Abstract
Eukaryotic E2Fs are conserved transcription factors playing crucial and antagonistic roles in several pathways related to cell division, DNA repair, and differentiation. In plants, these processes are strictly intermingled at the growing zone to produce postembryonic development in response to internal signals and environmental cues. Of the six AtE2F proteins found in Arabidopsis (Arabidopsis thaliana), only AtE2Fa and AtE2Fb have been clearly indicated as activators of E2F-responsive genes. AtE2Fa activity was shown to induce S phase and endoreduplication, whereas the function of AtE2Fb and the interrelationship between these two transcription factors was unclear. We have investigated the role played by the AtE2Fb gene during cell cycle and development performing in situ RNA hybridization, immunolocalization of the AtE2Fb protein in planta, and analysis of AtE2Fb promoter activity in transgenic plants. Overexpression of AtE2Fb in transgenic Arabidopsis plants led to striking modifications of the morphology of roots, cotyledons, and leaves that can be ascribed to stimulation of cell division. The accumulation of the AtE2Fb protein in these lines was paralleled by an increased expression of E2F-responsive G1/S and G2/M marker genes. These results suggest that AtE2Fa and AtE2Fb have specific expression patterns and play similar but distinct roles during cell cycle progression.
The identification of various components of the plant cell cycle machinery has revealed remarkable similarities with the regulatory pathways found in animal cells, for which a key role is exerted by the E2F/DP family of transcription factors. The genome of the model plant Arabidopsis (Arabidopsis thaliana) contains eight genes of this family (six E2Fs and two DPs), whereas in mammalian cells 10 E2F/DP members have been discovered (eight E2Fs and two DPs; Attwooll et al., 2004; Christensen et al., 2005; Dimova and Dyson, 2005; Maiti et al., 2005). Most mammalian E2F proteins (E2F1–5) and three of the Arabidopsis members (AtE2Fa–c) show a similar domain organization, characterized by a highly conserved DNA-binding domain followed by a DP heterodimerization domain and a C-terminal transactivating domain, containing the pocket protein-binding region. The mammalian E2F6 lacks the carboxy-terminal transactivating region. Six mammalian E2Fs (E2F1–6) and three Arabidopsis E2F proteins (AtE2Fa–c) bind DNA by forming heterodimers with the distantly related DP proteins that contribute a second DNA-binding domain for binding to the consensus E2F cis-elements found in several E2F-responsive promoters. The remaining Arabidopsis E2Fs (AtE2Fd, e, and f/DEL2, 1, and 3) and the E2F7 and E2F8 proteins of mammalian cells only contain conserved duplicated DNA-binding domains. They cannot form heterodimers with DP proteins, but their duplicated DNA-binding domains allow autonomous binding to the consensus E2F sites (Mariconti et al., 2002; Kosugi and Ohashi, 2002a; de Bruin et al., 2003; Di Stefano et al., 2003; Christensen et al., 2005; Maiti et al., 2005).
E2F transcriptional regulation relies on activating or repressing functions that depend, in part, on the interaction between some E2Fs and the pocket proteins, known as the pRB/E2F pathway (Stevens and La Thangue, 2003). The mammalian E2F1 to 5 proteins can interact with hypophosphorylated pocket proteins and have been divided in two subclasses of activating and repressive factors, playing crucial and antagonistic roles in the regulation of several genes involved in DNA replication and expressed during late G1 and near the G1/S boundary (Trimarchi and Lees, 2002). E2F1 to 3 are potent activators of E2F-responsive genes and their overexpression can induce quiescent cells to reenter the cell cycle (Johnson et al., 1994; Shan and Lee, 1994; Singh et al., 1994; Xu et al., 1995). As judged by the changes in global gene expression induced by their overexpression, E2F1 to 3 play different activating roles during differentiation and development. This observation is supported by the analysis of mouse mutant strains characterized by the knocking out of these E2F genes (Field et al., 1996; Yamasaki et al., 1996; Humbert et al., 2000). In contrast, E2F4 and E2F5 expressed predominantly in quiescent cells and hence are thought to act mainly as repressors of cell cycle genes (Trimarchi and Lees, 2002). E2F6 has been shown to be a transcriptional repressor, whereas the E2F7 and E2F8 factors are believed to act as inhibitors of E2F transcriptional activity (Trimarchi et al., 2001; de Bruin et al., 2003; Di Stefano et al., 2003; Maiti et al., 2005). Similar to human E2F1 to 5, the homologous Arabidopsis AtE2Fa to c proteins have been classified as activating (AtE2Fa and b) or repressive factors (AtE2Fc) and shown to interact with plant pocket proteins (pRBR) in yeast two-hybrid and in vitro pull-down experiments (de Jager et al., 2001; del Pozo et al., 2002).
The physiological roles of AtE2Fa and AtE2Fc have been examined at the cellular and organism levels. Transient overexpression of AtE2Fa in Arabidopsis protoplasts from mature leaves induces these quiescent cells to progress into S phase (Rossignol et al., 2002). In transgenic Arabidopsis plants, AtE2Fa overexpression induces ectopic cell division, while overexpression of AtE2Fa in combination with AtDPa can either induce endoreduplication or cell proliferation depending on the cellular or developmental context, resulting in delayed differentiation and a striking block in development (De Veylder et al., 2002). Plants ectopically overexpressing AtE2Fa and AtDPa also up-regulate S-phase-specific genes, such as DNA polymerase α, cell division cycle 6 (AtCDC6), origin recognition complex 1 (AtORC1), and minichromosome maintenance 5 (AtMCM5). Similar results were obtained when AtE2Fa and AtDPa cDNAs were overexpressed in transgenic tobacco (Nicotiana tabacum) plants (Kosugi and Ohashi, 2003). Consistent with its role as an S-phase inducer, AtE2Fa is highly expressed in the shoot apical meristem (SAM), emerging leaf primordia, and vascular tissues of young leaf primordia (De Veylder et al., 2002). AtE2Fa is also expressed in the epidermis and cortex of the hypocotyls, which show a high level of endoreduplication (De Veylder et al., 2002). These observations are in agreement with reverse transcription (RT)-PCR results showing that AtE2Fa is maximally expressed in late G1 and early S phase (Mariconti et al., 2002). In contrast, AtE2Fc, which possesses all the features of activating factors but a truncated transactivation domain, is a poor transcriptional activator (Kosugi and Ohashi, 2002b) and down-regulates the early S-phase gene AtCDC6 through its interactions with pRBR, thereby acting as a repressor of cell proliferation (del Pozo et al., 2002).
Although structural features and transient expression data suggest a strong activating role for AtE2Fb, this factor has not been as thoroughly investigated as AtE2Fa and AtE2Fc. Only recently, it was reported that AtE2Fb overexpression in tobacco Bright Yellow-2 (BY-2) cells increases cell cycle rate and promotes cell division in the absence of auxin (Magyar et al., 2005). In this work, we analyzed the role played by AtE2Fb during cell cycle progression and development. Our results show that AtE2Fb is an activator of E2F-responsive G1/S and G2/M marker genes and suggest that, as in mammals, plant activating E2Fs play similar but distinct roles during cell cycle and development.
RESULTS
Expression of AtE2Fb during Development
It was previously reported that AtE2Fb is poorly transcribed in quiescent Arabidopsis suspension cells and is expressed in proliferating cells, with its RNA accumulating to slightly higher levels at the G1/S transition (de Jager et al., 2001; Mariconti et al., 2002). We used two different strategies to analyze the expression pattern of AtE2Fb during plant development. The first approach relied on the generation of transgenic Arabidopsis lines expressing the uidA (β-glucuronidase [gus]) reporter gene under the control of the putative AtE2Fb promoter (AtE2Fb∷uidA), while the second was the analysis of AtE2Fb transcript accumulation by in situ hybridization.
