Abstract
Exhibiting rapid polarized growth, the pollen tube delivers the male gametes into the ovule for fertilization in higher plants. To get an overall picture of gene expression during pollen germination and pollen tube growth, we profiled the transcription patterns of 1,536 pollen cDNAs from lily (Lilium longiflorum) by microarray. Among those that exhibited significant differential expression, a cDNA named lily ankyrin repeat-containing protein (LlANK) was thoroughly studied. The full-length LlANK cDNA sequence predicts a protein containing five tandem ankyrin repeats and a RING zinc-finger domain. The LlANK protein possesses ubiquitin ligase activity in vitro. RNA blots demonstrated that LlANK transcript is present in mature pollen and its level, interestingly contrary to most pollen mRNAs, up-regulated significantly during pollen germination and pollen tube growth. When fused with green fluorescent protein and transiently expressed in pollen, LlANK was found dominantly associated with membrane-enclosed organelles as well as the generative cell. Overexpression of LlANK, however, led to abnormal growth of the pollen tube. On the other hand, transient silencing of LlANK impaired pollen germination and tube growth. Taken together, these results showed that LlANK is a ubiquitin ligase associated with membrane-enclosed organelles and required for polarized pollen tube growth.
In higher plants, mature pollen grains land on the stigma and protrude tubes, which travel a long distance in the style. Eventually, one tube deposits two sperm cells into the ovule to achieve fertilization. Probably the fastest growth of plant cells, pollen tube growth is under tight control and elaborately modulated (Taylor and Hepler, 1997; Yang, 1998; Hepler et al., 2001; Feijó et al., 2004). The crucial players in this extreme type of polarized growth include Ca2+ (Franklin-Tong, 1999), Rop/Rac GTPase (Zheng and Yang, 2000; Fu et al., 2001; Gu et al., 2004), and phosphoinositide (Franklin-Tong et al., 1996). Recently, new molecules such as γ-amino butyric acid (Palanivelu et al., 2003) and nitric oxide (Prado et al., 2004) have emerged as important actors. Moreover, pollen growth responds to other factors that affect the pollen cell wall (Li et al., 1996, 2002; Zhou et al., 2004; Bosch et al., 2005). The cytoskeleton and its related proteins play essential roles in pollen tube growth (Vidali et al., 2001; Chen et al., 2003), and it is most likely that the majority of the signaling pathways regulate tip growth by directly or indirectly connecting to and remodeling the cytoskeleton (Feijó et al., 2004). It is noteworthy that protein turnover during pollen elongation is also important and the ubiquitin/proteasome pathway has been implicated as a major regulator of tip growth (Speranza et al., 2001; Scoccianti et al., 2003).
Decades of intensive investigation have given deep insight into pollen physiology, but the complex mechanisms underlying this tip growth are far from being clear. However, an overall understanding of the process could be accelerated significantly with the advent of high-throughput technologies such as gene expression profiling (Schena et al., 1995; Lockhart and Winzeler, 2000; Blohm and Guiseppi-Elie, 2001). Application of oligonucleotide chip to Arabidopsis (Arabidopsis thaliana) pollen has provided a comprehensive view of the male gametophytic transcriptome (Becker et al., 2003; Honys and Twell, 2003). However, it should be noted that the work of both groups was designed to compare with sporophytic transcriptomes rather than to dissect pollen germination and pollen tube growth. cDNA microarray, another type of gene expression profiling, can be prepared by printing anonymous cDNAs onto slides but sequencing some of them after hybridizations. This technique is thus applicable to study pollen from unsequenced plant species such as lily (Lilium longiflorum).
Here we report the application of a 1,536-cDNA microarray to profile the gene expression during lily pollen germination and tube growth. Sequencing of a subset of cDNA clones led to the identification of a number of unannotated genes. Through characterization of one of them, lily ankyrin repeat-containing protein (LlANK), we show that this protein is a ubiquitin ligase closely associated with membrane-enclosed organelles and required for pollen germination and pollen tube growth.
RESULTS
Lily Pollen cDNA Microarray
A 1,536-cDNA microarray was prepared to profile the gene expression from germinated and ungerminated lily pollen. The data have been deposited in the National Center for Biotechnology Information's Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) and are accessible through Gene Expression Omnibus Series accession number GSE2648. We then sequenced 100 cDNA clones that exhibited the highest germinated/ungerminated ratios and matched the data against GenBank. The result is available as supplemental information (Supplemental Table I).
Of the 100 sequenced cDNA clones, 60 were identified homologous to known genes in GenBank. In these sequences, three groups of genes are highly represented: pectin methylesterases, small GTPases, and proteases including ubiquitination-proteasome pathway components (Table I). Though the differential expression needs to be verified individually, the prominence of these genes supported their critical roles in pollen germination and pollen tube growth as characterized previously (Li et al., 1996; Speranza et al., 2001; Cheung et al., 2002; Li et al., 2002; Scoccianti et al., 2003; Bosch et al., 2005). In addition to those homologous to known genes, there were cDNAs representative of uncharacterized genes. One of them was a cDNA clone predicting a protein fragment featured with three imperfect ankyrin repeats as well as one RING zinc-finger domain. This cDNA, named LlANK, was chosen for detailed investigation.
Table I.
Three groups of genes are highly represented in the sequenced cDNAs differentially expressed during lily pollen germination and tube growth as monitored by cDNA microarray
Homologies (E value < 0.001) are shown as accession (species). Ad, Kiwifruit; At, Arabidopsis; Hs, Homo sapiens; Le, Lycopersicon esculentum (tomato); Os, rice (japonica cultivar group).
