Abstract
To date, the function of most genes in the Arabidopsis (Arabidopsis thaliana) genome is unknown. Here we present the first analysis of the novel, plant-specific BRX (BREVIS RADIX) gene family. BRX has been identified as a modulator of root growth through a naturally occurring loss-of-function allele. The biochemical function of BRX is enigmatic, however several domains in BRX are conserved in the proteins encoded by the related BRX-like (BRXL) genes. The similarity between Arabidopsis BRXL proteins within these domains ranges from 84% to 93%. Nevertheless, analysis of brx brx-like multiple mutants indicates that functional redundancy of BRXLs is limited. This results mainly from differences in protein activity, as demonstrated by assaying the propensity of constitutively expressed BRXL cDNAs to rescue the brx phenotype. Among the genes tested, only BRXL1 can replace BRX in this assay. Nevertheless, BRXL1 does not act redundantly with BRX in vivo, presumably because it is expressed at a much lower level than BRX. BRX and BRXL1 similarity is most pronounced in a characteristic tandem repeat domain, which we named BRX domain. One copy of this domain is also present in the PRAF (PH, RCC1, and FYVE)-like family proteins. The BRX domain mediates homotypic and heterotypic interactions within and between the BRX and PRAF protein families in yeast (Saccharomyces cerevisiae), and therefore likely represents a novel protein-protein interaction domain. The importance of this domain for BRX activity in planta is underscored by our finding that expression of the C-terminal fragment of BRX, comprising the two BRX domains, is largely sufficient to rescue the brx phenotype.
Genetic redundancy is a common phenomenon in both plants and animals (Pickett and Meeks-Wagner, 1995). Although occasionally observed between nonhomologous genes (e.g. Weigel et al., 1992; Shannon and Meeks-Wagner, 1993), it generally occurs between members of the same gene family. In Arabidopsis (Arabidopsis thaliana), chromosomal segments have been duplicated several times during evolution (Vision et al., 2000; Simillion et al., 2002; Blanc et al., 2003), contributing to the creation and extension of gene families. Only a fraction of those gene families have been experimentally assigned functions (Somerville and Dangl, 2000), mainly by analysis of single loss-of-function mutants. However, this approach is limited, since the majority of gene knockouts do not result in phenotypes (Bouche and Bouchez, 2001), presumably due to genetic redundancy. Thus, it has been suggested that more research be devoted to study the evolutionary and functional divergence of gene families (Somerville and Dangl, 2000; Bouche and Bouchez, 2001; Hirschi, 2003). This is particularly important for characterizing the approximately 45% of Arabidopsis genes that can neither be assigned biological nor biochemical function, because of a lack of homology to functionally defined genes or protein domains (Somerville and Dangl, 2000). With this study we contribute to the exploration of plant gene families by presenting the first comprehensive analysis of the novel, plant-specific BREVIS RADIX (BRX) gene family.
We previously identified a regulator of root growth, the BRX gene (Mouchel et al., 2004), through a naturally occurring allele in the accession Uk-1. Initially, BRX was annotated incorrectly, because of its low expression and particular intron-exon structure. The same was true for BRX homologs, with exception of one gene, for which a full-length cDNA clone was available. Based on this clone and genomic homology searches, the correct intron-exon structure of BRX was determined and three more BRX homologs were identified. Together, the five genes, BRX and BRX-like (BRXL) 1 to 4, constitute the BRX gene family. The respective proteins are highly similar and contain four highly conserved domains of unknown function.
In this study, we present a broader characterization of the BRX gene family in plants. We determine the functional overlap between Arabidopsis BRX family genes, and we assign a putative function to the conserved repeat domain, which is a prominent feature of BRX family proteins.
RESULTS
BRXL Genes Form a Highly Conserved, Plant-Specific Gene Family
We previously isolated BRX (Mouchel et al., 2004), the founding member of the novel family of plant-specific BRXL genes. Arabidopsis BRXL genes are transcribed at low levels (Meyers et al., 2004); however, expressed sequence tags (ESTs) have by now been found for all five family members, with full-length clones confirming our annotations of four genes. The open reading frames of all BRX family genes are composed of five exons, whose lengths are conserved between genes (Fig. 1A). The encoded proteins are 35 to 40 kD in size and show extensive conservation of domain structure and primary sequence. The overall similarity between Arabidopsis BRXL proteins is at least 50% and can be as high as 81%. Thus, BRXL proteins are remarkably well conserved.
Figure 1.