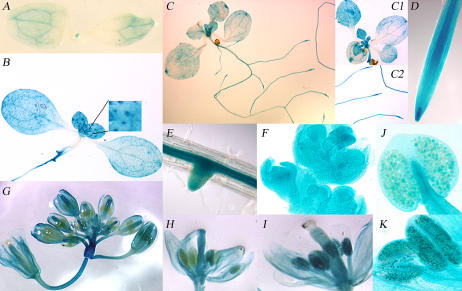
For the promoter expression analysis, histochemical staining for GUS activity was investigated in the T2 progeny of 19 AtE2Fb∷uidA transgenic plants using 4-, 7-, 18-d-old seedlings and flowering plants. In 4-d-old seedlings, GUS staining was observed in the SAM and in cotyledonary vascular tissues (Fig. 1A). In older plantlets (7 and 18 d old), GUS staining was intense and generalized in young leaves, while it was weaker or limited to tips in old leaves and cotyledons (Fig. 1, B, C, and C1). GUS staining was detected also in cells other than the vascular tissue, and in young leaves a strong signal was found at the base of trichomes (Fig. 1B). The AtE2Fb promoter appeared to be highly active in the central cylinder of both primary and secondary roots (Fig. 1, B, C2, D, and E). In 18-d-old primary roots, GUS activity was strong in the root tip, particularly at the elongation zone (Fig. 1D), whereas in secondary roots the staining appeared associated with the development of lateral root primordia (Fig. 1E). In young inflorescence meristems, GUS staining was widespread (Fig. 1F) while in a maturing inflorescence was maintained at different extents in sepals, petals, and styles (Fig. 1G). The staining of pistils and stamens clearly showed differences between immature and mature florets. In the immature flowers, the pistils were entirely blue while the anthers were unstained (Fig. 1H), whereas GUS staining of pistils disappeared and a strong GUS activity was observed in anthers of mature flowers (Fig. 1I). GUS staining of anthers appears to be due to the expression of AtE2Fb in maturing pollen grains (Fig. 1, J and K).
Figure 1.
Histochemical localization of GUS activity in transgenic Arabidopsis plants carrying the chimeric AtE2Fb∷uidA gene. A, Four-day-old seedling. B, Seven-day-old seedling. Inset in B, Magnification of young leaves, which shows GUS staining at the base of trichome cells and primary root tip. C, Eighteen-day-old Arabidopsis plants. Leaves at different stages of development and root tips are shown in C1 and C2, respectively. D, Primary root tip and elongation zone of an 18-d-old Arabidopsis plant. E, Eighteen-day-old seedling with developing lateral root primordia. F, Flower meristems. G, Developing and mature flowers. H, Pistils and stamens in immature florets. Note the strong staining of the stigma that disappears in mature florets (see I). I, Staining of pistils and stamens in mature florets. Note the strong staining of anthers that is not present in immature florets (see H). J, Anthers in immature florets. K, Anthers with mature pollen grains.
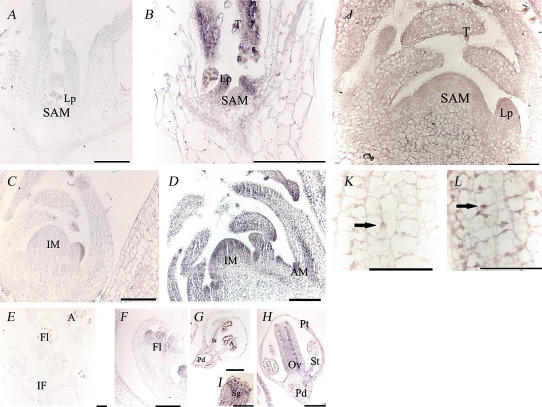
For in situ hybridization, a fragment corresponding to the 3′ untranslated region of the AtE2Fb mRNA was used as a probe to minimize cross-hybridization with other AtE2F transcripts. The shoot apex of a 10-d-old Arabidopsis plant continuously generates organs at its flanks, and, consequently, the SAM exhibits various kinds of tissues at different proliferating and differentiating stages. AtE2Fb transcripts accumulated at high levels in cells of the shoot apex and leaf primordia (Fig. 2, A and B). In the apex of 20-d-old plants, the hybridization signal was also visible in the main veins of the leaves and in the axillary meristems (Fig. 2, C and D). The signal was particularly strong in actively dividing tissues such as leaf primordia (Fig. 2, B and D) and the tips of young leaves (Fig. 2B). Interestingly, a very strong signal was also observed in trichomes of young leaves (Fig. 2B). In Arabidopsis inflorescences, AtE2Fb mRNAs were localized in floret primordia (Fig. 2F) and in petals, stigma, styles, and ovaries upon complete flower differentiation (Fig. 2, H and I). The hybridization signal was also detectable in the stamens and the flower pedicels (Fig. 2, G and H). No signal was detected in microspores or anther locules of flower buds.
Figure 2.
A to I, Localization of AtE2Fb transcripts by in situ hybridization in Arabidopsis plantlet and inflorescence sections. The hybridization signal is represented by the purple to blue staining. A, C, and E show sections hybridized with the sense probe (negative controls); B, D, F, G, H, and I show sections hybridized with the antisense probe. In A and B, longitudinal sections through the plantlets were obtained from plants 10 d after germination; in C and D, from plants 20 d after germination. In E to I, longitudinal sections were obtained from inflorescences. Bars = 100 μm. J to L, Immunolocalization of AtE2Fb in Arabidopsis plantlet longitudinal sections, 20 d after germination. J, A longitudinal section through a plant apex and leaf primordia. K and L show a higher magnification of a leaf primordium. In K, the treatment with the secondary antibody was omitted (negative control). Bars = 50 μm. Arrows indicate nuclei. A, Anther; AM, axillary meristem; IM, inflorescence meristem; IF, inflorescence; Fl, flower; Lp, leaf primordium; Ov, ovary; Pd, pedicel; Pt, petal; Sg, stigma; St; stamen; T, trichome.
Accumulation and Localization of the AtE2Fb Protein
To evaluate the in vivo accumulation and subcellular localization of AtE2Fb protein, immunolocalization experiments were carried out using longitudinal sections of the shoot apex of 15-d-old Arabidopsis plants. Results of these experiments showed that the AtE2Fb is localized primarily in the SAM and leaf primordia. This pattern of accumulation is very similar to that of the relevant transcripts (Fig. 2J). In the cells of young leaves, the protein was located in both the nucleus and cytoplasm (Fig. 2L). Interestingly, a strong signal was observed in leaf trichomes (Fig. 2J) also in both the nucleus and cytoplasm. Transient expression assay using the chimeric fusion construct Cauliflower mosaic virus 35S (CaMV35S)∷AtE2Fb-green fluorescent protein (GFP) in BY-2 protoplasts confirmed both cytoplasmic and nuclear localizations of the AtE2Fb-GFP fusion protein as previously reported (Kosugi and Ohashi, 2002b). Remarkably, all the cells containing AtE2Fb-GFP featured two nuclei, suggesting that expression of the fusion protein stimulated cell cycle progression toward mitosis. In contrast, control cells that accumulated the truncated inactive NtKIS1b fused to the GFP as a nuclear marker (Jasinski et al., 2002) and mock transfected cells were characterized by the presence of only one nucleus (Supplemental Fig. 1).
AtE2Fb-Overexpressing Plants Are Characterized by an Altered Phenotype and Up-Regulation of S-Phase Genes
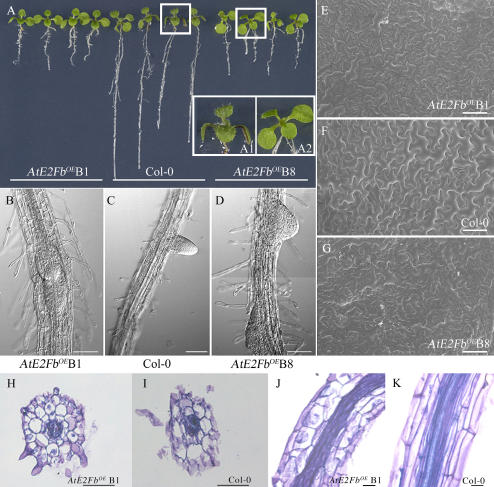
To assess the effect of an increased level of AtE2Fb on cell cycle and development, we generated transgenic Arabidopsis plants carrying the AtE2Fb cDNA under the control of the double CaMV 35S promoter. Of the 15 transgenic lines obtained, most showed striking morphological effects. Compared to untransformed control plants, the most evident alteration in 16-d-old AtE2FbOE (“OE” for overexpressing) seedlings was a shortening of the primary root, which featured an almost 3-fold reduction in length (Fig. 3A) combined with a closeness of the lateral root primordia and a higher density of thicker root hairs (Fig. 3, B–D). Hypocotyls were also shorter compared to untransformed controls (data not shown). A reduced size of AtE2FbOE plants was particularly evident in 4-d-old seedlings germinated in the dark. In these plants, hypocotyls were about 60% the size of those of untransformed controls, while roots were reduced in length to less than one-third (Supplemental Fig. 2).
Figure 3.