| GenBank Accession | Name | Homologies in Other Species | ||
|---|---|---|---|---|
| Pectin methylesterase (inhibitor) | ||||
| DN985103 | Pectin methylesterase | XP_479611 (Os) | AAF26136 (At) | BAC42986 (At) |
| DN985143 | Pectin esterase inhibitor | NP_182256 (At) | BAC54964 (Ad) | BAC54965 (Ad) |
| DN985149 | Pectin esterase inhibitor | NP_182256 (At) | BAC54964 (Ad) | BAC54965 (Ad) |
| Small GTPase | ||||
| DN985094 | Ras/Rab-like GTP-binding protein | BAB10106 (At) | AAT77401 (Os) | BAA97069 (At) |
| DN985121 | Ras/Rab-like GTPase | CAB90933 (At) | BAD30623 (Os) | AAT39172 (Os) |
| DN985152 | GTPase regulator | AAD15318 (At) | AAG00551 (Hs) | |
| Protease and ubiquitination-proteasome pathway components | ||||
| DN985099 | Cathepsin B-like Cys protease | AAN60355 (At) | AAX11351 (Os) | CAB77732 (At) |
| DN985111 | Cathepsin B-like Cys protease | AAN60355 (At) | AAX11351 (Os) | CAB77732 (At) |
| DN985130 | Subtilisin-like Ser protease | CAB67120 (Le) | ABA97963 (Os) | CAA07250 (Le) |
| DN985072 | F-box family protein | NP_565401 (At) | NP_565403 (At) | NP_176753 (At) |
| DN985135 | Ubiquitin-conjugating enzyme | AAM11574 (At) | AAY44867 (At) | NP_851115 (At) |
| DN985163 | Proteasome subunit 4 like | BAB78491 (Os) | CAB79662 (At) | AAF22522 (At) |
LlANK Protein Contains Five Ankyrin Repeats and One RING Zinc-Finger Domain
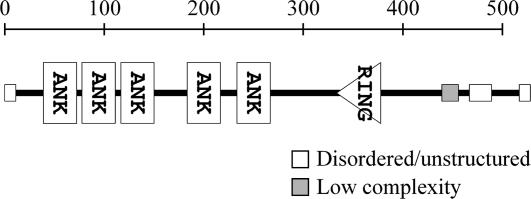
The original LlANK cDNA was a truncated form lacking its 5′ end coding region. Using 5′ RACE, we obtained an additional 522-bp upstream cDNA sequence. The new longer cDNA could be readily retrieved by reverse transcription (RT)-PCR from pollen at all stages of in vitro growth. Sequence analysis indicated that LlANK cDNA contains a complete coding sequence for a 516-amino acid protein in which the SMART program (http://smart.embl-heidelberg.de/) confidently predicted five tandem ankyrin repeats and a canonical C3HC4-type RING zinc-finger domain (Fig. 1). Both ankyrin repeat and RING zinc finger are common protein-protein interaction motifs. Since ankyrin repeat appears conserved in structure rather than in function (Bork, 1993), little could be inferred about the function of LlANK. Similarly, RING zinc finger has been implicated in a range of diverse biological processes. But recently, ubiquitin ligase activity has been found intrinsic to some RING-containing proteins (Lorick et al., 1999; Hardtke et al., 2002; Stone et al., 2005) and is likely to be a general function of this domain. In addition, disordered/unstructured segments and a region of low complexity were predicted near both termini, but the functional significance of these regions remains unknown. In Arabidopsis and rice (Oryza sativa), there are proteins that share considerable homology with LlANK (Table II). The rice homologies are all hypothesized proteins without further characterization. Some of them are similar to proteins binding to a rice disease resistance gene product XA21 (Song et al., 1995), which, however, can hardly be related to pollen physiology. In Arabidopsis, the two homologies, XBAT32 (At5g57740) and At5g07270, were recently found to possess ubiquitin ligase activity in vitro (Nodzon et al., 2004; Stone et al., 2005). Particularly, the XBAT32 protein is capable of ubiquitinating itself. It is thus reasonable to test if LlANK possesses this activity.
Figure 1.
Predicted structure of LlANK protein, generated by the SMART program (http://smart.embl-heidelberg.de/).
Table II.
Proteins homologous to LlANK in existing databases
At, Arabidopsis. Os, rice (japonica cultivar group).
| Score | Accession | Protein Description | Confirmed Function | Reference |
|---|---|---|---|---|
| 1,779 | AAM92304 | Hypothetical protein (Os) | ||
| 1,711 | AAZ14070 | At5g07270 (At) | Ubiquitin ligase activity in vitro | Stone et al. (2005) |
| 1,152 | XP_468209 | Putative receptor-like kinase Xa21-binding protein 3 (Os) | ||
| 1,132 | AAS76759 | XBAT32 (At5g57740; At) | Ubiquitin ligase activity in vitro; regulates lateral root development | Nodzon et al. (2004) |
| 1,101 | XP_481070 | Receptor-like kinase Xa21-binding protein 3 like (Os) | ||
| 1,071 | AAY88733 | XB3-related protein (Os) | ||
| 760 | NP_910374 | Similar to mouse ankyrin 3 (AC005727; Os) |
LlANK Possesses Ubiquitin Ligase Activity in Vitro
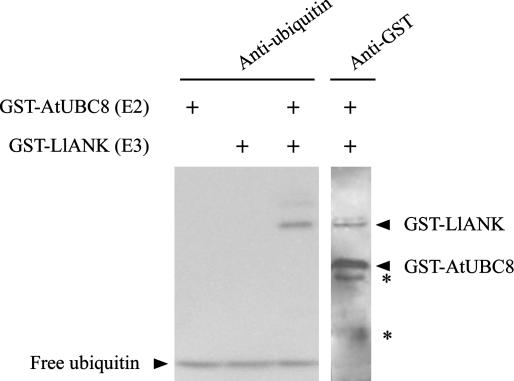
Conjugating of ubiquitin to a target protein requires at least three kinds of enzymes: Ubiquitin-activating enzyme (E1) creates an activated ubiquitin that is transferred to ubiquitin-conjugating enzyme (E2) and subsequently, depending on ligase enzyme (E3), to a target protein. The activities and specificities of E2 are controlled by E3, which binds to substrate (Pickart, 2001). To test if LlANK possesses ubiquitin ligase activity, we set up ubiquitination assays consisting of recombinant human E1 enzyme, recombinant Arabidopsis E2 enzyme glutathione-S-transferase (GST)-AtUBC8, an ATP-regenerating system, and GST-LlANK fusion protein. To monitor protein ubiquitination, we probed the assays by western blot using either anti-ubiquitin or anti-GST antibody (Fig. 2). The appearance of a ubiquitinated product of equal size to GST-LlANK indicated that LlANK itself is ubiquitinated in the presence of E1 and E2. Products of higher Mr were also visible in the upper lane, probably indicating the formation of a polyubiquitin chain. Omission of E2 or LlANK, however, resulted in a loss of ubiquitination. Thus, we concluded that LlANK possesses ubiquitin ligase activity in vitro.
Figure 2.
Autoubiquitination of the LlANK fusion protein. Complete in vitro ubiquitination assays contained recombinant human 6× His-tagged E1 enzyme, recombinant Arabidopsis E2 enzyme GST-AtUBC8, recombinant GST-LlANK, and ubiquitin. Assay products were analyzed with western blot using either anti-ubiquitin or anti-GST antibodies. Omission of GST-AtUBC8 or GST-LlANK protein from the assay, as indicated above the lanes, resulted in a loss of protein ubiquitination. Asterisk (*), GST or GST-fusion fragments presumably due to partial proteolysis during protein purification and incubation.
Expression of LlANK Is Up-Regulated during Pollen Germination and Pollen Tube Growth
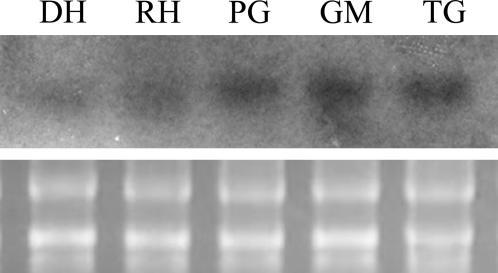
Since the gene expression profile of cold-stored pollen differs from that of fresh pollen because of selected protein/mRNA degradation during long-term storage (Wang et al., 2004), we performed RNA gel blots to evaluate LlANK expression in fresh pollen. As shown in Figure 3, LlANK transcript is present in fresh mature pollen and up-regulated significantly during germination and tube growth. Since it is widely accepted that pollen mRNAs are presynthesized and stored in pollen at the time of maturation but utilized during germination and tube growth (Mascarenhas, 1993; Taylor and Hepler, 1997; McCormick, 2004), it is interesting to find that LlANK, in contrast, is continuously synthesized and even up-regulated during germination and tube growth.
Figure 3.
RNA gel-blot evaluation of LlANK expression during pollen germination and pollen tube growth. DH, Dehydrated; RH, rehydrated; PG, pregermination; GM, germinating; TG, tube growth. Bottom, 28 and 18 s rRNA.