Structure of BRX family genes and proteins in Arabidopsis, poplar, and rice. A, Intron-exon structure of confirmed and predicted BRXL genes in Arabidopsis (AtBRXL), poplar (PtBRXL), and rice (OsBRXL), drawn to scale. Numbers indicate the size of features in nucleotides. If available, 5′ and 3′ untranslated regions are included. Gray boxes indicate exon, untranslated; black boxes indicate exon, coding; white boxes indicate intron. Patterned boxes in OsBRXL3 indicate predicted coding exons, which are, however, missing in the only available cDNA clone for this gene. B, Amino acid alignment of conserved domains in the N terminus of BRXL proteins. Numbers indicate amino acid position for the first and last residue shown. C, As in B, showing alignment of the first BRX repeat domain. D, As in B, showing alignment of the second BRX repeat domain.
BRXL genes are found in all higher plants for which data are available, but not in unicellular organisms and animals. Based on EST and genomic searches, we defined the full set of BRXL genes in the entirely sequenced plant genomes of poplar (Populus trichocarpa) and rice (Oryza sativa; Fig. 1A). In both species, as in Arabidopsis, five BRXL genes can be found, which we named PtBRXL1 to 5 and OsBRXL1 to 5 in poplar and rice, respectively, ordered according to their similarity to Arabidopsis BRX (AtBRX). ESTs exist for two poplar and four rice genes (Table I). The intron-exon structure of PtBRXL and OsBRXL genes is mostly similar to AtBRXL genes (Fig. 1A). An exception is OsBRXL5, which appears to be the result of transposition of a BRXL cDNA and has acquired numerous nonsynonymous mutations in absolutely conserved amino acid residues (see below). Except OsBRXL5, all rice and poplar BRXL proteins are well conserved. Compared to AtBRX, overall amino acid similarity ranges from 55% to 76% in poplar and 39% to 56% in rice. In summary, BRXL genes are highly conserved both in dicotyledons and monocotyledons, indicating their possible orthologous origin.
Table I.
BRXL genes from different plants
Table of BRXL genes identified in Arabidopsis (At), poplar (Pt), and rice (Os), with respective gene identification (ID) numbers and existence of ESTs indicated.
| Gene Name | Gene ID | ESTs Exist? |
|---|---|---|
| AtBRX | At1g31880 | Yes |
| AtBRXL1 | At2g35600 | Yes |
| AtBRXL2 | At3g14000 | Yes |
| AtBRXL3 | Fuse At1g54180 + At1g54190 | Yes |
| AtBRXL4 | At5g20540 | Yes |
| PtBRXL1 | C-scaff-290001 | Yes |
| PtBRXL2 | LG_III000279 | No |
| PtBRXL3 | LGIII1601 | Yes |
| PtBRXL4 | LGVI000394 | No |
| PtBRXL5 | LGXVIII000005 | No |
| OsBRXL1 | Os08g36020 | Yes |
| OsBRXL2 | Os02g47230 | Yes |
| OsBRXL3 | Os04g51170 | Yes |
| OsBRXL4 | Os03g63650 | Yes |
| OsBRXL5 | Os12g09080 | No |
Domain Structure of BRXL Proteins
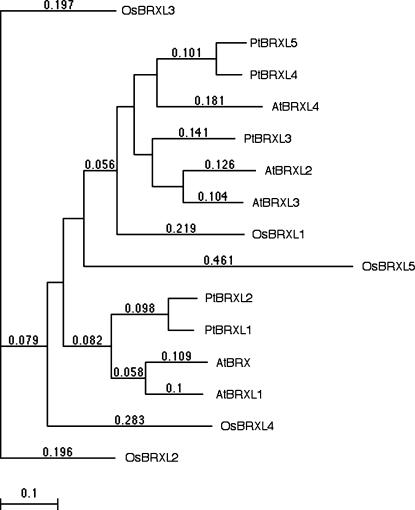
Four regions of high conservation can be distinguished in BRX family proteins. So far however, no function has been described for these domains. The homology among BRX family proteins within and between species is especially conserved in these regions, with amino acid similarity to AtBRX ranging from 84% to 93% in Arabidopsis, 86% to 96% in poplar, and 84% to 87% in rice (OsBRXL5 excluded). At the N terminus, short stretches of approximately 10 and 25 amino acids are conserved (Fig. 1B). In most BRX family proteins, they contain conserved Cyss, whose spacing is indicative of a potential zinc-binding motif. The middle region of BRX family proteins contains a highly conserved domain of approximately 55 amino acids, with 33 invariant positions (Fig. 1C). Following a variable spacer of approximately 100 to 150 amino acids, another highly conserved domain of approximately 55 amino acids is present, with 27 invariant positions (Fig. 1D). This domain is homologous to the middle domain (56% amino acid similarity). Therefore, the two domains constitute a novel type of tandem repeat, which is the main characteristic of BRX family proteins. Thus, we named the repeat domain the BRX domain. Amino acid similarity between any two BRX domains is at least 84% for the N-terminal domains (Fig. 1C), and 81% for the C-terminal ones (Fig. 1D). This indicates that selection pressure maintains the structural integrity of BRX domains, since they are nearly invariant in proteins from distantly related species that can be clearly separated in a phylogenetic tree (Fig. 2).
