, comparison of our Arabidopsis npcRNA genes with their neighboring 5′ and 3′ intergenic sequences showed large S biases. This was paralleled with a significantly high GC content of these regions. Therefore, the npcRNA sequences contained in our set are strongly biased, suggesting that they are submitted to selection processes leading to the observed compositional properties. Such biases can reflect the presence of sequence motifs involved in RNA-protein interactions and/or RNA secondary structure elements. Interestingly, potential RNA secondary structures seem to be present in several npcRNAs, as monitored using Z-score values. This criterion has been used for other genes of this class (Crespi et al., 1994) or for the prediction of miRNA hairpins (Bonnet et al., 2004; Jones-Rhoades and Bartel, 2004; Wang et al., 2004) and particular secondary RNA structures have been shown to play diverse roles in mRNA translation, stability, and localization (Eddy, 2002). Finally, the step-like profiles evidenced at both gene extremities of npcRNAs suggest that the ends of the sequences correspond, within the limits of the window size, to the true 5′ and 3′ gene extremities.
Regulatory Roles for npcRNAs
We classified the npcRNAs as putative peptide-coding RNAs (sORF-RNAs) or pfRNAs, although this classification is at best tentative, and further biological analyses are required to define their active gene products. Nevertheless, the presence of conserved sORFs in homologous transcripts strongly argues for the classification as peptide-coding RNAs. Peptide signaling is an emerging field in plants (Lindsey et al., 2002), and our collection of npcRNAs offers interesting candidates for further analysis. However, translation into peptides does not preclude an additional role of the npcRNAs per se, as has been shown for the enod40 transcript in legumes (Sousa et al., 2001). Being directed to the ribosome and translated by the protein synthetic machinery, whatever the size of the sORFs they contain, is likely to be the default pathway for capped and polyadenylated transcripts. Consequently, certain npcRNAs may regulate translation, a function that might stem from their default localization in active translation sites (Zalfa et al., 2003). Such a regulatory role would be akin to that of miRNAs in animal cells, which are believed to act mainly through translation repression by binding to target 3′-UTRs (He and Hannon, 2004). Conservation at the nucleotide level, independent of encoded peptides, as well as the presence of statistically significant secondary structures strongly support a role for the npcRNA molecules. These conserved or structured RNA domains could also be functional elements that play a role in posttranscriptional regulation of these npcRNAs, as has been shown for conserved 5′- or 3′-UTRs of protein-coding genes. Integration into RNP complexes seems to be a common theme in the action of certain npcRNAs. A small number of examples, such as the meiRNA in yeast (Saccharomyces cerevisiae) and enod40 in legume plants, have shown that npcRNAs may be required for correct localization of RNP particles (Yamashita et al., 1998; Franco-Zorrilla et al., 2002; Campalans et al., 2004). Furthermore, miRNAs confer specificity to the action of the RNP known as the RNA-induced silencing complex (Bartel, 2004; He and Hannon, 2004).
The tissue-specific expression patterns of several npcRNAs from our set support a developmental role for these transcripts. Three npcRNAs identified in this study are regulated by growth conditions that alter root architecture, widening the set of npcRNAs responsive to endogenous and external cues in roots, as shown for At4 and enod40 (Franco-Zorrilla et al., 2002; Campalans et al., 2004). Hence, npcRNAs may frequently play regulatory roles in root responses that require high plasticity.
Identification of an Alternatively Spliced Transcript Containing the miR162a Sequence
Among our dataset, we identified a miRNA primary transcript, pri-miR162a/npcRNA 78. Our study provides a detailed description showing that a plant miRNA can be encoded within an intron of an npcRNA. There is accumulating evidence of miRNAs that are intron-derived from noncoding transcripts in mammals. Recently, 27 mammalian miRNAs were located within introns of long npcRNAs (Rodriguez et al., 2004). Our finding indicates that this class of miRNAs is not restricted to the animal kingdom.