Phenotype of AtE2FbOE 16-d-old seedlings. A, AtE2FbOE line B1, left; untransformed control (Col-0), middle; and AtE2FbOE line B8, right. B to D, Scanning electron micrographs of roots: B and D, AtE2FbOE lines; and C, Col-0. E to G, Scanning electron micrographs of cotyledons: E and G, AtE2FbOE lines; and F, Col-0. H to K, Microscopic analysis of root sections of AtE2FbOE plants, line B1 and Col-0: H and I, same magnification, bar = 20 μm, radial sections; and J and K, same magnification, bar = 100 μm, longitudinal sections.
AtE2FbOE seedlings were also characterized by young leaves essentially lacking trichomes (Fig. 3, insets A1 and A2). Like AtE2FaOE plants (De Veylder et al., 2002), AtE2FbOE cotyledons were slightly larger than controls, although a scanning electron microscope (SEM) analysis revealed a clear reduction of the size of cotyledonary epidermal cells (Table I) compensated by an increase in their number (Fig. 3, E–G). These data suggest a positive influence of AtE2Fb on cell proliferation. The microscopic analysis of root sections showed the presence of shorter isodiametric cortex cells in AtE2FbOE plants (Fig. 3, H and J) compared to elongated cells in untransformed controls (Fig. 3, I and K). Remarkably, this phenotype is very similar to that of AtE2FaOE plants that showed a marked radial growth (De Veylder et al., 2002). To evaluate any possible effect of AtE2Fb accumulation on endoreduplication, we measured the ploidy levels of nuclei extracted from leaves and hypocotyls/roots of wild-type and AtE2FbOE plants. Results of this analysis showed a modest increase of endoreduplicated cells in transgenic plants as compared to untransformed controls (Supplemental Fig. 3).
Table I.
Adaxial epidermal cell size in cotyledons of AtE2FbOE plants
Mean values are ±se. To warrant homogeneous growth, control and transgenic plants were grown on semisolid medium in petri dishes in a growth chamber. The reported results are the average of at least three different experiments.
| Line | Cotyledons 16 d after Sowing | Adaxial Epidermal Cells 16 d after Sowing |
|---|---|---|
| mm2 | μm2 | |
| Col-0 | 4.80 ± 0.49 | 4,761 ± 2,636 |
| AtE2FbOE line B1 | 5.22 ± 0.63 | 1,257 ± 693 |
| AtE2FbOE line B8 | 5.81 ± 1.19 | 1,861 ± 1,297 |
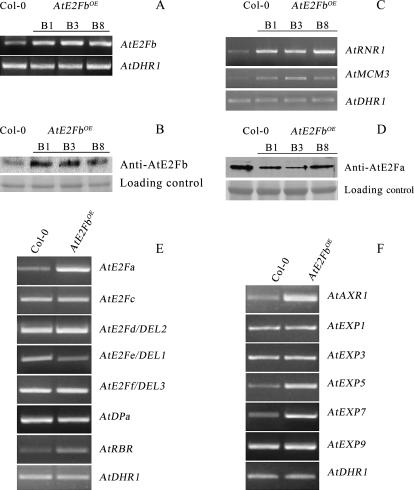
Semiquantitative RT-PCR of three of the most affected transgenic lines showed an increased steady-state level of AtE2Fb transcripts (Fig. 4A). A paralleled increase in the content of AtE2Fb protein was also revealed by immunoblot analyses performed using monospecific antibodies raised against the N terminus of AtE2Fb (Fig. 4B; see Supplemental Fig. 4 for the specificity of antibodies).
Figure 4.
Levels of activating AtE2F transcripts and relevant protein in AtE2FbOE plants. A, RT-PCR analysis using primers specific for AtE2Fb of 7-d-old seedlings of wild type (Col-0) and three AtE2FbOE lines (B1, B3, and B8). The level of AtDHR1 transcripts was used as the loading control. B, Immunoblot analysis of 15-d-old plants using antibodies against AtE2Fb. Wild type and AtE2FbOE lines are as in A. The molecular mass of the recognized AtE2Fb protein is 52 kD. A region of the filter stained with the Ponceau S is shown as the loading control. C, Semiquantitative RT-PCR of AtMCM3 and AtRNR1 using RNA extracted from 7-d-old AtE2FbOE seedlings. D, Immunoblot analysis of 15-d-old plants using antibodies against AtE2Fa. The molecular mass of the recognized AtE2Fa protein is 66 kD. E, RT-PCR analysis of wild type (Col-0) and AtE2FbOE line B8 using primers specific for genes of the AtE2F family and for AtRBR. F, RT-PCR analysis of wild type (Col-0) and AtE2FbOE line B8 using primers specific for AtAXR1 and for AtEXP genes.
To further characterize these AtE2FbOE lines, we analyzed expression of the E2F-responsive genes encoding AtMCM3 and the ortholog of tobacco ribonucleotide reductase 1b (AtRNR1; Chabouté et al., 2002; Stevens et al., 2002). Semiquantitative RT-PCR analysis showed that the overproduction of AtE2Fb results in up-regulation of these S-phase-specific genes, whereas the transcription of a gene encoding a dicer-related helicase (AtDRH1), which is not expected to be E2F responsive, was unchanged in the AtE2FbOE lines (Fig. 4C). Similarly, the AtCYCD3, AtPCNA (proliferating cell nuclear antigen), and AtCDC6 genes, which are also S phase specific and E2F responsive, were up-regulated in AtE2FbOE lines, while the expression of AtORC1, another E2F-responsive gene, was unchanged (Supplemental Fig. 5). The up-regulation of AtCYCD3, which is induced before the onset of S phase and does not contain E2F sites, could depend on the cellular context rather than on the accumulation of AtE2Fb.
Given the phenotypic similarities between AtE2FbOE and AtE2FaOE plants (De Veylder et al., 2002), we asked whether AtE2Fb overexpression results in an increase in AtE2Fa protein levels. Surprisingly, immunoblot analysis using monospecific antibodies raised against AtE2Fa revealed that the amount of AtE2Fa protein was reduced in plants overexpressing AtE2Fb (Fig. 4D). We then used RT-PCR to evaluate the steady-state mRNA levels of the other five AtE2F genes in AtE2FbOE plants. The promoters of the AtE2Fc, AtE2Fe/DEL1, AtE2Ff/DEL3, and AtRBR genes contain E2F cis-elements, while AtE2Fa and AtE2Fd/DEL2 promoters lack recognizable E2F consensus motifs. This experiment (Fig. 4E) showed that AtE2Fc, AtE2Fd/DEL2, AtE2Ff/DEL3, and AtDPa mRNA levels did not change in the AtE2FbOE plants compared to control plants. In contrast, AtE2Fe/DEL1 mRNA was reduced, while AtE2Fa and AtRBR were up-regulated. The up-regulation of AtE2Fa was unexpected because its promoter lacks E2F sites and AtE2Fa protein content decreased in AtE2FbOE plants. Together, these results strongly suggest that the expression of AtE2F genes is regulated by both transcriptional and posttranscriptional mechanisms. Previous studies have shown a strong influence of auxin over the regulation of AtE2F stability (del Pozo et al., 2002; Magyar et al., 2005). The presence of an E2F cis-element in the promoter of AXR1, the product of which is required for auxin response (del Pozo et al., 1998), prompted us to evaluate the level of AXR1 transcripts in an AtE2FbOE line. Results of RT-PCR showed that AXR1 is indeed up-regulated, thus indicating a possible link between AtE2Fb and auxin action.
The phenotype of AtE2FbOE plants also resembled AtE2FfOE plants (Ramirez-Parra et al., 2004) with respect to the reduced length of roots and hypocotyls compared to controls. However, AtE2Ff/DEL3 transcripts levels were similar in AtE2FbOE and untransformed control plants, suggesting that the AtE2FbOE morphology is not due to increased AtE2Ff/DEL3 expression. This hypothesis is consistent with the observation that AtEXP mRNA levels (AtEXP1, 3, 5, 7, and 9) were either unchanged (AtEXP1, 3, and 9) or up-regulated (AtEXP5 and 7; Fig. 4F). Thus, this situation differs sharply from that of AtE2FfOE plants, in which the down-regulation of AtEXP3, 7, and 9 was deemed responsible for the phenotype characterized by short roots and hypocotyls (Ramirez-Parra et al., 2004).