Figure 2.
Phylogenetic tree of BRXL proteins. Unrooted phylogenetic tree diagram of BRXL proteins from Arabidopsis, poplar, and rice (see Fig. 1), based on full-length protein sequences.
Analysis of Multiple Mutants for Arabidopsis BRXL Genes
The high level of conservation within the BRX gene family could mean that these genes act redundantly, as frequently observed in other gene families (e.g. Kempin et al., 1995; Holm et al., 2002; Hardtke et al., 2004). Redundancy in a developmental context often requires overlapping domains of gene activity. Thus, we investigated whether BRXL genes are expressed in the root by quantitative real-time PCR (qPCR). In these assays, expression of all BRXLs was detected in seedling roots (see below), suggesting that they might indeed be partially redundant with BRX in root growth. To clarify this issue, we analyzed different combinations of brxl mutants.
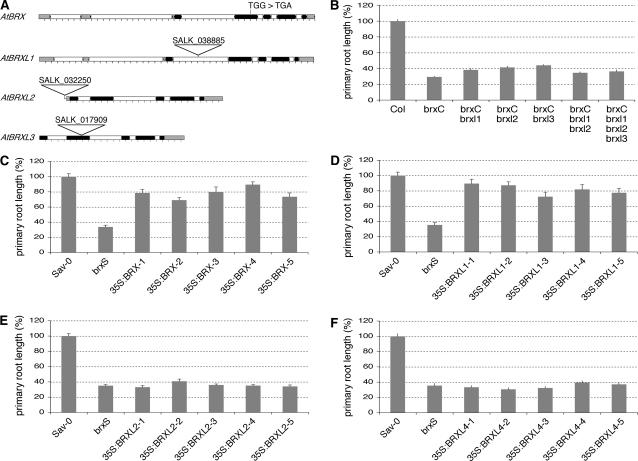
Many putative mutants are available for BRXL genes, however, the same T-DNA insertions are often annotated for in several genes because of their high sequence similarity. We analyzed the available SALK lines (Alonso et al., 2003) by PCR and Southern-blot analysis and confirmed T-DNA insertions in BRXL1, 2, and 3 (Fig. 3A; Mouchel et al., 2004). No insertions were confirmed for BRXL4. SALK line 038885 carries a T-DNA insert in an intron of BRXL1. The T-DNA in line 032250 is located a few nucleotides 5′ to the transcription start of BRXL2. Finally, line 017909 carries a T-DNA in an exon of BRXL3. Wild-type transcript was not detectable for any of the mutant loci (data not shown), suggesting that the mutants represent partial or total loss-of-function alleles. brxl1, brxl2, and brxl3 did not show any conspicuous root system defect, suggesting that, unlike BRX, these genes have no role in primary root growth. However, potential synergistic action of BRX and BRXL genes in the root might only become evident in a brx background. Therefore, we generated multiple mutants using the T-DNA mutants and the original brx allele from Uk-1, introgressed into ecotype Columbia of Arabidopsis (Col) background (brxC; Mouchel et al., 2004).
Figure 3.
Redundancy of BRXL genes of Arabidopsis. A, Schematic representation of Arabidopsis mutants for different BRXL genes. The stock numbers of respective SALK T-DNA insertion lines are indicated. B to F, Relative primary root length of 9-d-old seedlings grown in tissue culture with respect to wild-type control. B, Different single and multiple brxl mutant lines. C to F, Values for independent transgenic lines each expressing a 35S:BRX (C), 35S:BRXL1 (D), 35S:BRXL2 (E), or 35S:BRXL4 (F) transgene in brxS background. Error bars indicate se of the mean.
As reported previously (Mouchel et al., 2004), double mutants between brxC and brxl1, 2, or 3 did not show an enhanced brxC root phenotype. The same is true for brxC brxl1 brxl2 triple mutants and brxC brxl1 brxl2 brxl3 quadruple mutants (Fig. 3B). Rather, all multiple mutant lines had slightly longer roots than brxC, likely because of insufficient introgression of the brx mutation into Col. Additional root system quantitative trait loci from the Uk-1 background retained in brxC (Mouchel et al., 2004) should have been largely lost in subsequent crosses for the creation of double and multiple mutants, leading to slightly longer roots. Irrespective of background influence, the data suggest that brxl1, brxl2, and brxl3 do not enhance the brxC root phenotype.