Expression analysis revealed alternative splicing events involving the miRNA 162a-containing region. Among the few plant miRNA primary transcripts characterized, other cases of alternative splicing have been reported. pri-miR172b and pri-miR163 are alternatively spliced, although both miRNA sequences are localized in exons (Aukerman and Sakai, 2003; Kurihara and Watanabe, 2004) and both Zm MIR166a and Zm MIR166b produce unspliced and spliced transcripts (Kidner and Martienssen, 2004).
Are miRNAs processed out of intron lariats, as previously reported for small nucleolar RNAs (Weinstein and Steitz, 1999)? In the case of the pri-miR162a, the miRNA may not derive from the excised intron but rather from the full-length unspliced transcript. Indeed, miRNA processing solely from the intron lariat would not be expected to lead to an increase in the levels of all RNA isoforms in the dcl1 mutant. The fact that spliced transcripts that do not contain miR162 accumulate in this mutant suggests that splicing and miRNA processing are acting on the same pool of unspliced RNA and could thus be competitive nuclear events. Furthermore, no changes in pri-miR162a levels were detected in hst mutants affected in the function of an exportin-5 homolog (Park et al., 2005), reinforcing the idea that miR162a production occurs in the nucleus.
Detection of increased transcript accumulation for both MIR162a and MIR162b genes in plants with reduced DCL1 activity suggests that both encode genuine miR162 primary transcripts. As previously suggested (Xie et al., 2003), DCL1 and miR162 may be involved in a negative feedback loop. Analysis of a SALK T-DNA insertion line in the MIR162a/npcRNA 78 gene revealed unaltered miR162 and DCL1 mRNA levels, suggesting functional redundancy or overlap between the MIR162a and MIR162b genes. Our results suggest that both pri-miR162a and b are substrates of DCL1 and contribute to DCL1 feedback regulation. Alternatively, accumulation of a particular pri-miRNA in a dcl1 mutant may be due to an indirect effect of the perturbation of DCL1 function. dcl1 mutations may induce pleiotropic effects via deregulation of miRNA-controlled transcription factor mRNAs (Rhoades et al., 2002). This in turn may increase transcription of pri-miRNA genes, although very little is currently known about pri-miRNA promoters or transcriptional control of these genes.
Two other pri-miRNAs for which ESTs are available, MIR171 and MIR172b, are npcRNAs (their longest ORFs are 153 and 159 nt long, respectively). Low abundance of pri-miRNAs (Juarez et al., 2004), contrasting with relatively high levels of miRNAs due to rapid processing, may explain their absence from most EST and cDNA libraries. The pri-miR162a may be an interesting exception, possibly due to the complex alternative splicing it undergoes, a feature that may be related to its function as a negative regulator of DCL1. Nevertheless, we cannot rule out that certain npcRNAs may be primary transcripts of so far unidentified miRNAs or endogenous tasiRNAs (Peragine et al., 2004; Vazquez et al., 2004b). Indeed, the npcRNA 41 (annotated as an expressed protein; Table I) has been shown to generate various tasiRNAs (Allen et al., 2005). More generally, much like mi/siRNA precursors, other types of npcRNAs may need to be processed to give rise to functional RNA products.
Elucidating the molecular mechanisms in which npcRNAs are involved is of major interest. Depending on their localization within the cell, npcRNAs may play a number of roles in transcription, RNA maturation, and translation, as well as chromatin structure, chromosomal silencing, and imprinting. Functional analyses of this collection of Arabidopsis npcRNAs should help us to better grasp the scope of npcRNA cellular roles.