LlANK Protein Is Associated with Membrane-Enclosed Organelles in Pollen
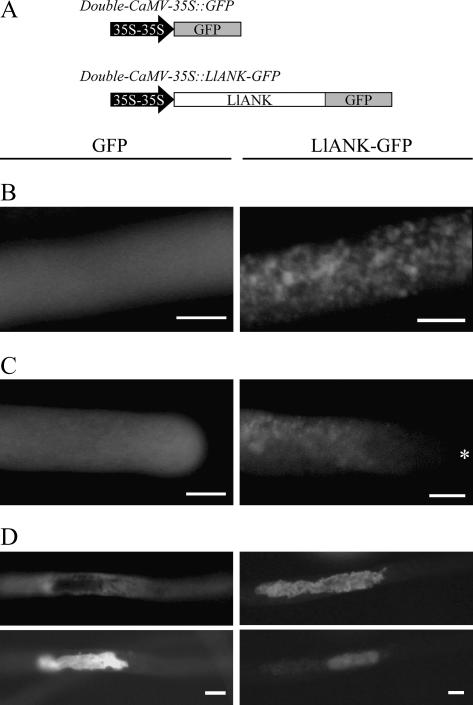
To determine the cellular localization of LlANK, we transiently expressed LlANK-green fluorescent protein (GFP) fusion protein in lily pollen via particle bombardment, a method that has been successfully applied in pollen (Chen et al., 2002; Cheung and Wu, 2004). LlANK-GFP fusion gene was put under the control of double-cauliflower mosaic virus (CaMV)-35S promoter, which works moderately in lily pollen (Nishihara et al., 1993). Using an increasing amount of the double-CaMV-35S∷GFP DNA, we found that a GFP signal was readily detectable when no less than 5 μg DNA was used per bombardment. The experimental settings were then used for cellular localization of LlANK-GFP. The results are shown in Figure 4.
Figure 4.
Cellular localization of transiently expressed LlANK-GFP fusion protein in lily pollen. A, Diagrammatic representation (not to scale) of both GFP control and LlANK-GFP fusion constructs. 35S-35S, Constitutive double-CaMV-35S promoter. B, LlANK-GFP associated with punctate or granular structures in pollen tube. C, Apical region of pollen tube free of the highlighted structure (asterisk (*), pollen apex). D, LlANK-GFP associated with a generative cell (Top, green fluorescence viewing; Bottom, generative nuclei visualized with Hoechst 33258). GFP control (left) and LlANK-GFP (right) images are juxtaposed for comparison. B and C are confocal images. Bar = 10 μm.
Transiently expressed control GFP was evenly distributed throughout the cytoplasm of tubes, while LlANK-GFP fusion protein was prominently associated with distinct structures that appeared to be membrane-enclosed organelles. One of the frequently observed phenomena was the punctate or granular dots in pollen tubes (Fig. 4B). In grains, such dots were also observed (data not shown). However, they were never found at the apical region of pollen tube, the so-called clear zone (Fig. 4C). The most notable structure with which LlANK-GFP was associated, however, was generative cell, which from a certain respect can be viewed as the largest membrane-enclosed organelle in the bicellular pollen (Fig. 4D). Furthermore, neither cell wall nor plasma membrane was found labeled with LlANK-GFP, indicating that LlANK was not targeted to these places. Observations above together led us to conclude that LlANK protein is associated with membrane-enclosed organelles in lily pollen.
Overexpression of LlANK Leads to Abnormal Tube Growth
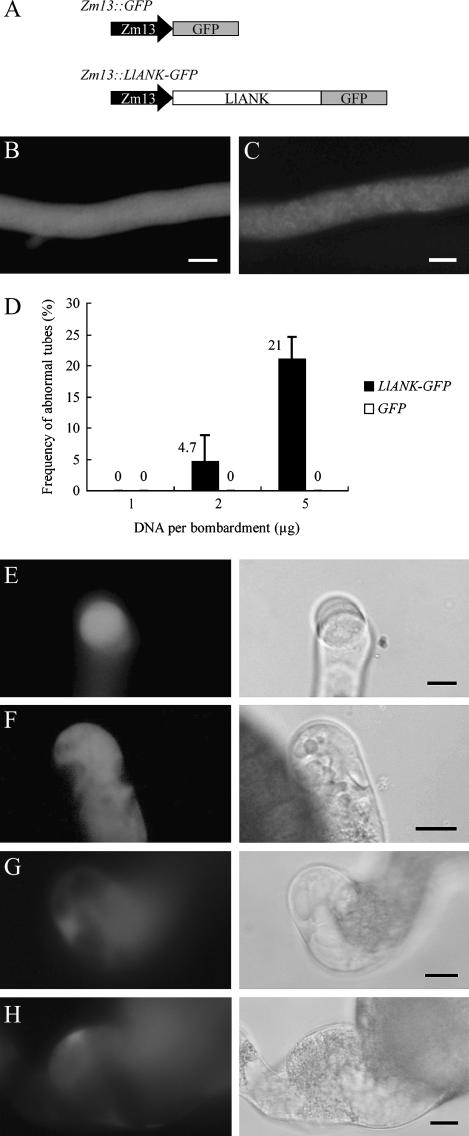
Zm13 is a pollen-specific gene from maize (Zea mays; Hamilton et al., 1989; Hanson et al., 1989) and its promoter has proved highly active in lily pollen (Morikawa et al., 1994; Miyoshi et al., 1995; Chen et al., 2002). To see the consequence of overexpressing LlANK, we constructed Zm13∷LlANK-GFP as well as the control Zm13∷GFP (Fig. 5A) and introduced them into lily pollen.
Figure 5.
Overexpression of LlANK in lily pollen. A, Diagrammatic representation (not to scale) of both GFP and LlANK-GFP constructs. Zm13, Pollen-specific strong promoter. B, Control GFP evenly distributed in pollen tube. C, Moderately expressed LlANK-GFP associated with granular structures. D, Increasing amount of LlANK-GFP DNA led to abnormal pollen tube growth. Bar = sd. Data were from three independent bombardments. Morphological abnormalities included budding (E), membrane invagination (F), tip ballooning (G), and tube swelling (H). In E to H, green fluorescence (left) and their corresponding transmitted images (right) are juxtaposed. Bar = 10 μm.
Unlike their double-CaMV-35S-driven counterparts, the Zm13-driven constructs were capable of producing a detectable signal using DNA as little as 1 μg per bombardment. Increasing the DNA amount enhanced GFP intensity, but no further improvement was observed when using more than 5 μg DNA per bombardment, presumably due to the coating limitation of gold particles. Overexpression of the control GFP invariably displayed uniform distribution throughout pollen tubes (Fig. 5B) and no apparent effect was found on germination and tube growth. In the case of Zm13∷LlANK-GFP transformation, the typical punctate or granular structures were present (Fig. 5C), while in a small fraction of tubes an excess of overexpressed LlANK-GFP was found dispersed into the cytosol and morphological abnormalities were usually observed when using a large amount of DNA (Fig. 5D). These abnormalities included budding (Fig. 5E), membrane invagination (Fig. 5F), tip ballooning (Fig. 5G), and tube swelling (Fig. 5H). We also measured the average length of transformed but normal tubes as well as that of the untransformed ones, but no significant difference was found (data not shown). These observations indicated that LlANK is involved in pollen tube growth but its overexpression is prone to interrupt the fine coordinance of the regulation and to induce abnormal tube growth.