The AtE2Fb Promoter Is Regulated by an Activating AtE2F Factor
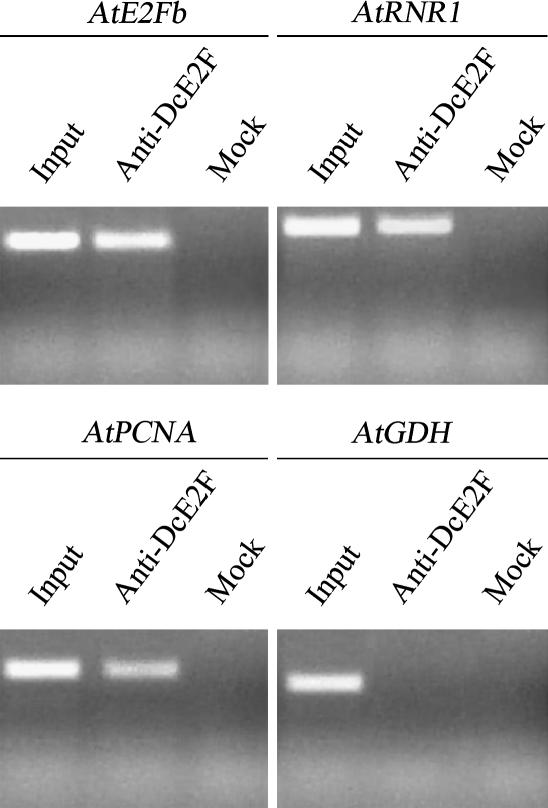
E2F consensus binding sites have been found in one or more copies in close proximity to the transcription start sites of several plant cell cycle-specific promoters. Using in silico analysis, we have identified three putative E2F cis-elements in the AtE2Fb promoter. To verify whether any of these E2F sites could be actually recognized by activating AtE2F factors, we applied the chromatin immunoprecipitation (ChIP) technique using polyclonal antibodies against the carrot (Daucus carota) DcE2F protein (Albani et al., 2000). These antibodies recognize only activating AtE2Fs with a greater preference for AtE2Fa (Supplemental Fig. 4). As a positive control to assess the efficiency of the ChIP assay, PCR reactions were performed on immunoprecipitated genomic fragments using primers specific for AtRNR1 and the AtPCNA promoters, which contain E2F sites and are well known E2F targets (Chabouté et al., 2002; Egelkrout et al., 2002). A mock reaction with no antibody was used as a negative control. To rule out nonspecific interactions, PCR was also performed using primers specific for the Glu dehydrogenase (AtGDH) promoter that is not predicted to be an E2F target. The results of this experiment (Fig. 5) revealed a positive amplification with the AtE2Fb primer set, indicating that the AtE2Fb promoter, like the AtRNR1 and AtPCNA promoters, is bound in vivo by an activating AtE2F and can be immunoprecipitated by antibodies against DcE2F. The specificity of this recognition was confirmed by the negative result obtained in a ChIP reaction performed using nonspecific antibodies against the epitope Flag 7 (data not shown).
Figure 5.
ChIP of the AtE2Fb promoter. The immunoprecipitation assay was performed using antibodies against DcE2F. Immunoprecipitated genomic DNA was amplified with primers specific for AtE2Fb and for two known E2F-responsive promoters of AtRNR1 and AtPCNA (positive controls). The mock reactions and the PCR on immunoprecipitated fragments, using primers specific for the E2F-nonresponsive promoter (AtGDH), were used as negative controls. As a further control, an aliquot of the input was examined by PCR using AtE2Fb-, AtRNR1-, AtPCNA-, and AtGDH-specific primers.
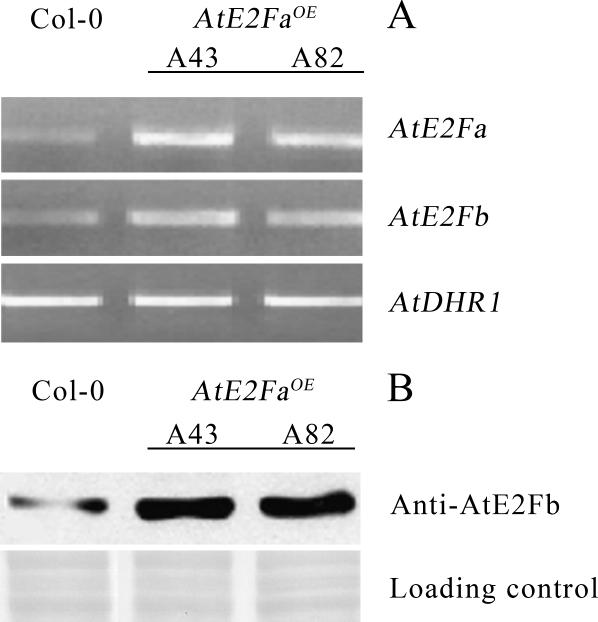
These results point to a positive regulation of the AtE2Fb promoter by an activating AtE2F, possibly AtE2Fa. To test this hypothesis, expression of AtE2Fb and the accumulation of the corresponding protein in AtE2FaOE plants versus untransformed controls were compared by semiquantitative RT-PCR and immunoblot analyses. These experiments (Fig. 6A) revealed that AtE2FaOE plants contain an increased level of AtE2Fb transcripts that is paralleled by an increase in the amount of the AtE2Fb protein (Fig. 6B), suggesting that AtE2Fb expression might actually be up-regulated by the AtE2Fa transcription factor.
Figure 6.
Levels of AtE2Fb transcripts and the relevant protein in AtE2FaOE plants. A, RT-PCR of AtE2Fb using RNA extracted from 7-d-old wild-type (Col-0) and AtE2FaOE (lines A43 and A82) seedlings. The level of AtDHR1 transcripts was used as the loading control. B, Immunoblot analysis of 15-d-old plants of control (Col-0) and AtE2FaOE (lines A43 and A82) using antibodies against AtE2Fb. The molecular mass of the recognized AtE2Fb protein is 52 kD. A region of the nitrocellulose filter stained with Ponceau S was used as the loading control.
AtE2FbOE Plants Also Up-Regulate Some G2/M Marker Genes
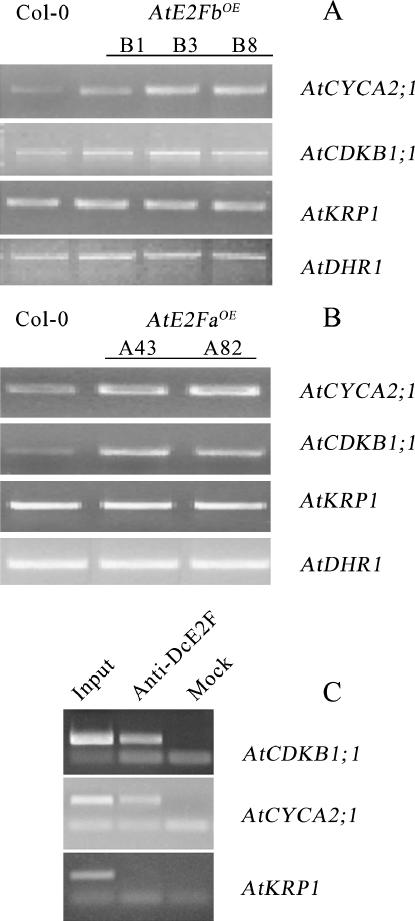
To evaluate whether overproduction of AtE2Fb affects the expression of cell cycle genes acting at a stage different from G1/S, the steady-state level of transcripts of selected G2/M marker genes was investigated in the AtE2FbOE plants. For this purpose, the expression of AtCYCA2;1 and AtCDKB1;1, both containing a E2F consensus site in their promoters, and AtKRP1, which lacks E2F sites (Mironov et al., 1999; Menges and Murray, 2002), was analyzed by RT-PCR. Results of this analysis (Fig. 7A) show that the steady-state level of AtCYCA2;1 transcripts and, to a lesser extent, of AtCDKB1;1 transcripts increased compared to control plants. In contrast, the transcript level of AtKRP1 was unchanged. Remarkably, when this analysis was conducted on plants overexpressing AtE2Fa, an increase in the expression of the two E2F-containing G2/M marker genes was again observed (Fig. 7B). To establish whether the up-regulation of these genes is due to a stimulation of cell cycle progression or a direct involvement of activating AtE2Fs, ChIP was performed using antibodies anti-DcE2Fs. Results of this analysis showed that the E2F sites present in these promoters were occupied in vivo by activating AtE2Fs (Fig. 7C). Another G2/M-specific marker, such as AtKRP2, was up-regulated in AtE2FbOE plants, while the expression of AtKRP3 was unchanged (Supplemental Fig. 5). It is worth noting that the promoter of AtKRP2 does not contain consensus E2F sites and its up-regulation might not be directly dependent on AtE2Fb overaccumulation.