Finally, because all BRX family genes are also expressed in all the shoot organs (data not shown), we also inspected the shoot morphology of the different mutants throughout their life cycle. However, we did not detect any apparent phenotypes.
BRXL1 Has BRX Activity
A lack of redundancy between BRX and BRXL genes could be due to nonoverlapping expression patterns or differences in protein activity. To determine whether differential expression patterns could play a role, we expressed BRX and BRXL1, BRXL2, and BRXL4 under control of the 35S promoter, which confers strong expression in root tissues (Benfey et al., 1990a, 1990b), in brxS plants and analyzed their root growth. 35S:BRX rescued the root length to approximately 78% of wild type (Sav-0; Fig. 3C), consistent with the quantitative trait locus nature of the BRX locus in the brxS background (Mouchel et al., 2004). Strikingly, rescue of the same magnitude was obtained with 35S:BRXL1 (Fig. 3D). By contrast, overexpression of BRXL2 (Fig. 3E) or BRXL4 (Fig. 3F) was not able to rescue brxS root length. Thus, BRXL1, but not BRXL2 or BRXL4, can replace BRX when expressed constitutively. Therefore, despite their high degree of similarity, BRX family proteins have functionally diversified.
The Original brx Allele Is a Loss of Function
One explanation for the lack of redundancy between BRX family genes in root development could be a dosage-dependent dominant negative effect of the brx allele in a homozygous state, similar for instance to the recessive interfering alleles of SLEEPY (Strader et al., 2004). Alternatively, the brx allele might simply be hypomorphic or null. If brx is null, this would facilitate discovery of possible redundancy. However, if it is hypomorphic, the residual activity could have masked the redundancy. Therefore, it is important to clarify the nature of the brx allele.
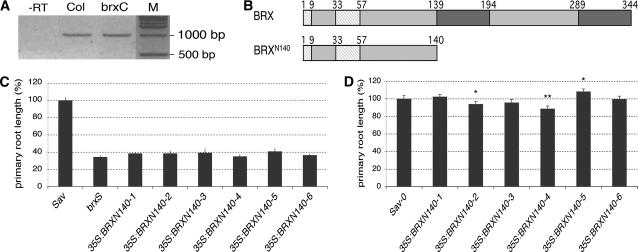
The brx allele carries a point mutation that creates an early stop codon at amino acid 141 (Mouchel et al., 2004). Full-length BRX mRNA can be detected in homozygous brxC seedlings (Fig. 4A), indicating that truncated BRX comprising the N-terminal 140 amino acids could be produced from the mutant allele (Fig. 4B). To test whether this BRXN140 fragment has residual activity in planta, we raised its quantity in brxS plants by expressing a respective cDNA fragment under control of the 35S promoter. However, this did not significantly rescue root growth (Fig. 4C). Next, we tested whether BRXN140 could act in a dominant negative fashion by introducing the construct into the wild-type control Sav-0. No consistent, and quantitatively only small effects were observed (Fig. 4D), arguing against a dominant negative effect of the BRXN140 fragment. In summary, our results suggest that the brx allele is a loss-of-function and most likely null allele.
Figure 4.
The original brx allele is a loss-of-function allele. A, RT-PCR of BRX mRNA, amplified from total RNA isolated from wild-type (Col) or brxC mutant seedlings. −RT, Control reaction without reverse transcriptase. B, Schematic presentation of BRX full-length and potential truncated mutant protein. Light-gray boxes indicate the position of conserved N-terminal domains in BRX (see Fig. 1B) and dark-gray boxes indicate the position of the conserved BRX domains (see Fig. 1, C and D). Respective amino acid positions are indicated. C to D, Relative primary root length of 9-d-old seedlings grown in tissue culture with respect to wild-type control (Sav-0). C, Values for independent transgenic lines expressing the 35S:BRXN140 transgene in brxS background. D, Values for independent transgenic lines expressing the 35S:BRXN140 transgene in Sav-0 background. Error bars indicate se of the mean. Asterisks indicate P values of Student's t test, with one asterisk signifying P < 0.05 and two asterisks signifying P < 0.01.