Silencing of LlANK Impairs Pollen Germination and Pollen Tube Growth
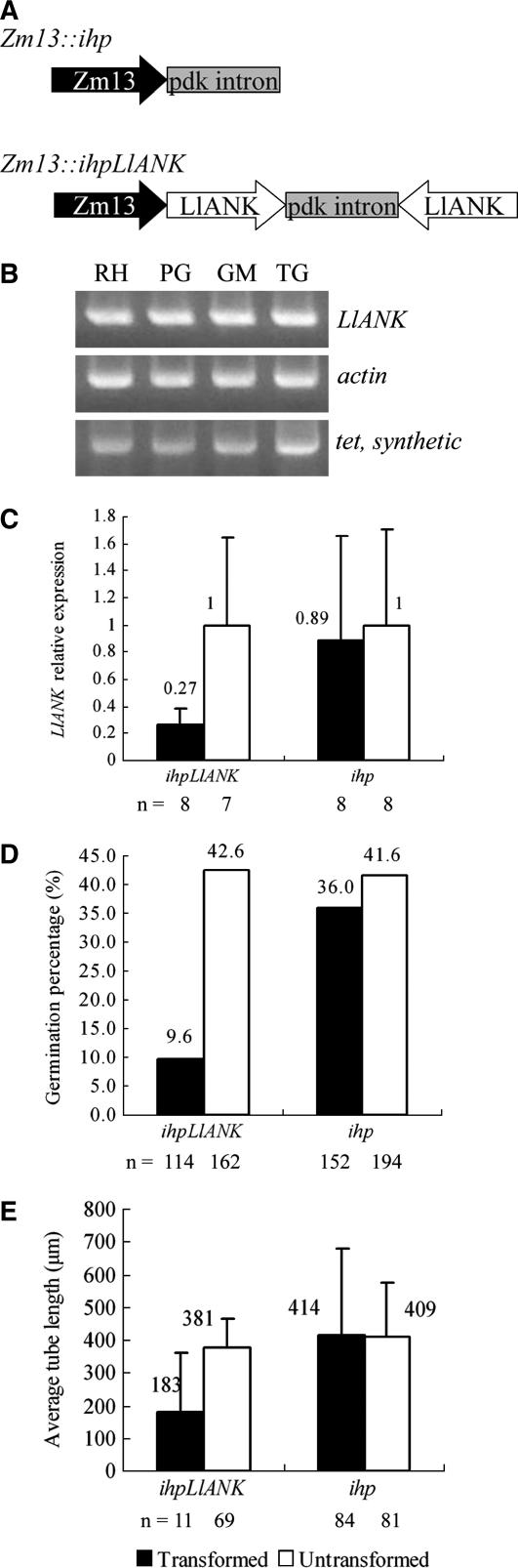
RNA-induced gene silencing is a powerful tool available for plant biologists in recent years (Burch-Smith et al., 2003; Waterhouse and Helliwell, 2003). This technique has also been exploited successfully in pollen (Gupta et al., 2002; Wang et al., 2003; Swain et al., 2004). To specifically silence LlANK while ensuring that other genes were not affected, we purposely selected the 3′ portion of LlANK cDNA to prepare the silencing construct Zm13∷ihpLlANK, which was designed to synthesize an intron-spliced hairpin RNA in vivo and promote the most effective silencing (Smith et al., 2000; Dykxhoorn et al., 2003). The empty hairpin vector Zm13∷ihp was used as control (Fig. 6A). To indicate successful transformation and expression, Zm13∷GFP was combined 1:1 (w/w) with either Zm13∷ihpLlANK or Zm13∷ihp. Since transformation frequency was low (<3% in our experiments) and sorting out transformed pollen grains/tubes on flow cytometer was not applicable, it was difficult to assess the effect of silencing by standard RNA gel blot. However, we found that LlANK could be readily amplified by RT-PCR from one single pollen (Fig. 6B), which provided the possibility that we perform quantitative real-time PCR to monitor LlANK expression level in an individual transformed pollen. Since there is no validating evidence that the widely accepted housekeeping gene actin expresses at a constant level during the rapid tip growth of pollen, we used an externally applied synthetic poly(A)+ RNA as normalizing standard in this case of single-pollen evaluation.
Figure 6.
Transient down-regulation of LlANK in lily pollen. A, Diagrammatic representation (not to scale) of both ihp and ihpLlANK constructs. Zm13, Pollen-specific strong promoter; pdk, a splicable intron. B, From one single pollen of different stage, LlANK, actin, and an exogenously applied synthetic tet transcript could be simultaneously amplified using conventional RT-PCR. RH, Rehydrated; PG, pregermination; GM, germinating; TG, tube growth. C, Quantitative real-time PCR evaluation of single pollen demonstrated the down-regulation of LlANK expression in ihpLlANK-transformed pollen (Student's t test, P = 0.01). Data are presented as the ratio in expression level normalized to tet and relative to untransformed pollen. D, Down-regulation of LlANK reduced in vitro pollen germination significantly. E, Effect of LlANK down-regulation on average tube length (Student's t test, P = 0.0046). For each pair of juxtaposed columns, data were from a same bombarded population. Numbers of grains/tubes scored are shown beneath each column. Bars = sd.
Quantitative comparative real-time PCR results demonstrated that the LlANK expression was down-regulated in pollen transformed with ihpLlANK (Fig. 6C). The calculated germination percentage for transformed pollens was 9.6%, which was significantly lower than the 42.6% in untransformed pollens from the same bombarded population (Fig. 6D). When it came to tube growth, the average length of all transformed tubes was 183 μm, much shorter than the 381 μm of the equal top proportion of tubes that received no DNA (Fig. 6E). Meanwhile, the control experiment using ihp demonstrated that bombardment as well as the subsequent exogenous expression had little effect on pollen germination and pollen tube growth (Fig. 6, D–E, right, respectively). Taken together, the results above indicated that down-regulation of LlANK impaired pollen germination and pollen tube growth. The inhibitory effect coming after LlANK down-regulation suggested the essential role that LlANK plays, directly or indirectly, in this polarized tip growth. However, it should be noted that no morphological abnormality was observed in the tubes of transformed pollens.
DISCUSSION
GFP fusion protein localization demonstrated that LlANK is associated with membrane-enclosed organelles. The nature of these organelles, however, remains to be determined. We tried a range of specific fluorescent probes such as Rhodamine 123 (for mitochondria) and ER-Tracer Blue-White DPX (for endoplasmic reticulum), but there was no convincing conclusion (data not shown). The appearance of these highlighted structures resembles that of Golgi bodies (Cheung et al., 2002), dispersed vacuoles (Hicks et al., 2004), and the so-called membrane-enclosed organelles (Cai et al., 2000), but the identity of these organelles awaits determination. Observations suggested that the association of LlANK with organelle is selective. For example, it seemed that not all the vesicles were fluorescently labeled (Fig. 4B). The tip area, where secretory vesicles accumulate (Derkson et al., 1999), was free of LlANK distribution (Fig. 4C). Moreover, vegetative nuclei were never found fluorescently labeled, though they were absolutely membrane-enclosed structures. Since analysis of LlANK amino acid sequence revealed no features characteristic of transmembrane region, the association selectivity may be attributed to specific interactions of LlANK, presumably via its ankyrin repeats, with certain membrane proteins that are unique to a subset of organelles.
Our data provided the biochemical evidence that LlANK possesses E3 ubiquitin ligase activity in vitro. We also showed that down-regulation of LlANK reduced pollen tube emergence and growth. This finding is consistent with the observation that ubiquitin/proteasome pathway has a direct role in regulating pollen tube emergence in kiwifruit (Actinidia deliciosa; Speranza et al., 2001; Scoccianti et al., 2003). Since the initial demonstration of ubiquitin/proteasome pathway, it was believed that its primary function was the rapid degradation of proteins with abnormal conformations and of many regulatory proteins whose short half lives have evolved to facilitate in the regulation of their activities (Weissman, 2001). Therefore, it is likely that LlANK affects pollen tube growth not directly by itself, but rather by regulating activities of certain effectors. Unlike inhibiting the overall pathway with specific inhibitor, suppressing LlANK did not produce any apparent morphological abnormality, probably because it stabilized only one or a few defined substrate(s) associated with certain biological process(es). On the other hand, it should be noted that degradation is not the only fate possible for ubiquitin-tagged proteins. Rather, ubiquitination touches upon all aspects of eukaryotic biology (Hicke, 2001; Pickart, 2001; Weissman, 2001). In our assay, LlANK protein ubiquitinated itself in vitro. The ubiquitination activity toward itself, however, does not rule out that LlANK has other defined substrate(s), as exemplified by Mdm2, a typical RING-type E3 that ubiquitinates itself as well as p53 (Fang et al., 2000).