Figure 7.
Levels of G2/M marker gene transcripts in plants overexpressing activating AtE2Fs. A, RT-PCR of AtCYCA2;1, AtCDKB1;1, and AtKRP1 using RNA extracted from 7-d-old wild type (Col-0) and AtE2FbOE lines (B1, B3, and B8). B, RT-PCR of AtCYCA2;1, AtCDKB1;1, and AtKRP1 using RNA extracted from 7-d-old wild type (Col-0) and AtE2FaOE lines (A43 and A82). The level of AtDHR1 transcripts was used as the loading control. C, ChIP of AtCDKB1;1, AtCYCA2;1, and AtKRP1 promoters. Experimental conditions were as described in the legend for Figure 5.
DISCUSSION
Of the six E2Fs of Arabidopsis, only two (AtE2Fa and AtE2Fb) possess all the structural and functional features of typical activating animal and plant E2Fs (del Pozo et al., 2002; Kosugi and Ohashi, 2002b; Mariconti et al., 2002). AtE2Fa is an activator of E2F-responsive genes (Kosugi and Ohashi, 2002b; Mariconti et al., 2002) that induces quiescent leaf cells to enter S phase (Rossignol et al., 2002). The ectopic AtE2Fa overexpression in Arabidopsis plants stimulates cotyledonary cells to proliferate and delays differentiation (De Veylder et al., 2002). In AtE2Fa-DPaOE plants, the decision to engage in proliferation or endoreduplication depends on the cellular context through the action of other cell cycle regulators, such as AtCDKB1;1 and the repressing E2F factor AtE2Fe/DEL1 (Boudolf et al., 2004; Vlieghe et al., 2005). In this work, we have demonstrated that AtE2Fb also is a stimulator of cell cycle progression and that its overexpression affects plant morphology.
According to in situ hybridization analyses, immunolocalizations, and the analysis of promoter activity in transgenic plants, AtE2Fb appears to be mainly expressed in proliferating cells. However, there is also a clear expression at the base of trichomes and in several differentiated tissues. Altogether, these data point to a relationship between AtE2Fb expression and cell division without excluding a possible involvement of this transcription factor in endoreduplication and differentiation.
Experiments of transient expression of a chimeric AtE2Fb∷GFP construct in BY-2 tobacco suspension cells are also supporting a role of this factor in cell proliferation as judged by the presence of two nuclei in cells accumulating the AtE2Fb∷GFP protein. Remarkably, the analysis of Arabidopsis plants ectopically overexpressing AtE2Fb showed slightly enlarged cotyledons containing almost twice the number of smaller epidermal cells. These plants also showed up-regulation of the E2F-responsive S-phase genes AtRNR1 and AtMCM3 (Chabouté et al., 2002; Stevens et al., 2002) as well as the G2/M marker genes AtCYCA2;1 and AtCDKB1;1. Altogether, these results indicate that AtE2Fb overexpression induces G1/S transition and cell division similar to what was observed following overexpression of AtE2Fa (De Veylder et al., 2002; Rossignol et al., 2002; Kosugi and Ohashi, 2003). This observation agrees with the experiments conducted very recently by Magyar et al. (2005) in tobacco BY-2 cells, in which accumulation of either AtE2Fa or AtE2Fb, together with their partner AtDPa, could sustain cell division, and AtE2Fb also could be effective in the absence of 2,4-dichlorophenoxyacetic acid.
The overexpression of AtE2Fb in transgenic plants led to an up-regulation of AtE2Fa and AtRBR and a down-regulation of AtE2Fe/DEL1, whereas the transcript levels of the other AtE2Fs and of the AtDPa gene remained unchanged. However, contrary to the increased level of AtE2Fa transcripts, the accumulation of AtE2Fa protein was reduced in AtE2FbOE plants. Since the AtE2Fa promoter lacks E2F-binding sites, its up-regulation could be ascribable to an overall stimulation of cell cycle progression. In contrast, the low level of AtE2Fa could be due to its higher turnover rate as a result of hyperphosphorylation by AtCDKB1;1 as suggested by Magyar et al. (2005). It is worth noting that the AtE2Fe/DEL1 and AtRBR promoters contain E2F cis-elements and could be direct targets of AtE2Fb. On the other hand, AtE2Fc and AtE2Ff/DEL3, which contain E2F cis-elements in their promoters, were not up-regulated in AtE2FbOE plants, whereas they were strongly up-regulated in AtE2Fa-DPaOE plants (Vandepoele et al., 2005). This suggests that the activating AtE2Fs are likely to have different DNA-binding specificity. In this respect, it was previously observed that E2F sites could be differentially recognized by different E2F factors (Egelkrout et al., 2002; Ramirez-Parra et al., 2004).
The presence of putative E2F cis-elements in the AtE2Fb promoter suggests that AtE2Fb gene expression could be regulated by other AtE2F factors. In fact, the up-regulation of AtE2Fb and the overaccumulation of the relevant protein in AtE2FaOE plants indicate that AtE2Fa might up-regulate the AtE2Fb gene. This is also supported by the results of ChIP experiments showing that one or more putative E2F cis-elements of the AtE2Fb promoter are actually occupied in vivo by an activating AtE2F. Up-regulation of AtE2Fb in plants overexpressing AtE2Fa and AtDPa also has been detected recently by microarray analysis (Vandepoele et al., 2005). Nevertheless, the fact that AtE2FbOE plants showed a reduction of the AtE2Fa protein content suggests a complex interplay between these two transcription factors and/or a sequential scheduling of these AtE2Fs during cell cycle progression. Whether the expression of AtE2Fb might actively contribute to reduce the accumulation of AtE2Fa or rather enhance a scheduled physiological change in the stability of the two proteins remains to be established.
A remarkable result concerning the phenotype of AtE2FbOE seedlings was the considerably shorter length of the primary roots (about 3-fold shorter than controls) that appeared to correlate with a reduction of the length of root cells. This suggests that accumulation of AtE2Fb antagonizes cell elongation and might delay root cell differentiation. A similar phenotype also was seen in plants overexpressing AtE2Ff/DEL3. However, overexpression of AtE2Fb led to increased content of AtEXP5 and AtEXP7 transcripts and did not decrease the expression of other AtEXP genes (AtEXP3 and AtEXP9) shown to be down-regulated in AtE2FfOE plants (Ramirez-Parra et al., 2004). Therefore, the altered root phenotype of AtE2FbOE and AtE2FfOE plants is likely to depend on different regulatory mechanisms.
Another striking feature of AtE2FbOE seedlings was the absence (or almost complete absence) of trichomes in leaves. Remarkably, this phenotype is not seen in AtE2FaOE plants that in absence of a concomitant overexpression of the AtDPa partner did not show major developmental abnormalities (De Veylder et al., 2002). A strong inhibition of trichome development also has been found recently in Nicotiana benthamiana plants upon disruption of RBR function by virus-induced gene silencing. Confirming the expected role of RBR as a negative regulator of activating plant E2Fs, the reduction of RBR levels in these plants also resulted in induction of ectopic cell division, increased endoreduplication, and delay in cell differentiation (Park et al., 2005). Thus, it appears that in particular cellular contexts, overexpression of AtE2Fb could lead to inhibition of cell differentiation.
Whereas ectopic overexpression of AtE2Fa-DPa or AtE2Fe/DEL1 leads to changes in ploidy levels in several plant organs (De Veylder et al., 2002; Vlieghe et al., 2005), AtE2Fb overexpression did not induce major changes in ploidy levels in AtE2FbOE plants. This suggests that activation of S phase by AtE2Fb is almost completely counterbalanced by a comparable activation of M phase. This latter effect is likely ascribable to the up-regulation of AtCDKB1;1, the product of which was shown to inhibit the endocycle, thus acting as the mitosis-inducing factor (De Veylder et al., 2002; Boudolf et al., 2004).
In summary, our results suggest an interplay between the activating factors AtE2Fa and AtE2Fb in controlling the balance of cell division, endoreduplication, and differentiation in Arabidopsis plants. Both factors appear to stimulate cell proliferation and inhibit differentiation. This similarity of roles most likely results from activation of AtE2Fb expression by the AtE2Fa protein. Although in different contexts these two factors could act independently from each other and exert specific roles, the emerging framework of cell cycle progression in plants suggests a hierarchical organization of the E2F players. AtE2Fa is likely to be implicated in the control of early cell cycle genes and activates the expression of AtE2Fb, which in turn could be the primary E2F factor responsible for direct stimulation of cell proliferation as also suggested by Magyar et al. (2005) in a heterologous system.