Expression Levels of BRXL Genes in the Root
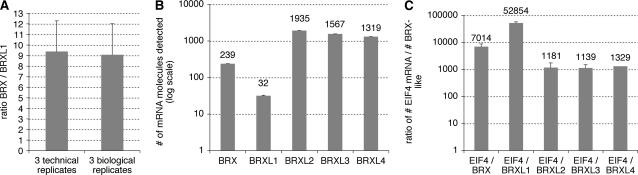
Lack of redundancy between BRX and BRXL1 in planta despite functional equivalence of the proteins could be due to nonoverlapping expression patterns or differences in transcription level of the two genes. In fact, both genes are expressed at very low levels (Meyers et al., 2004; Mouchel et al., 2004). Moreover, qPCR experiments show that BRX is expressed at a 9 to 10 times higher level than BRXL1 in roots (Fig. 5A). Thus, BRXL1 activity might be too low to compensate a loss of BRX activity in vivo. Notably, the qPCR experiments confirmed the very low expression levels of the two genes (Fig. 5B), even compared to the other BRXLs, which can be detected in the same samples at much higher levels (5- to 8-fold as compared to BRX). However, in general all BRX family genes are poorly expressed, as demonstrated by their relative expression as compared to a housekeeping gene, eIF4 (Fig. 5C).
Figure 5.
Quantification of expression levels of BRXL genes in the root. Quantification of BRXL expression levels by qPCR using total RNA isolated from roots of 9-d-old seedlings. A, Relative expression of BRX compared to BRXL1 as determined in identical samples. B, Absolute number of mRNA molecules detected in 1 μg of total RNA. Average of three samples assayed for all five genes. C, Relative expression of BRXL genes as compared to housekeeping gene EIF4, in the samples analyzed in B. Error bars indicate se of the mean.
The BRX Domain Is a Novel Protein-Protein Interaction Domain
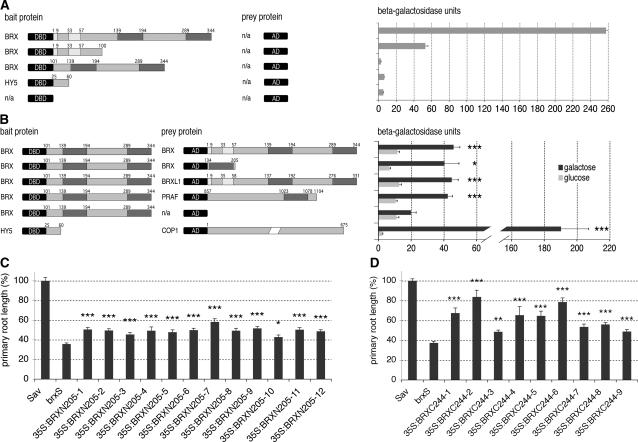
Since our data indicate diverse functions of BRX family proteins, we sought to characterize them in more detail to define functionally relevant domains. Previously, secondary structure predictions identified α-helical regions, which are characteristic of DNA binding and protein-protein interaction domains, within the conserved domains (Mouchel et al., 2004). Interestingly, many known protein-protein interaction domains mediate homodimerization (Fan et al., 1997; Ulmasov et al., 1999; Holm et al., 2002). We tested whether the BRX domain mediates homodimerization using a yeast (Saccharomyces cerevisiae) two-hybrid assay. Full-length BRX and C-terminally truncated versions display a high transactivation potential (Mouchel et al., 2004) and could thus not be used as baits. However, a bait containing the BRX C terminus, starting at amino acid 101, was stably expressed and did not result in significant background activation (Fig. 6A). We used this bait to test interactions with prey fusion proteins of BRX and BRXL1. Strikingly, highly significant interactions were observed for both BRX and BRXL1 (Fig. 6B), indicating that these proteins can homodimerize and heterodimerize. Further, a truncated prey protein containing only the first BRX domain of BRX interacted as well, suggesting that interaction is mediated by the BRX domain.
Figure 6.
The BRX domain is a protein-protein interaction domain and functionally significant in planta. A, Autoactivation of BRX-derived bait proteins fused to lexA DNA binding domain in yeast. Amino acids of BRX are indicated. Light-gray boxes indicate the position of conserved N-terminal domains in BRX (see Fig. 1B) and dark-gray boxes indicate the position of the conserved BRX domains (see Fig. 1, C and D). B, Yeast two-hybrid interaction assays of BRX-derived bait protein fused to lexA DNA binding domain and different prey proteins fused to an activation domain. Light-gray boxes indicate the position of conserved N-terminal domains in BRX and dark boxes indicate the position of the conserved BRX domains in BRX, BRXL1, or PRAF. lacZ reporter gene activity measured in liquid assays is indicated in Miller units. C and D, Relative primary root length of 9-d-old seedlings grown in tissue culture with respect to wild-type control (Sav-0). C, Values for independent transgenic lines expressing the 35S:BRXN205 transgene (see text) in brxS background. D, Values for independent transgenic lines expressing the 35S:BRXC244 transgene in brxS background. Error bars indicate se of the mean. Asterisks indicate P values of Student's t test, with one asterisk signifying P < 0.05, two asterisks signifying P < 0.01, and three asterisks signifying P < 0.001.