Based on the results obtained and the implications mentioned above, it is logical to launch into isolating and identifying the protein(s) that LlANK recognizes and binds to presumably via its ankyrin repeat domain. In such an attempt, we expressed recombinant GST-LlANK and used the fusion protein as a bait to pull down (Simpson, 2002) its interacting proteins from pollen extract. Using mass spectrometry, all major captured protein spots turned out to be truncated LlANK fragments (data not shown), probably due to the partial proteolysis during protein capturing, whereas the partner protein pursued remains evasive to date. Currently, additional efforts are being made to improve the affinity capturing. However, considering that LlANK is capable of ubiquitinating itself (Fig. 2), it is possible that the fragments were the true reflection of ubiquitin-mediated degradation of LlANK protein in pollen extract. In this sense, it appears that the fragments strongly bind to the bait LlANK protein, raising the possibility that LlANK protein undergoes dimerization and, subsequently, ubiquitinates each other in a trans manner. Though attractive, all these possibilities need to be tested with additional experiments.
We have shown that LlANK is closely associated with membrane-enclosed organelles and required for pollen germination and tube growth in lily. Of the proteins homologous to LlANK (Table II), XBAT32 from Arabidopsis is the only one that has been comprehensively studied (Nodzon et al., 2004). Transgenic study demonstrated that high XBAT32∷GUS activity was observed in the vascular system of primary roots, particularly in the zone where lateral root initiation occurs. The authors also noticed that XBAT32∷GUS expression was found in the anthers. However, they did not further examine the pollen inside. When it comes to At5g070270, another Arabidopsis gene that shares the highest homology with LlANK, only an in vitro E3 ubiquitin ligase activity was reported for this protein in a genome-wide analysis of RING domain-containing proteins in Arabidopsis (Stone et al., 2005). In addition to At5g07270 and XBAT32, there are three other Arabidopsis genes that are predicted to encode proteins containing both ankyrin repeats and RING zinc finger (Becerra et al., 2004; Nodzon et al., 2004) but none of them has been characterized. Recently from the Arabidopsis Biological Resource Center (http://Arabidopsis.org/abrc) we have obtained a mutant line carrying a T-DNA insertion in the At5g07270 gene. Turning to the model plant species, we are now embarking on characterization of the LlANK-like genes not only in pollen but also in whole plant. It is expected that these structurally related genes have overlapping but distinct functions, but it is certain that the future will hold many surprises.
MATERIALS AND METHODS
Plant Material and Culture Condition
Plants of lily (Lilium longiflorum) were grown in a green house under natural day length. Anther was harvested from young flowers, induced to dehisce by light, and dehydrated in a desiccator. Pollen was then collected and, if not to be used immediately, stored at −20°C. Prior to use, cold-stored pollen was brought gradually to room temperature. After that, like fresh pollen, the stored pollen was rehydrated in a moisture chamber for 3 h. Rehydrated pollen was then resuspended in Dickenson medium [0.03% Ca(NO3)2·4H2O, 0.01% KNO3, 0.001% H3BO4, and 10% Suc] at a density of 4 to 5 mg mL−1 with gentle shaking at 25°C. Generally, pollen bulk germinated and took on active tube growth 1 and 3 h after culture initiation, respectively. For practical convenience, in vitro germination and tube growth were divided into the following stages: (1) dehydrated pollen freshly collected from desiccated anther, (2) rehydrated pollen (culture initiation), (3) pregermination pollen (no visible tube), (4) germinating pollen (tube protruding), and (5) tube growth (>100 μm in length). Aliquot of pollen suspension can be withdrawn for microscopic observation at any time point as desired. For RNA/protein extraction, a large number of pollen grains/tubes were harvested from the culture medium using 10-μm nylon membrane.
cDNA Microarray
Total RNA was isolated from microspores, ungerminated, and germinated pollen using Qiagen RNAeasy plant mini kit. mRNA was isolated from total RNA with FastTrack 2.0 mRNA isolation kit (Invitrogen). From the pooled mRNAs, a cDNA library was constructed using PCR cDNA library construction kit (Stratagene). As soon as it was produced, the cDNA library was used for microarray preparation. Since then, the work was done by Genetix and is only briefly described here. A total of 1,536 clones were picked randomly from the cDNA library and the cDNA inserts were PCR amplified directly from clones in culture using the M13 universal primers. PCR products were purified with Montage PCR96 cleanup kit (Millipore), transferred to 384-well microtitres, and vacuum dried. After redissolved, the PCR products were spotted (diameter: 250 μm) onto microscope glass slides in adjacent duplicate using Genetix Qarrayer, UV cross-linked, blocked, and denatured. For preparation of the labeled cDNA targets, RNA samples were extracted from the pollen harvested at culture initiation (ungerminated) as well as after 3 h (germinated) as described above, labeled with Cy3-dUTP or Cy5-dUTP, respectively, according to previously reported protocols (Hedge et al., 2000). Two labeled targets were mixed together and simultaneously hybridized to the same microarray slides at 42°C overnight in Genpak genHYB hybridization buffer (Genetix). After being thoroughly washed, microslides were scanned twice at two different sensitivity levels using Packard Bioscience ScanArray 4000 (PerkinElmer) and analyzed on QuantArray (PerkinElmer). Systematic errors due to spotting and scanning were reduced by subtracting background intensity and taking into account the dimension of the spot. Raw data were normalized and Cy5/Cy3 ratio calculated by adjusting the mean to 1. Some outliers were discarded from analysis. Following normalization, confidence intervals as defined by Chen et al. (1997) were used to identify differentially expressed genes. We compared the two spots with the identical probe basing truly differential expression on consensus. Only the clones that presented a statistically higher ratio in both spots were considered for sequencing.
5′-RACE and RNA Gel Blot
5′-RACE was performed by following the standard strategy (Sambrook and Russell, 2001) with slight modifications: The method described by (Carninci et al. 1996, 2000) was used for second-strand cDNA synthesis, and a nested-PCR approach with two specific primers for final amplification. RT-PCR for full-length LlANK cDNA was done as instructed in standard protocols. In both RACE and RT-PCR experiments, SuperScript III reverse transcriptase (Invitrogen) was used for first-strand cDNA synthesis and Ex Taq DNA polymerase (TaKaRa) for second-strand cDNA synthesis and PCR amplification. RNA was prepared from fresh pollen with TRIzol Reagent (Invitrogen) as instructed, dissolved in formamide, separated on formaldehyde-containing 1.0% agarose gel, alkaline blotted onto Hybond-XL membrane (GE Healthcare), probed with [α-32P]CTP-labeled RNA (Riboprobe system, Promega) at 68°C in aqueous solution overnight, washed, and autographed as described (Sambrook and Russell, 2001).