MATERIALS AND METHODS
Plant Material and Cell Suspension Lines
All Arabidopsis (Arabidopsis thaliana) transgenic lines in this study were generated by the floral dip method (Clough and Bent, 1998) using Agrobacterium tumefaciens GV3101/pMP90 and HBA105 strains. The transgenic T1 seeds were selected on 0.5× Murashige and Skoog medium (Duchefa) containing kanamycin (50 mg/L) or hygromycin (40 mg/L) as required. Wild-type and transformed plants were transferred to soil and grown to maturity in a greenhouse or in a growth chamber. In both cases, the growing conditions were 16 h of light (23°C ± 3°C) and 8 h of dark (18°C ± 3°C) with 70% relative humidity. For phenotypic analysis, AtE2FbOE T2 seeds were germinated on semisolid Murashige and Skoog salt medium without kanamycin.
The Arabidopsis cell line T87 (Axelos et al., 1992) was grown under dim light conditions at 23°C on a rotary shaker (130 rpm) in B5 Gamborg's medium (Duchefa), pH 5.8, supplemented with 30 g/L Suc and 1 μm naphthylacetic acid. Suspension cells were subcultured weekly transferring 5 mL into 100 mL of fresh medium.
In Situ Hybridization and Immunolocalization
In situ hybridization experiments were carried out as described (Varotto et al., 2003). In brief, plant materials (seedlings and plantlets 10, 15, and 20 d after germination) were fixed in 4% paraformaldehyde, 0.2% glutaraldehyde in 0.1 m phosphate buffer, pH 7.2, for 16 h at 4°C and embedded in Paraplast Plus (Sigma-Aldrich). Sections (7–10 μm) were cut using a microtome (RM 2135; Leica) and collected on xylane-coated slides. Slides were deparaffinized, treated with 5 μg/mL Proteinase K, and hybridized with sense and antisense riboprobes in 50% formamide at 50°C overnight. The sections were hybridized with an AtE2Fb-specific, digoxigenin-labeled probe corresponding to 380 bp of the 3′ untranslated region cloned into the pBS II KS plasmid (Stratagene). T7 and Sp6 polymerases were used for the synthesis of sense and antisense labeled transcripts. Sense transcripts were used as negative control in the hybridization experiments. After hybridization, the slides were washed extensively in 2× SSC at 50°C and treated with 20 μg/mL RNaseA (Roche). Digoxigenin detection and signal visualization were carried out using 4-nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate, 4-toluidine (Roche) following the manufacturer's instructions. Images were acquired using a Leica DC 300F camera.
For immunolocalization experiments, sections were deparaffinized in xylene, rehydrated in a graded series of ethanol, and finally rinsed in Tris-buffered saline (Sambrook et al., 1989). Before the antibody reactions, sections were briefly treated with 5 μm/mL of proteinase K. Primary antibodies against AtE2Fb were used at a 1:200 dilution. Slides were incubated for 2 h at 37°C with primary antibodies as described below and with mouse monoclonal anti-rabbit IgGs conjugated with alkaline phosphatase (Sigma) for 1 h at room temperature. Digoxigenin detection and signal visualization were carried out as described above for in situ hybridization experiments.
Construction of the CaMV35S∷AtE2Fb∷GFP Fusion Plasmid
A full-length AtE2Fb cDNA (1.4 kb; GenBank accession no. AF242580) was amplified by PCR. The primers were designed to mutate the stop codon as well as to introduce BamHI and SalI cloning sites. The primer sequences were MWD9-5 (5′-GGGGATCCTTATGTCTGAAGAAGT) and MWD9-3 (5′-TTGTCGACGCTACCTGTAGGTGATCT). The 1.4-kb PCR product was digested with BamHI-SalI, purified by gel electrophoresis, and inserted upstream of the GFP sequence in a modified pBI121 vector (CLONTECH) in which the uidA sequence was replaced with GFP coding sequence. AtE2Fb-GFP fusion protein localization was determined by confocal microscopy (Leica TCS SP2) at a wavelength between 504 and 530 nm.
Isolation of the AtE2Fb Promoter Region and Construction of the AtE2Fb∷uidA Chimeric Plasmid
The promoter region of AtE2Fb was amplified from Arabidopsis genomic DNA using the primers Pro-5B (5′-GGGGTTCTTCTATTGTTGTCTC) and Pro-3B (5′-CAGCTGCCAATAAAGTCACCAA), which amplified the region spanning the stop codon of the upstream gene to the first intron of the AtE2Fb gene. The PCR fragment was cloned into pCR-Blunt (Invitrogen), checked by sequencing, and subcloned as a HindIII-XbaI fragment upstream the region encoding the gus (uidA) into the vector pTAK to create AtE2Fb∷uidA. The HindIII-EcoRI fragment of the AtE2Fb∷uidA plasmid containing the AtE2Fb promoter, the uidA coding region, and the terminator was then inserted into the binary vector pPZP111 (Hajdukiewicz et al., 1994) for the stable transformation of Arabidopsis plants.
Histochemical Determination of GUS Activity
Histochemical detection of GUS activity was performed on Arabidopsis transgenic plants at different developmental stages using 5-bromo-4-chloro-3-indolyl-β-d-glucuronide (Jefferson et al., 1987). Plants were incubated in the GUS staining solution (100 mm phosphate buffer, pH 7, 1 mg/mL X-Gluc A, 1 mm potassium ferricyanide) for at least 1 h at 37°C. After staining, the samples were transferred in 70% ethanol to remove the chlorophyll. Images were captured with a Zeiss (SV11) equipped with a Sony PowerHAD camera and AxioVision 1.01 software (Zeiss).
Construction of CaMV2x35S∷AtE2Fb and CaMV35S∷AtE2Fa
For the overexpression of AtE2Fb in transgenic plants, the corresponding cDNA (Mariconti et al., 2002) was inserted as a BamHI-XhoI fragment into the BamHI-SalI digested polylinker of the binary vector pGusNpt/FF19 that carries a T-DNA region containing the expression cassette of the pFF19 plasmid (Timmermans et al., 1990) and a bifunctional GUS/neomycin phosphotransferase marker gene (Datla et al., 1991). The resulting 2x35S∷AtE2Fb vector was introduced into the A. tumefaciens GV3101/pMP90 strain for plant transformation. The 35S∷AtE2Fa vector (Rossignol et al., 2002) was introduced into A. tumefaciens HBA105.
Optical and SEM Analyses
For SEM analyses, samples were slowly frozen at −18°C under a partial vacuum on the Peltier stage before observation under the environmental secondary electron detector mode (Hitachi 3-3000). Differential interference contrast microscopy was used to observe plantlets that had been fixed with FAA (50% ethanol, 5% acetic acid, and 10% formaldehyde) for 6 h, depigmented in increasing ethanol concentration, and cleared in chloral hydrate (8 g chloral hydrate, 1 mL glycerol, 2 mL water). Images were elaborated using the Olympus DP-Soft software (Olympus). For root histology, tissues were fixed and embedded as described for in situ hybridization. Deparaffinized sections were stained with toluidine blue, and pictures were taken with a Leica DC 300F camera.
Gene Expression Analysis
Total RNA was extracted from leaves, stems, roots, and flowers of wild-type (Col-0), AtE2FbOE (2x35S∷AtE2Fb), and AtE2FaOE (35S∷AtE2Fa) transgenic Arabidopsis plants using the RNeasy Plant Mini kit (Qiagen). cDNA reactions were performed using the SuperScript first-strand synthesis system (Invitrogen). The level of gene expression was determined by semiquantitative RT-PCR using 1 μg of total RNA in each reaction. RT-PCR was performed using the primers listed in Table II. All primers were designed on the basis of the relevant cDNA sequences.
Table II.