MATERIALS AND METHODS
Computational Analyses
Arabidopsis (Arabidopsis thaliana) ESTs (172,495 sequences) and mRNAs (24,985 sequences) were retrieved from the National Center for Biotechnology Information (NCBI), as were the genomic sequence and annotation data (August 2002). The detection procedure of the npcRNA genes was performed as follows. For the process of alignment, full-length cDNA sequences were used as such; for ESTs, a clustering step was performed to reconstruct mRNAs. To avoid artefacts due to microsatellites and repeated regions (which can lead to chimeric constructs), we used genome sequences as a guide for the clustering. ESTs/cDNAs were first tentatively assigned to few (or single) high-quality matches on the genome. This was performed by stringent validation thresholds (percent of nucleotide identity larger than 97% over at least 90% of the sequence) of the results of a BLAST 2.2.10 from NCBI against the genome, as well as by eliminating ESTs matching at multiple distant hits, which often characterize low-complexity sequences, regulatory, or mobile elements. ESTs/cDNAs were aligned with SIM4 (Ogasawara and Morishita, 2003) to determine the exon/intron organization. A sequence was retained if splice site sequences were canonical (GT/AG extremities) and if the size of each intron was smaller than 10 kb. When they did not overlap previously annotated genes, we retained as genes the genomic fragments corresponding to single mRNAs or to a cluster of two or more ESTs/cDNAs. Since 5′- and 3′-UTR annotations were lacking in a number of Arabidopsis genes, putative genes situated in close vicinity (<200 bp) of annotated genes were discarded in our initial screening. The strand orientation of candidate genes was determined using the majority rule in EST assignations. The longest possible ORF within each gene was then determined and only candidates that did not exhibit an ORF longer than 210 nt (70 amino acids) were retained. The probability of protein-coding capacity for these sORFs was evaluated with nonhomogeneous fifth-order Markov models of the three frames of all the coding sequences and all introns of Arabidopsis, and a Bayesian estimation with a prior probability of 0.3 to be coding. Signal peptides were detected with SignalP (Bendtsen et al., 2004). Final status (pf, putative functional RNA; sORF, short ORF-encoded peptide) was set to pf according to the following criteria applied in decreasing order: ORF length smaller than 100 nt, numerous ATGs before the start of the CDS, high noncoding probability, absence of a signal peptide, and ORF length less than 20% of the gene length. In the course of experiments, the candidates were mapped again on the TAIR genome (release January 2004).
The presence of statistically significant secondary structures was monitored using Z-score values as described by Crespi et al. (1994) and Bonnet et al. (2004). The secondary structures showing the lowest free energy of folding were calculated using the Vienna package (http://www.tbi.univie.ac.at/∼ivo/RNA). For each mRNA, the sequence was scanned every 10 nt using sliding windows of sizes ranging between 31 and 301 nt (by increments of 20 nt). For each window, 300 shuffled sequences (in mononucleotides) were generated to estimate the mean and sd of the free energy of folding for all possible sequences. The Z score is the number of sds between the actual free energy of folding of the sequence and the mean value of the energies of folding of the shuffled sequences (Crespi et al., 1994). This detection was performed on the npcRNA genes and, as a control, on a set of Arabidopsis protein-coding sequences that have a GC content similar to that of the npcRNA genes. The control set comprised the 1,488 coding sequences annotated in TAIR (excluding repeat elements and pseudogenes) with a GC content ranging between 35% and 40% and spanned an overall length of 1.5 Mb.
Plant Material and Growth Conditions
All experiments were performed on the Columbia (Col-0) ecotype of Arabidopsis. For all in vitro experiments, plants were grown in long days (16-h-light/8-h-dark photoperiod) with 150 μmol m−2 s−1 of supplemental fluorescent light at 23°C. Roots were collected on 8-d-old plants grown vertically on plates containing 0.5× Murashige and Skoog salts (Sigma), 1% Suc, and 0.8% agar. For cytokinin treatments, BA was added to the autoclaved medium at a 0.1 μm final concentration. For phosphate starvation assays, plants were grown on 0.1× Murashige and Skoog, 0.5% Suc, and 0.8% agar plates supplemented with 5 or 500 μm NaH2PO4 and roots were collected on 13-d-old plants. Salt stress experiments were performed on 3-week-old plants grown in liquid 0.5× Murashige and Skoog, 1% Suc medium. The plants were transferred to 150 mM NaCl-containing medium for 2 h.