Interestingly, one copy of the BRX domain is also present in the PRAF (PH, RCC1, and FYVE) protein family of Arabidopsis, although this has not been noticed so far (van Leeuwen et al., 2004). Thus, we also tested interaction with a prey construct that contained the C terminus (amino acids 857–1104) of PRAF1 (At1g76950) (Jensen et al., 2001). This fragment includes the BRX domain but otherwise shows no homology to BRX. Again, highly significant interaction was observed (Fig. 6B), indicating that the BRX domain is necessary and sufficient for this interaction. Thus, our results suggest that the BRX domain is a novel protein-protein interaction domain, which likely mediates homodimerization and heterodimerization within and/or between the BRXL and PRAF-like protein families.
The BRX Domains Are Largely Sufficient for BRX Activity
The early stop codon in the brx loss-of-function allele is located at amino acid 141, immediately in front of the first BRX domain, suggesting that the BRX domains are absolutely required for BRX function. To test whether this is indeed the case, we expressed transgenes of BRX fragments under control of the 35S promoter in a brxS background, and assayed their ability to normalize root growth. As a control, the BRXN140 transgene was assayed in parallel and again had no significant effect on brxS root length. By contrast, expression of the BRX N terminus, including the first BRX domain (35S:BRXN205), partially rescued brxS root length, and this rescue was highly significant (P value < 0.001; Fig. 6C). On average, root length was restored to approximately 49% (±1.1%) of the wild-type control, as compared to a value of approximately 34% for the brxS background line. Strikingly, expression of the C-terminal two-thirds of BRX including both BRX domains (35S:BRXC244) was even more effective, restoring root length to approximately 63% (±4.2%) of the wild-type control. Notably, in a few lines root growth was restored to the level of full rescue (as observed with full-length BRX, approximately 78% [±3.4%] of control, see above). Thus, our data suggest a minor role for the N terminus in BRX activity and support the notion that the presence of two BRX domains is of critical importance for BRX function in planta.
DISCUSSION
Genetic Redundancy in Plant Development
Rearrangements of genetic material during species evolution can result in gene duplication, which, if occurring repeatedly in a phylogeny, leads to the creation of gene families. Genome analyses indicate that this has been the case in Arabidopsis (Vision et al., 2000; Simillion et al., 2002; Blanc et al., 2003). In fact, all BRX family genes, except BRXL4, are located on duplicated chromosome segments, suggesting that they are true paralogs. In principle, gene duplication creates functionally identical copies, which should be fully redundant. However, positive selection pressure on one of the copies can decrease, thus providing a template for the evolution of a gene with novel function. In fact, models of evolutionary selection argue against the maintenance of full genetic redundancy (Tautz, 1992; Weintraub, 1993). Indeed, partial redundancy is often observed among gene family members (e.g. Holm et al., 2002; Hardtke et al., 2004), indicating that duplicated genes can evolve new, mutually exclusive roles while preserving a shared set of functions. Interestingly, shared functions often become unequally distributed between duplicated genes, ultimately allowing one of them to predominantly perform new functions and eventually leading to diversification of protein activity (e.g. Hardtke et al., 2004). This might have happened in the BRX gene family.
Genetic Redundancy in Arabidopsis BRXL Genes
In the context of redundancy, it was important to clarify the nature of the so far only available brx allele (Mouchel et al., 2004). Our finding that this allele likely is a loss of function excludes the possibility of full genetic redundancy between BRX and BRXL genes. Further, we did not observe root phenotypes in brxl1, brxl2, and brxl3 mutants. Since even quadruple brx brxl1 brxl2 brxl3 mutants do not display an enhanced brx phenotype, these BRXL genes likely do not play a role in modulating root growth. This should also be true for BRXL4, since it cannot replace BRX when expressed constitutively (see below). Whether BRX family genes have a role in shoot development remains to be determined. Although the genes do not act redundantly in the root, this might be different in the shoot. However, no shoot phenotypes were observed in the quadruple mutant. Thus, a so far unavailable quintuple mutant of all five genes would be necessary to clarify this issue.
We also employed a gain-of-function approach to compare BRXL genes, by assaying their ability to rescue the brx phenotype. We show that BRXL1, but not BRXL2 or BRXL4, when expressed under control of the 35S promoter, can rescue as well as BRX itself. Thus, BRXL1 in principle possesses BRX activity, suggesting that a lack of redundancy between the genes results from insufficient BRXL1 activity in tissues where BRX is required. Therefore, BRX and BRXL1 must have diverged due to the evolution of differential expression, rather than differential protein activity. In light of our qPCR results, a likely explanation for the lack of redundancy between BRX and BRXL1 in planta is the 8-to-9-fold lower level of BRXL1 expression in roots. This level might not be able to saturate the threshold of required BRX activity. Alternatively, BRX and BRXL1 expression patterns might be mutually exclusive. To address this issue, the in situ expression patterns of BRX and BRXL1 will have to be determined, which is hampered by the very low expression level of these genes.