Primers used for RT-PCR analyses
| Name | Target | Sequence |
|---|---|---|
| A | AtE2Fa | TCTTTAGGTCTCCTTACAAAA |
| AGAAGTACAATGGGACCTAT | ||
| B | AtE2Fb | TCAACATCTGGTCTCCCTGA |
| AGCGTGGTCTTGATCAATG | ||
| C | AtE2Fc | CAGGCGAAGAT CCGACTC |
| GCCATTCGCCATTCGTT | ||
| D | AtE2Fd/DEL2 | ACCGGACGTGAAGAATTTTG |
| TCGTTGTAATGCGCAAAAAG | ||
| E | AtE2Fe/DEL1 | AGTGAGGCGGCTTTATGA |
| TCCAGATTCTCAACATCAAAAG | ||
| F | AtE2Ff/DEL3 | GTTAGAAGACTTTACGACATTGC |
| CCTCGATCTCTAGTAACCTTCC | ||
| RB | AtRBR | AAGGTGTAGACTTGGTTGCAT |
| TTGTCATTGCTGTGCTCACT | ||
| DP | AtDPa | AACCCTCACGCAGTAGTC |
| GCGAGTATCAATGGATCC | ||
| RN | AtRNR1 | GGGCTTAGCAGTGACCATTGTGA |
| TTCTGGTACCATGGAGCCGCCACAGCATCAG | ||
| M | AtMCM3 | TTCTGGTACCATGGAGCCGCCACAGCATCAG |
| TCTTGGAGCTCCTAGTTCAGACGTAGCTCAAG | ||
| CY | AtCYCA2;1 | ATTCTCGATTCCGGTTTA |
| AACGTAGTTTACTGCCAAAT | ||
| CD | AtCDKB1;1 | GGTGGTGACATGTGGTCTGTT |
| CGCAGTGTGGAAACACC | ||
| KRa | AtKRP1 | AGCTAAAGGAATTGTAGAAGC |
| ACTTTACCCATTCGTAACG | ||
| AX | AtAXR1 | GATTTGGGGGGAGGTAGG |
| CTTTACAGAGATGCGAACAAACC | ||
| EXa | AtEXP1 | ATGGTCTAAGT TGTGGTGCTT |
| AAAGACCAGCCTGCGTT | ||
| EXb | AtEXP3 | TATACCGTGCAGGCATTGTC |
| AGTGATTGGCCCGATGAGAAC | ||
| EXc | AtEXP5 | AGGACTTAGT TGTGGCGC |
| GTGGGTGGTGCAACATTA | ||
| EXd | AtEXP7 | ACGCCACTTTCTACGGTGAC |
| TAGGAGGGCAAAGATTGGTG | ||
| EXe | AtEXP9 | TCAAGCTAGCGACAATGGTG |
| AGCTCCGGCTACGTTAGTGA | ||
| DR | AtDRH1 | AAGAGGAGCAGATATCGTGGTTG |
| CGACGAGATATGTACTCTTGTT |
Production of a Recombinant Polypeptide Corresponding to the N-Terminal Domain of the AtE2Fb Protein
To raise AtE2Fa- and AtE2Fb-specific antibodies, the cDNA sequences corresponding to the divergent N-terminal 165 and 127 amino acids of AtE2Fa and AtE2Fb, respectively, were expressed in Escherichia coli. The corresponding region of the AtE2Fa and AtE2Fb cDNA were amplified by PCR using primers AtE2Fa/5Bam and AtE2Fb/5Bam (Mariconti et al., 2002), and ΔF11F19 (5′-TAACTGCAGCTTCCTGATGGAGTAAGT) and ΔMWD9 (5′-ATCCTGCAGTACCGGCCTGTGCAAAG). The PCR fragments were digested with BamHI and PstI and cloned in pRSET (A and B, respectively, for AtE2Fa and AtE2Fb; Invitrogen) digested with the same restriction enzymes. The resulting plasmids were then introduced into E. coli BL21 (DE3) pLysE for the production of the recombinant N-terminal portion of the two AtE2Fs. The HIS-AtE2Fa-(1-165) and HIS-AtE2Fb-(1-127) polypeptides were purified by metal-affinity chromatography on nickel-nitrilotriacetic acid resin (Qiagen) using phosphate buffer containing 8 m urea. The analysis by SDS-PAGE of the eluted protein revealed a single polypeptide of the expected dimensions.
Antisera and Immunoblotting
Four rabbit polyclonal antisera were used in this study. The first antiserum was raised against the carrot (Daucus carota) DcE2F, which recognizes AtE2Fa and, to a much lesser extent, AtE2Fb (Albani et al., 2000; Supplemental Fig. 2). The monospecific antisera against AtE2Fa and AtE2Fb were obtained by the immunization of rabbits using the dialyzed HIS-AtE2Fa-(1-165) and His-AtE2Fb-(1-127) polypeptides eluted under denaturing conditions. These antibodies showed a low cross-reactivity. The fourth antiserum was against the AtE2Fb C-terminal oligopolypeptide DQDHAGPSDNKILE conjugated to a carrier protein. To determine the amount of AtE2F protein in suspension cell cultures and transgenic plants, a 100-mg aliquot of each sample was frozen in liquid nitrogen, ground in a mortar to a fine powder, which was dissolved in 500 μL of extraction buffer containing 100 mm Tris-HCl, pH 7.8, 1 mm EDTA, 200 mm NaCl, 0.2% Triton X-100, and 1× Complete Mini protein inhibitors (Roche), and centrifuged for 10 min to remove debris. Protein concentration was estimated by the Bradford assay. Protein extracts (30 μg) were subjected to SDS-PAGE in a 12% polyacrylamide gel and transferred to a nitrocellulose membrane using standard techniques (Sambrook et al., 1989). Immunodetection was performed using the indicated polyclonal antibodies at 1:1,000 (v/v) dilutions. Goat anti-rabbit alkaline phosphatase-conjugated secondary antibody was used at 1:15,000 (v/v) dilutions and detection was performed using ECL chemiluminescence detection reagents (Amersham).
ChIP
ChIP assays were performed using nuclei extracted from suspension-cultured Arabidopsis T87 cells. Nuclei, extracted as described previously (Albani et al., 2000), were treated with 1% formaldehyde at 22°C for 10 min, and the cross-linking was stopped by the addition of 0.125 m Gly. Fixed nuclei were resuspended in SDS buffer (50 mm Tris-HCl, pH 8.0, 10 mm EDTA, and 1% SDS) and sonicated to shear DNA to 600- to 1,000-bp fragment size. To reduce false positives, sonicated chromatin samples were preincubated with 20 μL of preimmune serum for 1 h at 4°C with gentle mixing, transferred to a new tube with 20 μL of protein A-Sepharose (50% slurry in 15 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1 mm EDTA, and 1% Triton X-100), and incubated with gentle mixing for 1 h at 4°C. Samples were centrifuged at 13,000 rpm for 2 min at 4°C. The resulting supernatant was specifically immunoprecipitated with 20 μL of anti-DcE2F serum and further incubated for 2 h at 4°C with gentle mixing. Immunocomplexes were recovered using 20 μL of protein A-Sepharose (50% slurry) for 2 h at room temperature with gentle mixing, extensively washed, and eluted from beads. The immunoprecipitated chromatin was incubated for 5 h at 65°C, added with two volumes of ethanol, and centrifuged. The resulting pellet was incubated with proteinase K (18.5 mg/mL) for 2 h at 42°C and extracted with phenol/chloroform. Upon ethanol precipitation, DNA was resuspended in 10 μL of water, and 1 μL was used for PCR analysis using the following primers adjacent to promoter E2F cis-elements: B5 (5′-TCCCCTCAATCTCAAGGAAA-3′) and B3 (5′-AAGAACGAATCTCGATAAAA-3′) for AtE2Fb; P5 (5′-GAGACAAGACTCACAGATGA-3′) and P3 (5′-GGTTAGAGTGTGAATCGA-3′) for AtPCNA; R5 (5′-AATGGGCTTTAACTCTCTAA-3′) and R3 (5′-AAGGGATTTGAAGATTTG-3′) for AtRNR1; CD5 (5′-TAACTCGTGAAGAATTTGAA-3′) and CD3 (5′-TTCTGAGAGGTTTCGTAAAA-3′) for AtCDKB1;1; Cy5 (5′-GGAAATCAATGCTGAAAGAG-3′) and Cy3 (5′-TGAGAGAGAGAGATCTTGAA-3′) for AtCYCA2;1; KR5 (5′-GTTTCGCGTAATGGCAAAT-3′) and KR3 (5′-GCGTGAAGTCACAATCT-3′) for AtKRP1; and D5 (5′-TCTCAAATTTTAGGCAAGTT-3′) and D3 (5′-GGCTTCTTCTTCTTCAACTT-3′) for AtGDH.
Supplementary Material
Acknowledgments
We gratefully acknowledge Linda Hanley-Bowdoin for critically reading the manuscript, Marta Erra Pujada for her help with the production of transgenic Arabidopsis plants, Paolo Longoni for his help with RT-PCR assays, Rita Cantoni for technical support, and Severine Domenichini for microscope pictures.
This work was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (grant nos. RBNE01TYZF–004 and PRIN 2004).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Rino Cella (cella@ipvgen.unipv.it).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.106.077990.