For all other purposes, plants were grown in vitro for 1 week and then transferred to the greenhouse (16-h-light/8-h-dark photoperiod with a minimum of 150 μmol m−2 s−1 of light ensured by supplemental fluorescent tubes, 23°C, 60% relative humidity). Rosette and cauline leaves as well as stems were collected from 3-week-old plants and inflorescences from 1-month-old plants. Cell suspensions were maintained under continuous light at 23°C and samples collected on 5-d-old cultures.
The mir162a T-DNA mutant (SALK_107598) was identified in the collection of SALK mutants (Alonso et al., 2003). PCR amplification and sequencing confirmed the presence of the T-DNA insert within the miR162a sequence using the following primers: LBb1 (5′-GCGTGGACCGCTTGCTGCAACT-3′) located on the left border of the T-DNA, and primers located in the MIR162a/npcRNA 78 gene (78E2F, 5′-GTCTGCAGATGCATGTGTGT-3′ and 78E4R, 5′-AAATCCTCAGCTTTCCCAGA-3′).
Seeds of silencing-related mutants ago1-26, dcl1-9, hen1-5, hst-1, and hst-6, respectively described by Morel et al. (2002), Vazquez et al. (2004a, 2004b), and Telfer and Poethig (1998), were kindly provided by Hervé Vaucheret. The dcl1-9 mutant in Col-0 was obtained by five backcrosses of the original dcl1-9 mutant (Jacobsen et al., 1999) to Col-0, as described by Vazquez et al. (2004b). All analyses were performed on homozygous mutants and their wild-type siblings.
5′-RACE
Total RNA was extracted using TRIzol reagent (Invitrogen) followed by column purification (RNeasy mini kit; Qiagen). 5′-RACE was carried out using the FirstChoice RLM-RACE kit (Ambion). The RNA ligase-mediated 5′-RACE procedure is selective for transcripts that contain a 5′ cap. 5′-RACE was performed on total RNA from roots (for npcRNAs 113, 311, and 351) or flowers (for npcRNAs 155, 156, and 375) according to the manufacturer's instructions. The gene-specific reverse primers used in the RACE-PCR reactions are as follows: 113-outer, 5′-CAACCATCGTACTCGCTTCATCT-3′; 113-inner, 5′-GCCATGTGTGGAGGAGCTATAAT-3′; 155-outer, 5′-GCTCCTTGTTGAAGCCAACCAT-3′; 155-inner, 5′-AACGTTGGTTGCGATCATCGT-3′; 156-outer, 5′-AAGCTGGCCAACGCTCCTTATAGA-3′; 156-inner, 5′-ATCACAACTCCGGAAGTCGGAGA-3′; 311-outer, 5′-GACACATGAGCAAACATAGTCCAA-3′; 311-inner, 5′-TCATGGCCAAGCTAAACAAACTGT-3′; 351-outer, 5′-GAGACTCCACCACCGATTACA-3′; 351-inner, 5′-CGGTAACAGAAGATCCGATATGT-3′; 375-outer, 5′-CAACCACGAATTCTCTGTCTTCT-3′; and 375-inner, 5′-TGTCCAACAAGCAAGGAATGT-3′ (where “inner” designates primers used in the initial amplification step and “outer” primers used in the second, nested PCR). PCR products from the 5′-RACE reactions were cloned using the pGEM-T Easy system (Promega). Between five and 10 randomly chosen clones were sequenced for each RACE product.
Semiquantitative and Real-Time RT-PCR
Total RNA was extracted from plant tissues using the RNeasy plant mini kit (Qiagen). Residual genomic DNA was removed by on-column DNAse I digestion, using the RNase-free DNase set (Qiagen). RT was performed on 2 μg of total RNA using SuperScript II reverse transcriptase (Invitrogen) and (T)16 A/G/C oligonucleotides. RT-PCR was carried out using the primer pairs listed in Supplemental Tables I and II. Primer design was performed with SPADS (http://genoplante-info.infobiogen.fr/spads/spads.html), Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), or by careful analysis of the sequence.