Plasmid Construction
Standard methods (Sambrook and Russell, 2001) were followed for all molecular-cloning manipulations. Only strategic flow is described here. For GFP fusion protein expression we first constructed two sister vectors, double-CaMV-35S∷GFPv and Zm13∷GFPv, in which a modified GFP coding sequence (Reichel et al., 1996) was put under the control of double-CaMV-35S promoter and Zm13 promoter, respectively. In both constructs, a translation enhancer element from Tobacco etch virus was inserted between promoter and coding region to improve expression efficiency. NcoI and XhoI sites were introduced flanking LlANK's open reading frame, which was subsequently inserted in frame into the two sister vectors via NcoI-SalI. For gene silencing a 700-bp 3′ portion of LlANK cDNA was inserted head to head into silencing vector pHANNIBAL as described (Wesley et al., 2001; Helliwell and Waterhouse, 2003). The XhoI-XbaI selfcomplementary hairpin RNA encoding region was then subcloned into Zm13∷GFPv via SalI-XbaI to make Zm13∷ihpLlANK. The control construct Zm13∷ihp was prepared in a similar way. For expression of LlANK fusion protein in Escherichia coli, a new NcoI site was introduced into pGEX-6P-1 (GE Healthcare) so that both LlANK and the LlANK-GFP open reading frames could be in-frame fused with the GST coding sequence via NcoI-XhoI insertion without altering other features. All recombinant clones were verified by sequencing.
Recombinant Protein Expression and in Vitro Ubiquitination Assay
Recombinant plasmid constructs based on GST Gene Fusion system (GE Healthcare) were used for expression and purification of GST-LlANK and GST-AtUBC8 from bacteria, as instructed by the manufacturer. Ubiquitination reactions were done as described (Hardtke et al., 2002) in a total volume of 25 μL in conjugation buffer (50 mm Tris-HCl pH 7.5, 10 mm MgCl2, 0.05 mm ZnCl2, 1 mm ATP, 0.2 mm dithiothreitol, and 10 mm phosphocreatine) supplemented with 500 ng of recombinant 6× His-E1, 0.1 unit of creatine kinase (Sigma), 2 μg ubiquitin (Sigma), approximately 500 ng of GST-AtUBC8, and approximately 100 ng of GST-LlANK. Reactions were incubated at 30°C for 2 h and stopped by adding an equal amount of 2× sample buffer (100 mm Tris-HCl pH 6.8, 20% [v/v] glycerol, 4% [w/v] SDS, 200 mm β-mercaptoethanol, and 0.2% [w/v] bromphenol blue). Western blots using either anti-ubiquitin (Sigma) or anti-GST (Santa Cruz) antibody were performed by following standard protocols (Sambrook and Russell, 2001).
Particle Bombardment and Fluorescence Observation
Pollen was cultured as described above. For particle bombardment, pollen grain suspended in Dickenson medium was applied evenly onto filter paper put in a 90-mm petri dish. Enough medium was supplied to keep the paper wet but not to immerse pollen. Plasmid DNA was purified by resin (BioDev). For each bombardment, 0.5 mg gold particles (1.0 μm) were coated with 5 μg double-CaMV-35S∷(ANK-)GFP DNA for cellular localization or 1 to 5 μg Zm13∷(ANK-)GFP DNA for overexpression as described in the “Results” section. In silencing experiments, either Zm13∷ihpLlANK (effector) or Zm13∷ihp (control) was combined 1:1 (w/w) with Zm13∷GFP (indicator) and 5 μg DNA mixture used for each bombardment. Bombardment was performed with model PDS-1000/He Biolistic Particle Delivery system (Bio-Rad) as instructed by the manufacturer. Settings: 29-inch Hg vacuum, 4-cm gap distance, and 7-cm particle flight distance. To each sample, three consecutive bombardments were performed to increase transformation frequency. After bombardments, pollen was rinsed down from filter paper with the medium, cultured at a density of 4 to 5 mg mL−1 with gentle shaking at 25°C. Generally, GFP signal was detectable after 3 h. In cellular localization, over-, and down-expression experiments, pollen was observed 4 h after culture initiation. Pollen culture was applied on a microslide without any fixation. For nucleic acid staining, pollen culture was gently mixed 1:1 (v/v) with a mount solution, which was prepared by combining phosphate-buffered saline (140 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 1.8 mm KH2PO4, pH 7.3) 3:7 (v/v) with glycerol. Hoechst 33258 (Invitrogen) was supplemented to a final concentration of 100 ng mL−1 and incubated for 10 min at room temperature. Conventional fluorescence observation (filter: B-2A and UV-2A for GFP483/510 and Hoechst 33258352/461, respectively) and laser-scanning confocal microscopic observation (using the instrumental default settings for enhanced GFP) were performed on Nikon Eclipse E400 (Nikon Instech) and Olympus FluoView FV1000, respectively, and recorded with the software packaged therein. Pollen tube lengths were measured using Photoshop 7.0 software (Adobe); a layer was created above an original image, lines were drawn along pollen tubes and bars with the same width, pixel numbers were recorded, data was exported to Excel software (Microsoft), and eventually they were converted to tube lengths.
Single-Pollen Quantitative Real-Time PCR
Performances from single pollen isolation to PCR product detection were conducted consecutively as described below. Sixty microliters of pollen culture suspension was diluted in 1 mL Dickenson medium in one 35-mm cell culture dish (Corning), sited open on one microslide on Nikon Eclipse E400, and observed with only 10× objective lens. Monitored in the microscopic field, single pollen was carefully pipetted out with an Eppendorf 2.5 μL pipette, and immediately put into 0.5 mL TRIzol Reagent. To each single-pollen RNA extraction system, approximately 10 pg synthetic tetracyclin resistant gene (tet) poly(A)+ mRNA (1.4 kb, in vitro transcribed from tet of plasmid pBR322; TaKaRa) was added as a normalizing standard. RNA extraction using TRIzol Reagent was carried out as instructed, with 20 μg glycogen (Sigma) added as carrier prior to precipitating RNA with isopropyl alcohol. During RNA washing in 70% ethanol, a master mix of cDNA synthesis reactions was prepared that contained 20 nm oligo(dT)15 primer, 0.3 units μL−1 of moloney murine leukemia virus reverse transcriptase (Promega), and other standard components but with RNA template omitted. At the end of RNA extraction procedure, RNA pellet was directly dissolved in 30 μL aliquot of the cDNA synthesis master mix and incubated at 42°C for 1 h. In the meantime, PCR master mixes for LlANK, actin, and tet, respectively, were prepared. The master mixes contained respective primers, EX Taq, and other standard components but omitted deoxynucleotide triphosphates and cDNA template. When the cDNA synthesis was completed, the reaction was divided into three 10-μL aliquots, each of which was subsequently combined with a 10-μL aliquot of one of the three PCR master mixes and amplification was performed on a TGradient thermocycler (Biometra). Three pairs of 25-mer primers were designed to amplify the 500-bp regions of 3′ portion of the three gene transcripts, respectively. Thermal profile settings were 94°C 1 min, and then 94°C 15 s, 59°C 20 s, 72°C 20 s, for 40 cycles. For comparison, conventional PCR products of three genes were separated and stained in the same agarose gel. Quantitative real-time PCR using SYBR Green I Dye (Bio-V) was performed in ABgene microtubes with ultra clear cap (ABgene) on Mx3000P (Stratagene). To minimize the formation of nonspecific amplification products, total primer concentrations for LlANK, actin, and tet were empirically optimized to 800 nm, 400 nm, and 400 nm, respectively. Amplification efficiencies of three amplicons were confirmed similar using 2−ΔΔCt method (Livak and Schmittgen, 2001). Quantitative real-time PCR used a two-segment thermal profile: The amplification segment was the same as that used in conventional PCR amplification mentioned above, while the following dissociation segment was ramping up from 55°C to 95°C at a rate of 0.2°C s−1. Fluorescence data were collected during elongation steps within the cycles as well as the final dissociation period. When the real-time PCR program was completed, data were exported to Excel software. In analyzing the relative LlANK expression data using 2−ΔΔCt method (Livak and Schmittgen, 2001), two specific considerations were taken within the practical and biological context. First, the cDNA prepared from one single pollen was sufficient to set up three PCR reactions but not enough for further replication within each amplification. Alternatively, a number of single-pollen evaluations were performed in parallel and the data compiled for analysis. Second, actin expression was also analyzed and used as a quality control. Though disadvantageous to external applied tet in acting as the normalizing standard, internal actin expression was presumed to be relatively stable. Simultaneous increase of both actin and LlANK over 2-fold was regarded as an indication of accidentally isolating more than one single pollen during micromanipulation and, in this case, the corresponding data were discarded from analysis.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DN985072 to DN985171 (sequenced 100 cDNA clones after microarray), AY950617 (LlANK cDNA), and M15239 (tobacco etch virus translational enhancement element).