High Degree of Conservation of BRXL Proteins
Our results demonstrate that BRX and BRXL1 have equivalent activity, which is lost in BRXL2 and BRXL4. This functional diversification contrasts with the high degree of structural and amino acid similarity between BRXL proteins. Generally, one would expect novel protein functions to arise from mutations in functionally relevant domains. In the case of BRXL proteins, these are presumably the conserved domains found in BRX family proteins of all species investigated. Considerable positive selection acts on these domains, implying that they are essential for the function of BRXL proteins, and also that BRX family genes are important for plant development. Although the lack of apparent morphological phenotypes in investigated brxl mutants argues against the latter conclusion, one has to consider that plants have evolved mechanisms that are critical for survival in the wild, but not necessarily needed in a laboratory environment (e.g. Devlin et al., 1998). Thus, important roles for BRXL genes might be revealed in future assays that monitor physiological and environmental responses. Whether functional diversity among BRX family proteins is due to amino acid substitutions in the conserved domains remains to be seen. Compared to BRX and BRXL1, the conserved domains of BRXL2 and BRXL4 contain only eight nonsynonymous substitutions, out of 143 amino acids covered by the domains. Thus, either these amino acids are critical for the specialization of domain function, or differential activity of BRXL proteins is due to features in their more variable, nonconserved regions.
Interactions Mediated by the BRX Domain
Our finding that the BRX domain likely is a novel protein-protein interaction domain is a first hint toward the biochemical function of BRXL proteins. The strength of interaction between BRX domains is in the range of the interactions among indole-3-acetic acid and auxin response factor (ARF) proteins (Hardtke et al., 2004), and could reflect rather transient interaction. Also, because of the high sequence similarity of BRX domains, it appears likely that in yeast interaction occurs between any two BRX domains, again similar to interactions among indole-3-acetic acid and ARF proteins. Thus, whether BRX family proteins and PRAF family proteins interact in vivo remains to be determined. Nevertheless, the observation that the BRX domain also occurs in PRAF family proteins is of particular interest. PRAF family proteins contain other conserved domains, such as the pleckstrin homology (PH) and FYVE zinc-finger domains, which are assumed to bind phosphoinositides and target proteins to the plasma membrane. In vitro phosphoinositide binding of PRAF proteins has been demonstrated (Jensen et al., 2001), however, the in vivo relevance of this observation is not clear. In fact, most PH or FYVE domain-containing proteins are not targeted to membranes and PRAF family proteins are thought to be nuclear localized (Drobak and Heras, 2002; Hayakawa et al., 2004; Lemmon, 2004). PRAF-like proteins also contain regulator of chromosome condensation 1 (RCC1) repeats, which are implicated in multiple cellular processes (Renault et al., 1998). RCC1 repeats often provide guanine nucleotide exchange activity, which has also been demonstrated for PRAF1 (Jensen et al., 2001). Clearly, the notion that BRX domain-mediated interaction between BRXL proteins and PRAF family proteins could be relevant in planta is an attractive working hypothesis. Analysis of single and multiple mutants in PRAF-like genes, combined with verification of BRXL-PRAF interaction in vivo, will enable us to determine whether this hypothesis is valid and might help us to elucidate the biochemical and cell biological activity of BRXL proteins.
Relevance of the BRX Domains for BRX Function
The BRX domains are the most conspicuous feature of BRXL proteins and therefore it is not surprising that they play a role in BRX function. Their importance is already evident from the fact that the Uk-1 brx allele could at best direct synthesis of a truncated protein lacking the BRX domains. Our finding that adding one BRX domain to the respective BRXN140 fragment results in partial rescue emphasizes, however, that both BRX domains are absolutely required for BRX activity. Nevertheless, it is surprising that transgenic expression of the BRX C terminus, including both BRX domains rescues the brx phenotype in many lines just as well as full-length BRX. Therefore, the conserved N-terminal domains of BRX family proteins might only have a minor functional role. The importance of the BRX domains notwithstanding, the specific activity of BRX and BRXL1 as opposed to other BRXL proteins seems less likely to be localized in the BRX domains, since they are so highly conserved (see above). Rather, it might be the region between the BRX domains that differentiates those two proteins from the others. The entire C terminus is quite conserved between BRX and BRXL1, but has diverged in the other BRXL proteins. Clearly, extensive domain swapping and site-directed mutagenesis experiments will be required to determine the functionally important residues within the BRXL proteins, shedding more light on the structural requirements of this novel, highly conserved protein domain of higher plants.