References
- Albani D, Mariconti L, Ricagno S, Pitto L, Moroni C, Helin K, Cella R (2000) DcE2F, a functional plant E2F-like transcriptional activator from Daucus carota. J Biol Chem 275: 19258–19267 [DOI] [PubMed] [Google Scholar]
- Attwooll C, Lazzerini Denchi E, Helin K (2004) The E2F family: specific functions and overlapping interests. EMBO J 23: 4709–4716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelos M, Curie C, Mazzolini L, Bardet C, Lescure B (1992) A protocol for transient gene expression in Arabidopsis thaliana protoplasts isolated from cell suspension cultures. Plant Physiol Biochem 30: 123–128 [Google Scholar]
- Boudolf V, Vlieghe K, Beemster GT, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabouté ME, Clement B, Philipps G (2002) S phase and meristem-specific expression of the tobacco RNR1b gene is mediated by an E2F element located in the 5′ leader sequence. J Biol Chem 277: 17845–17851 [DOI] [PubMed] [Google Scholar]
- Christensen J, Cloos P, Toftegaard U, Klinkenberg D, Bracken AP, Trinh E, Heeran M, Di Stefano L, Helin K (2005) Characterization of E2F8, a novel E2F-like cell-cycle regulated repressor of E2F-activated transcription. Nucleic Acids Res 33: 5458–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Datla RS, Hammerlindl JK, Pelcher LE, Crosby WL, Selvaraj G (1991) A bifunctional fusion between beta-glucuronidase and neomycin phosphotransferase: a broad-spectrum marker enzyme for plants. Gene 101: 239–246 [DOI] [PubMed] [Google Scholar]
- de Bruin A, Maiti B, Jakoi L, Timmers C, Buerki R, Leone G (2003) Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 278: 42041–42049 [DOI] [PubMed] [Google Scholar]
- de Jager SM, Menges M, Bauer UM, Murray JA (2001) Arabidopsis E2F1 binds a sequence present in the promoter of S-phase-regulated gene AtCDC6 and is a member of a multigene family with differential activities. Plant Mol Biol 47: 555–568 [DOI] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GTS, de Almeida Engler J, Ormenese S, Maes S, Naudts M, Van Der Schueren E, Jacqmard A, Engler G, Inzé D (2002) Control of proliferation, endoreduplication and differentiation by the Arabidopsis E2Fa-DPa transcription factor. EMBO J 21: 1360–1368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Boniotti MB, Gutierrez C (2002) Arabidopsis E2Fc functions in cell division and is degraded by the ubiquitin-SCF(AtSKP2) pathway in response to light. Plant Cell 14: 3057–3071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Pozo JC, Timpte C, Tan S, Callis J, Estelle M (1998) The Ubiquitin-related protein RUB1 and auxin response in Arabidopsis. Science 280: 1760–1763 [DOI] [PubMed] [Google Scholar]
- Di Stefano L, Jensen MR, Helin K (2003) E2F7, a novel E2F featuring DP-independent repression of a subset of E2F-regulated genes. EMBO J 22: 6289–6298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova DK, Dyson NJ (2005) The E2F transcriptional network: old acquaintances with new faces. Oncogene 24: 2810–2826 [DOI] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Cella R, Robertson D, Hanley-Bowdoin L (2002) Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. Plant Cell 14: 3225–3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Livingston DM, Orkin SH, Greenberg ME (1996) E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell 85: 549–561 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P, Svab Z, Maliga P (1994) The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol Biol 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Humbert PO, Verona R, Trimarchi JM, Rogers C, Dandapani S, Lees JA (2000) E2f3 is critical for normal cellular proliferation. Genes Dev 14: 690–703 [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N (2002) Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol 130: 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefferson RA, Kavanagh TA, Bevan MW (1987) GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 61: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DG, Cress WD, Jakoi L, Nevins JR (1994) Oncogenic capacity of the E2F1 gene. Proc Natl Acad Sci USA 91: 12823–12827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002. a) E2Ls, E2F-like repressors of Arabidopsis that bind to E2F sites in a monomeric form. J Biol Chem 277: 16553–16558 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2002. b) Interaction of the Arabidopsis E2F and DP proteins confers their concomitant nuclear translocation and transactivation. Plant Physiol 128: 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y (2003) Constitutive E2F expression in tobacco plants exhibits altered cell cycle control and morphological change in a cell type-specific manner. Plant Physiol 132: 2012–2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bako L, Inzé D, Bogre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti B, Li J, de Bruin A, Gordon F, Timmers C, Opavsky R, Patil K, Tuttle J, Cleghorn W, Leone G (2005) Cloning and characterization of mouse E2F8, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem 280: 18211–18220 [DOI] [PubMed] [Google Scholar]
- Mariconti L, Pellegrini B, Cantoni R, Stevens R, Bergounioux C, Cella R, Albani D (2002) The E2F family of transcription factors from Arabidopsis thaliana. Novel and conserved components of the retinoblastoma/E2F pathway in plants. J Biol Chem 277: 9911–9919 [DOI] [PubMed] [Google Scholar]
- Menges M, Murray JA (2002) Synchronous Arabidopsis suspension cultures for analysis of cell-cycle gene activity. Plant J 30: 203–212 [DOI] [PubMed] [Google Scholar]
- Mironov V, De Veylder L, Van Montagu M, Inzé D (1999) Cyclin-dependent kinases and cell division in plants: the nexus. Plant Cell 11: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JA, Ahn JW, Kim YK, Kim SJ, Kim JK, Kim WT, Pai HS (2005) Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J 42: 153–163 [DOI] [PubMed] [Google Scholar]
- Ramirez-Parra E, López-Maatas MA, Frűndt C, Gutierrez C (2004) Role of an atypical E2F transcription factor in the control of Arabidopsis cell growth and differentiation. Plant Cell 16: 2350–2363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossignol P, Stevens R, Perennes C, Jasinski S, Cella R, Tremousaygue D, Bergounioux C (2002) AtE2F-a and AtDP-a, members of the E2F family of transcription factors, induce Arabidopsis leaf cells to re-enter S phase. Mol Genet Genomics 266: 995–1003 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Shan B, Lee WH (1994) Deregulated expression of E2F-1 induces S-phase entry and leads to apoptosis. Mol Cell Biol 14: 8166–8173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Wong SH, Hong W (1994) Overexpression of E2F-1 in rat embryo fibroblasts leads to neoplastic transformation. EMBO J 13: 3329–3338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C, La Thangue NB (2003) E2F and cell cycle control: a double-edged sword. Arch Biochem Biophys 412: 157–169 [DOI] [PubMed] [Google Scholar]
- Stevens R, Mariconti L, Rossignol P, Perennes C, Cella R, Bergounioux C (2002) Two E2F sites in the Arabidopsis MCM3 promoter have different roles in cell cycle activation and meristematic expression. J Biol Chem 277: 32978–32984 [DOI] [PubMed] [Google Scholar]
- Timmermans MC, Maliga P, Vieira J, Messing J (1990) The pFF plasmids: cassettes utilising CaMV sequences for expression of foreign genes in plants. J Biotechnol 14: 333–344 [DOI] [PubMed] [Google Scholar]
- Trimarchi JM, Fairchild B, Wen J, Lees JA (2001) The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci USA 98: 1519–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trimarchi JM, Lees JA (2002) Sibling rivalry in the E2F family. Nat Rev Mol Cell Biol 3: 11–20 [DOI] [PubMed] [Google Scholar]
- Vandepoele K, Vlieghe K, Florquin K, Hennig L, Beemster GTS, Gruissem W, Van de Peer Y, Inze D, De Veylder L (2005) Genome-wide identification of potential plant E2F target genes. Plant Physiol 139: 316–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varotto S, Locatelli S, Canova S, Pipal A, Motto M, Rossi V (2003) Expression profile and cellular localization of maize Rpd3-type histone deacetylases during plant development. Plant Physiol 133: 606–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlieghe K, Boudolf V, Beemster GT, Maes S, Magyar Z, Atanassova A, de Almeida Engler J, De Groodt R, Inzé D, De Veylder L (2005) The DP-E2F-like gene DEL1 controls the endocycle in Arabidopsis thaliana. Curr Biol 15: 59–63 [DOI] [PubMed] [Google Scholar]
- Xu G, Livingston DM, Krek W (1995) Multiple members of the E2F transcription factor family are the products of oncogenes. Proc Natl Acad Sci USA 92: 1357–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ (1996) Tumor induction and tissue atrophy in mice lacking E2F-1. Cell 85: 537–548 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.