Supplementary Material
Acknowledgments
We thank Dr. Alice Y. Cheung (Department of Biochemistry and Molecular Biology, University of Massachusetts, Amherst, MA) for her precious advice in experimental design and critical reviewing of the manuscript. The Zmc13 promoter was originally a gift from Prof. Joseph Mascarenhas (Department of Biological Sciences, State University of New York, Albany, NY). We thank Prof. Xingwang Deng (Department of Molecular, Cellular, and Developmental Biology, Yale University, New Haven, CT) and Renfeng Li (Department of Biological Sciences and Biotechnologies, Tsinghua University, Beijng, China) for kindly providing the recombinant E2 AtUBC8 plasmid and recombinant human E1 enzyme protein, respectively. We also thank Prof. Naoki Sakurai (Faculty of Integrated Arts and Sciences, Hiroshima University, Hiroshima, Japan) for help in optimizing particle bombardment.
This work was supported by the National Natural Science Foundation of China (grant no. 30370134).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yiqin Li (liyq@mail.tsinghua.edu.cn).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074922.
References
- Becerra C, Jahrmann T, Puigdomènech P, Vicient CM (2004) Ankyrin repeat-containing proteins in Arabidopsis: characterization of a novel and abundant group of genes coding ankyrin-transmembrane proteins. Gene 340: 111–121 [DOI] [PubMed] [Google Scholar]
- Becker JD, Boavida LC, Carneiro J, Haury M, Feijó JA (2003) Transcriptional profiling of Arabidopsis tissues reveals the unique characteristics of pollen transcriptome. Plant Physiol 133: 713–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blohm DH, Guiseppi-Elie A (2001) New development in microarray technology. Curr Opin Biotechnol 12: 41–47 [DOI] [PubMed] [Google Scholar]
- Bork P (1993) Hundreds of ankyrin-like repeats in functionally diverse proteins: mobile modules that cross phyla horizontally? Proteins 17: 363–374 [DOI] [PubMed] [Google Scholar]
- Bosch M, Cheung AY, Hepler P (2005) Pectin methylesterase, a regulator of pollen tube growth. Plant Physiol 138: 1334–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burch-Smith TM, Miller JL, Dinesh-Kumar SP (2003) PTGS approaches to large-scale functional genomics in plants. In GJ Hannon, ed, RNAi: A Guide to Gene Silencing. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, pp 243–264
- Cai G, Romagnoli S, Moscatelli A, Ovidi E, Gambellini G, Tiezzi A, Cresti M (2000) Identification and characterization of a novel microtubule-based motor associated with membranous organelles in tobacco pollen tubes. Plant Cell 12: 1719–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kvam C, Kitamura A, Ohsumi T, Okazaki Y, Itoh M, Kamiya M, Shibata K, Sasaki N, Izawa M, et al (1996) High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37: 327–336 [DOI] [PubMed] [Google Scholar]
- Carninci P, Shibata Y, Hayatsu N, Sugahara Y, Shibata K, Itoh M, Konno H, Okazaki Y, Muramatsu M, Hayashizaki Y (2000) Normalization and substraction of cap-trapper-selected cDNAs to prepare full-length cDNA libraries for rapid discovery of new genes. Genome Res 10: 1617–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Cheung AY, Wu H (2003) Actin-depolymerizing factor mediates Rac/Rop GTPase-regulated pollen tube growth. Plant Cell 15: 237–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Wong EI, Vidali L, Estavillo A, Hepler PK, Wu H, Cheung AY (2002) The regulation of actin organization by actin-depolymerizing factor in elongating pollen tubes. Plant Cell 14: 2175–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Dougherty ER, Bittner ML (1997) Ratio-based decisions and the quantitative analysis of cDNA microarray images. J Biomed Opt 2: 364–374 [DOI] [PubMed] [Google Scholar]
- Cheung AY, Chen C, Glaven RH, de Graaf BH, Vidali L, Hepler PK, Wu H (2002) Rab2 GTPase regulates vesicle trafficking between endoplasmic reticulum and the Golgi bodies and is important to pollen tube growth. Plant Cell 14: 945–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AY, Wu H (2004) Overexpression of an Arabidopsis formin stimulates supernumerary actin cable formation from pollen tube cell membrane. Plant Cell 16: 257–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derkson J, van Amstel ANM, Rutten ALM, Knuiman B, Li YQ, Pierson ES (1999) Pollen tubes: cellular organization and control of growth. In C Clément, E Pacini, J-C Audran, eds, Anther and Pollen: From Biology to Biotechnology. Springer, Berlin, pp 161–174
- Dykxhoorn DM, Novina CD, Sharp PA (2003) Killing the messenger: short RNAs that silence gene expression. Nat Rev Mol Cell Biol 4: 457–467 [DOI] [PubMed] [Google Scholar]
- Fang S, Jensen JP, Ludwig RL, Vousden KH, Weissman AM (2000) Mdm2 is a RING finger-dependent ubiquitin protein ligase for itself and p53. J Biol Chem 275: 8945–8951 [DOI] [PubMed] [Google Scholar]
- Feijó JA, Costa SS, Prado AM, Becker JD, Certal AC (2004) Signalling by tips. Curr Opin Plant Biol 7: 589–598 [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE (1999) Signaling in pollination. Curr Opin Plant Biol 2: 490–495 [DOI] [PubMed] [Google Scholar]
- Franklin-Tong VE, Drøbak BK, Allan AC, Trewavas AJ (1996) Growth of pollen tubes of Papaver rhoeas is regulated by a slow moving calcium wave propagated by inositol 1,4,5-trisphosphate. Plant Cell 8: 1305–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Wu G, Yang Z (2001) Rop GTPase-dependent dynamic of tip-localized F-actin controls tip growth in pollen tubes. J Cell Biol 152: 1019–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y, Wang Z, Yang Z (2004) ROP/RAC GTPase: an old new master regulator for plant signaling. Curr Opin Plant Biol 7: 527–536 [DOI] [PubMed] [Google Scholar]
- Gupta R, Ting JT, Sokolov LN, Johnson SA, Luan S (2002) A tumor suppressor homolog, AtPTEN1, is essential for pollen development in Arabidopsis. Plant Cell 14: 2495–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton DA, Bashe DM, Stinson JR, Mascarenhas JP (1989) Characterization of a pollen-specific genomic clone from maize. Sex Plant Reprod 2: 208–212 [Google Scholar]
- Hanson DD, Hamilton DA, Travis JL, Bashe DM, Mascarenhas JP (1989) Characterization of pollen-specific cDNA clone from Zea mays and its expression. Plant Cell 1: 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Okamoto H, Stoop-Myer C, Deng XW (2002) Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J 30: 385–394 [DOI] [PubMed] [Google Scholar]
- Hedge P, Qi R, Abernathy K, Gay C, Dharap S, Gaspard R, Hughes JE, Snesrud E, Lee N, Quackenbush J (2000) A concise guide to cDNA microarray analysis. Biotechniques 29: 548–562 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Waterhouse PM (2003) Constructs and methods for high-throughput gene silencing in plants. Methods 30: 289–295 [DOI] [PubMed] [Google Scholar]