For tissue expression experiments, RT-PCR reactions were performed at least three times using various cDNA dilutions and cycle numbers (between 23 and 30 cycles) to characterize the abundance of each transcript. Semiquantitative RT-PCR was performed on two biological replicates for each root treatment. PCR products were analyzed on BET-stained agarose gels. Real-time RT-PCR was performed on the Roche Light Cycler instrument using SYBR Green I dye (LightCycler FastStart DNA MasterPLUS SYBR Green I; Roche). The pGEM-T Easy system (Promega) was used for cloning prior to sequencing of PCR products.
Analysis of miRNA Expression
Total RNA was isolated from inflorescences of homozygous mir162a (SALK_107598) plants and their wild-type siblings using TRIzol reagent (Invitrogen). The same material as that used for DCL1 expression analyses was used for miRNA detection. Thirty micrograms of each RNA were subjected to electrophoresis on a denaturing 17% polyacrylamide gel and electroblotted onto Hybond-N+ filter paper (GE Healthcare) using the mini-PROTEAN II system (Bio-Rad). The blot was probed with an end-labeled locked nucleic acid (LNA)-modified oligonucleotide (Exiqon; Valoczi et al., 2004) complementary to miRNA 162 (5′-CTGGATGCAGAGGTTTATCGA-3′). The same filter was then successively stripped and reprobed with an LNA-modified miR171 antisense probe (5′-GATATTGGCGCGGCTCAATCA-3′) and a U6 antisense probe (5′-GCAGGGGCCATGCTAATCTTCTCTGTATCGT-3′). Probes were prepared by end labeling 20 pmol of the oligonucleotide with T4 polynucleotide kinase and [γ32P]ATP.
Supplementary Material
Acknowledgments
We thank Sakari Kaupinen (Exiqon, Denmark) for the kind gift of locked nucleic acid-modified oligonucleotides for detection of miRNAs, Hervé Vaucheret (INRA, Versailles, France) for all RNAi-related Arabidopsis mutants and useful advice, Scott Poethig (University of Pennsylvania, Philadelphia) for the hst-1 and hst-6 mutants, as well as Florian Frugier (ISV-CNRS, Gif sur Yvette, France) for a careful reading of the manuscript.
This work was supported by the GENOPLANTE Program (project no. Bi2001029) and in part by the European Community FP6 RIBOREG project (LSHG–CT–2003503022).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Martin Crespi (crespi@isv.cnrs-gif.fr).
The online version of this article contains Web-only data.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC (2005) microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121: 207–221 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H (2003) Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 15: 2730–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CC, Sieber P, Wellmer F, Meyerowitz EM (2005) The early extra petals1 mutant uncovers a role for microRNA miR164c in regulating petal number in Arabidopsis. Curr Biol 15: 303–315 [DOI] [PubMed] [Google Scholar]
- Barakat A, Szick-Miranda K, Chang IF, Guyot R, Blanc G, Cooke R, Delseny M, Bailey-Serres J (2001) The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol 127: 398–415 [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S (2004) Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340: 783–795 [DOI] [PubMed] [Google Scholar]
- Bonnet E, Wuyts J, Rouze P, Van de Peer Y (2004) Evidence that microRNA precursors, unlike other non-coding RNAs, have lower folding free energies than random sequences. Bioinformatics 20: 2911–2917 [DOI] [PubMed] [Google Scholar]
- Campalans A, Kondorosi A, Crespi M (2004) Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell 16: 1047–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casson SA, Chilley PM, Topping JF, Evans IM, Souter MA, Lindsey K (2002) The POLARIS gene of Arabidopsis encodes a predicted peptide required for correct root growth and leaf vascular patterning. Plant Cell 14: 1705–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crespi MD, Jurkevitch E, Poiret M, d'Aubenton-Carafa Y, Petrovics G, Kondorosi E, Kondorosi A (1994) enod40, a gene expressed during nodule organogenesis, codes for a non-translatable RNA involved in plant growth. EMBO J 13: 5099–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N (2002) Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Eddy SR (2002) Computational genomics of noncoding RNA genes. Cell 109: 137–140 [DOI] [PubMed] [Google Scholar]
- Erdmann VA, Barciszewska MZ, Szymanski M, Hochberg A, de Groot N, Barciszewski J (2001) The non-coding RNAs as riboregulators. Nucleic Acids Res 29: 189–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martin AC, Solano R, Rubio V, Leyva A, Paz-Ares J (2002) Mutations at CRE1 impair cytokinin-induced repression of phosphate starvation responses in Arabidopsis. Plant J 32: 353–360 [DOI] [PubMed] [Google Scholar]
- Frank AC, Lobry JR (1999) Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238: 65–77 [DOI] [PubMed] [Google Scholar]
- Grigoriev A (1998) Analyzing genomes with cumulative skew diagrams. Nucleic Acids Res 26: 2286–2290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ (2004) MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet 5: 522–531 [DOI] [PubMed] [Google Scholar]
- Jacobsen SE, Running MP, Meyerowitz EM (1999) Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126: 5231–5243 [DOI] [PubMed] [Google Scholar]
- Jones-Rhoades MW, Bartel DP (2004) Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol Cell 14: 787–799 [DOI] [PubMed] [Google Scholar]
- Joyce GF (2002) The antiquity of RNA-based evolution. Nature 418: 214–221 [DOI] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC (2004) microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature 428: 84–88 [DOI] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA (2004) Spatially restricted microRNA directs leaf polarity through ARGONAUTE1. Nature 428: 81–84 [DOI] [PubMed] [Google Scholar]
- Kiss T (2002) Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell 109: 145–148 [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Watanabe Y (2004) Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc Natl Acad Sci USA 101: 12753–12758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovitch MP, Nguyen VC, Gross MS, Solhonne B, Leibovitch SA, Bernheim A (1991) The human ASM (adult skeletal muscle) gene: expression and chromosomal assignment to 11p15. Biochem Biophys Res Commun 180: 1241–1250 [DOI] [PubMed] [Google Scholar]
- Leighton PA, Ingram RS, Eggenschwiler J, Efstratiadis A, Tilghman SM (1995) Disruption of imprinting caused by deletion of the H19 gene region in mice. Nature 375: 34–39 [DOI] [PubMed] [Google Scholar]
- Lindsey K, Casson S, Chilley P (2002) Peptides: new signalling molecules in plants. Trends Plant Sci 7: 78–83 [DOI] [PubMed] [Google Scholar]
- Lobry JR (1995) Properties of a general model of DNA evolution under no-strand-bias conditions. J Mol Evol 40: 326–330 [DOI] [PubMed] [Google Scholar]
- MacIntosh GC, Wilkerson C, Green PJ (2001) Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol 127: 765–776 [PMC free article] [PubMed] [Google Scholar]
- Manrow RE, Berger SL (1993) GAG triplets as splice acceptors of last resort. An unusual form of alternative splicing in prothymosin alpha pre-mRNA. J Mol Biol 234: 281–288 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Morel JB, Godon C, Mourrain P, Beclin C, Boutet S, Feuerbach F, Proux F, Vaucheret H (2002) Fertile hypomorphic ARGONAUTE (ago1) mutants impaired in post-transcriptional gene silencing and virus resistance. Plant Cell 14: 629–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrazek J, Karlin S (1998) Strand compositional asymmetry in bacterial and large viral genomes. Proc Natl Acad Sci USA 95: 3720–3725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Numata K, Kanai A, Saito R, Kondo S, Adachi J, Wilming LG, Hume DA, Hayashizaki Y, Tomita M (2003) Identification of putative noncoding RNAs among the RIKEN mouse full-length cDNA collection. Genome Res 13: 1301–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara J, Morishita S (2003) A fast and sensitive algorithm for aligning ESTs to the human genome. J Bioinform Comput Biol 1: 363–386 [DOI] [PubMed] [Google Scholar]