- Hepler PK, Vidali L, Cheung AY (2001) Polarized cell growth in higher plants. Annu Rev Cell Dev Biol 17: 159–187 [DOI] [PubMed] [Google Scholar]
- Hicke L (2001) Protein regulation by monoubiquitin. Nat Rev Mol Cell Biol 2: 195–201 [DOI] [PubMed] [Google Scholar]
- Hicks GR, Rojo E, Hong S, Carter DG, Raikhel NV (2004) Germinating pollen has tubular vacuoles, displays highly dynamic vacuole biogenesis, and requires VACUOLESS1 for proper function. Plant Physiol 134: 1127–1139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honys D, Twell D (2003) Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol 132: 640–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YQ, Mareck A, Faleri C, Moscatelli A, Liu Q, Cresti M (2002) Detection and localization of pectin methylesterase isoforms in pollen tubes of Nicotiana tabacum L. Planta 214: 734–740 [DOI] [PubMed] [Google Scholar]
- Li YQ, Zhang HQ, Pierson ES, Huang FY, Linskens HF, Hepler PK, Cresti M (1996) Enforced growth-rate fluctuation causes pectin ring formation in the cell wall of Lilium longiflorum pollen tubes. Planta 200: 41–49 [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature 405: 827–836 [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM (1999) RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc Natl Acad Sci USA 96: 11364–11369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascarenhas JP (1993) Molecular mechanisms of pollen tube growth and differentiation. Plant Cell 5: 1303–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick S (2004) Control of male gametephyte development. Plant Cell 16: S142–S153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi H, Usami T, Tanaka I (1995) High level of GUS gene expression driven by pollen-specific promoters in electroporated lily pollen protoplasts. Sex Plant Reprod 8: 205–209 [Google Scholar]
- Morikawa H, Nishihara M, Seki M, Irifune K (1994) Bombardment-mediated transformation of plant cells. J Plant Res 107: 117–123 [Google Scholar]
- Nishihara M, Ito M, Tanaka I, Kyo M, Ono K, Irifune K, Morikawa H (1993) Expression of the β-glucuronidase gene in pollen of lily (Lilium longiflorum), tobacco (Nicotiana tabacum), Nicotiana rustica, and peony (Paeonia lactiflora) by particle bombardment. Plant Physiol 102: 357–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodzon LA, Xu W-H, Wang Y, Pi L-Y, Chakrabarty PK, Song W-Y (2004) The ubiquitin ligase XBAT32 regulates lateral root development in Arabidopsis. Plant J 40: 996–1006 [DOI] [PubMed] [Google Scholar]
- Palanivelu R, Brass L, Edlund AF, Rreuss D (2003) Pollen tube growth and guidance is regulated by POP2, an Arabidopsis gene that controls GABA levels. Cell 114: 47–59 [DOI] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Prado AM, Porterfield DM, Feijó JA (2004) Nitric oxide is involved in growth regulation and re-orientation of pollen tubes. Development 131: 2707–2714 [DOI] [PubMed] [Google Scholar]
- Reichel C, Mathur J, Eckes P, Langenkemper K, Koncz C, Schell J, Reiss B, Maas C (1996) Enhanced green fluorescence by the expression of an Aequorea victoria green fluorescent protein mutant in mono- and dicotyledonous plant cells. Proc Natl Acad Sci USA 93: 5888–5893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW (2001) Molecular Cloning: A Laboratory Manual, Ed 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW (1995) Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270: 467–470 [DOI] [PubMed] [Google Scholar]
- Scoccianti V, Ovidi E, Taddei AR, Tiezzi A, Crinelli R, Gentilini L, Speranza A (2003) Involvement of the ubiquitin/proteasome pathway in the organisation and polarised growth of kiwifruit pollen tubes. Sex Plant Reprod 16: 123–133 [Google Scholar]
- Simpson RJ (2002) Proteins and Proteomics: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
- Smith NA, Singh SP, Wang M-B, Stoutjesdijk PA, Green AG, Waterhouse PM (2000) Total silencing by intron-spliced hairpin RNAs. Nature 407: 319–320 [DOI] [PubMed] [Google Scholar]
- Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, Gardner J, Wang B, Zhai WX, Zhu LH, et al (1995) A receptor kinase-kike protein encoded by the rice disease resistance gene Xa21. Science 270: 661–667 [DOI] [PubMed] [Google Scholar]
- Speranza A, Scoccianti V, Crinelli R, Calzoni GL, Magnani M (2001) Inhibition of proteasome activity strongly affects kiwifruit pollen germination: involvement of the ubiquitin/proteasome pathway as a major regulator. Plant Physiol 126: 1150–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone SL, Hauksdóttir H, Troy A, Herschleb J, Kraft E, Callis J (2005) Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol 137: 13–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain SM, Muller AJ, Singh DP (2004) The gar2 and rga alleles increase the growth of gibberellin-deficient pollen tubes in Arabidopsis. Plant Physiol 134: 694–705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor LP, Hepler PK (1997) Pollen germination and tube growth. Annu Rev Plant Physiol Plant Mol Biol 48: 461–491 [DOI] [PubMed] [Google Scholar]
- Vidali L, McKenna ST, Hepler PK (2001) Actin polymerization is essential for pollen tube growth. Mol Biol Cell 12: 2534–2545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Xia Q, Xie W, Datla R, Selvaraj G (2003) The classical Ubisch bodies carry a sporophytically produced structural protein (RAFTIN) that is essential for pollen development. Proc Natl Acad Sci USA 100: 14487–14492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M-L, Hsu C-M, Chang L-C, Wang C-S, Su T-H, Huang Y-JJ, Jiang L, Jauh G-Y (2004) Gene expression profiles of cold-stored and fresh pollen to investigate pollen germination and growth. Plant Cell Physiol 45: 1519–1528 [DOI] [PubMed] [Google Scholar]
- Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nat Rev Genet 4: 29–38 [DOI] [PubMed] [Google Scholar]
- Weissman AM (2001) Themes and variations on ubiquitination. Nat Rev Mol Cell Biol 2: 169–178 [DOI] [PubMed] [Google Scholar]
- Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al (2001) Construct designs for efficient, effective and high-throughput gene silencing in plants. Plant J 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Yang Z (1998) Signaling tip growth in plants. Curr Opin Plant Biol 1: 525–530 [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z (2000) The Rop GTPase switch turns on polar growth in pollen. Trends Plant Sci 5: 298–303 [DOI] [PubMed] [Google Scholar]
- Zhou W, Takeda H, Liu XZ, Nakagawa N, Sakurai N, Huang J, Li YQ (2004) A novel endo-1,4-beta-glucanase gene (LlpCel1) is exclusively expressed in pollen and pollen tubes of Lilium longiflorum. Acta Bot Sin 46: 142–147 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.