- Park MY, Wu G, Gonzalez-Sulser A, Vaucheret H, Poethig RS (2005) Nuclear processing and export of microRNAs in Arabidopsis. Proc Natl Acad Sci USA 102: 3691–3696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS (2004) SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev 18: 2368–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart BJ, Weinstein EG, Rhoades MW, Bartel B, Bartel DP (2002) MicroRNAs in plants. Genes Dev 16: 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades MW, Reinhart BJ, Lim LP, Burge CB, Bartel B, Bartel DP (2002) Prediction of plant microRNA targets. Cell 110: 513–520 [DOI] [PubMed] [Google Scholar]
- Riano-Pachon DM, Dreyer I, Mueller-Roeber B (2005) Orphan transcripts in Arabidopsis thaliana: identification of several hundred previously unrecognized genes. Plant J 43: 205–212 [DOI] [PubMed] [Google Scholar]
- Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A (2004) Identification of mammalian microRNA host genes and transcription units. Genome Res 14: 1902–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C, Johansson C, Charon C, Manyani H, Sautter C, Kondorosi A, Crespi M (2001) Translational and structural requirements of the early nodulin gene enod40, a short-open reading frame-containing RNA, for elicitation of a cell-specific growth response in the alfalfa root cortex. Mol Cell Biol 21: 354–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor MS, Devon RS, Millar JK, Porteous DJ (2003) Evolutionary constraints on the Disrupted in Schizophrenia locus. Genomics 81: 67–77 [DOI] [PubMed] [Google Scholar]
- Telfer A, Poethig RS (1998) HASTY: a gene that regulates the timing of shoot maturation in Arabidopsis thaliana. Development 125: 1889–1898 [DOI] [PubMed] [Google Scholar]
- Tenson T, DeBlasio A, Mankin A (1996) A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA 93: 5641–5646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolstrup N, Rouze P, Brunak S (1997) A branch point consensus from Arabidopsis found by non-circular analysis allows for better prediction of acceptor sites. Nucleic Acids Res 25: 3159–3163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touchon M, Arneodo A, d'Aubenton-Carafa Y, Thermes C (2004) Transcription-coupled and splicing-coupled strand asymmetries in eukaryotic genomes. Nucleic Acids Res 32: 4969–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valoczi A, Hornyik C, Varga N, Burgyan J, Kauppinen S, Havelda Z (2004) Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res 32: e175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Gasciolli V, Crete P, Vaucheret H (2004. a) The nuclear dsRNA binding protein HYL1 is required for microRNA accumulation and plant development, but not posttranscriptional transgene silencing. Curr Biol 14: 346–351 [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P (2004. b) Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol Cell 16: 69–79 [DOI] [PubMed] [Google Scholar]
- Wang XJ, Reyes JL, Chua NH, Gaasterland T (2004) Prediction and identification of Arabidopsis thaliana microRNAs and their mRNA targets. Genome Biol 5: R65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein LB, Steitz JA (1999) Guided tours: from precursor snoRNA to functional snoRNP. Curr Opin Cell Biol 11: 378–384 [DOI] [PubMed] [Google Scholar]
- Wen J, Lease KA, Walker JC (2004) DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J 37: 668–677 [DOI] [PubMed] [Google Scholar]
- Xie Z, Allen E, Fahlgren N, Calamar A, Givan SA, Carrington JC (2005) Expression of Arabidopsis MIRNA genes. Plant Physiol 138: 2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Kasschau KD, Carrington JC (2003) Negative feedback regulation of Dicer-Like1 in Arabidopsis by microRNA-guided mRNA degradation. Curr Biol 13: 784–789 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yamashita A, Watanabe Y, Nukina N, Yamamoto M (1998) RNA-assisted nuclear transport of the meiotic regulator Mei2p in fission yeast. Cell 95: 115–123 [DOI] [PubMed] [Google Scholar]
- Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C (2003) The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112: 317–327 [DOI] [PubMed] [Google Scholar]
- Zhou J, Goldsbrough PB (1994) Functional homologs of fungal metallothionein genes from Arabidopsis. Plant Cell 6: 875–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.