MATERIALS AND METHODS
Bioinformatic Analyses
ESTs and genomic sequences of BRXL genes were identified using BLAST (www.ncbi.nlm.nih.gov/BLAST/) in the indices of The Institute for Genomic Research (www.tigr.org) and the poplar (Populus trichocarpa) genome database (genome.jgi-psf.org/Poptr1/Poptr1.home.html; versions as of February 22nd, 2005). BRXL genes of rice (Oryza sativa) and poplar were further annotated and analyzed using splice site prediction programs and tools included in MacVector (Accelrys, version 7.2.2).
Plant Material and Tissue Culture
Seedlings were grown at 22°C under constant illumination on culture medium (0.5× Murashige and Skoog salts, 0.5 g/L MES, 0.9% agar, 1% Suc, pH 5.8). Light intensity was approximately 90 μE.
Nucleic Acid Isolation, Reverse Transcription-PCR, and qPCR
Nucleic acids were prepared using RNeasy and DNeasy kits from Qiagen according to the manufacturer's instructions. Reverse transcription (RT)-PCR reactions were performed according to standard procedures with Superscript II reverse transcriptase (Invitrogen). qPCR analysis was performed with a Stratagene MPx3000 instrument according to the standard curve method (Rutledge and Cote, 2003) using SyberGreen mix (Stratagene) and the reference dye ROX. In all biological and technical replicates, two standard curves were done concurrently with unknown samples to minimize experimental and machine variance. Ct (concentration of DNA molecules in moles multiplied by time) values obtained for each amplicon tested were plotted onto relevant standard curves to infer initial amounts (Rutledge and Cote, 2003).
Creation and Analysis of Transgenic Plants
Plasmids were created by amplification of open reading frame fragments of BRXL genes from cDNA templates with Pfu polymerase (Fermentas), followed by cloning into binary vector pTCSH1 (Hardtke et al., 2000). For truncated fragments, stop codons were introduced through the respective oligonucleotides. All constructs were verified by sequencing.
Binary constructs were transformed into brxS plants via floral dip using Agrobacterium, and transgenic lines were selected by screening seed progeny for glufosinate ammonium resistance (15 mg/L) on medium containing 0.3% Suc. Because rescue of the brxS root phenotype by 35S:BRX is not dosage dependent (Mouchel et al., 2004) and because of frequent problems with transgene silencing in subsequent generations, the T2 generation was chosen for analysis. Independent transgenic lines segregating single-locus insertions were selected. A minimum of 20 seedlings per line was investigated. In rescue experiments, one quarter of the sample with root lengths in the range of the brxS control was typically discounted to eliminate nontransgenic seedlings from the analyses. Transgene (over) expression was verified by RT-PCR, using 3′ untranslated region-specific primers to differentiate transgenes from endogenous genes. To determine primary root length, seedlings were grown on vertical plates and scanned on a flat bed scanner to produce image files suitable for analysis in ImageJ software (version 1.3). Seedlings were analyzed 9 d after germination. To compare lines measured in separate experiments, control lines were grown for each experiment and relative root lengths were calculated with respect to wild type (Sav-0 or Col) as a 100% reference.
Yeast Two-Hybrid Interaction Assays
BRX-derived baits were produced in vector pEG202. Prey plasmids were constructed by cloning cDNA fragments of BRX, BRXL1, and PRAF1 into vector pJG4-5. HY5 and COP1 control constructs have been described (Hardtke et al., 2000). Plasmids were introduced into yeast (Saccharomyces cerevisiae) strain EGY48, together with reporter construct pSH18-34. Transformants were grown in liquid culture overnight on selective medium. Cultures were diluted in the morning and prey expression was induced in one subsample. Cultures were incubated for 6 more hours before lacZ activity was measured by standard liquid assay. A minimum of eight independent transformants was assayed for each construct combination. Stable expression of all bait proteins was confirmed by western-blot analysis.
Acknowledgments
G.C.B. contributed results for Figures 1, 3 (A, B, E, and F), 4, 5, and 6. C.F.M. contributed results for Figure 3 (C and D). C.S.H. gathered data for Figures 1 and 2. C.S.H. and G.C.B. conceived this study and wrote the manuscript. We thank the Arabidopsis Biological Resource Center for providing T-DNA insertion lines, the RIKEN Plant Division for providing full-length cDNA clones of Arabidopsis BRXL genes, Dr. K. Osmont for helpful comments on the manuscript, and Drs. K.F.X. Mayer and T. Hindemitt for advice on bioinformatics issues.
This work was supported by a National Sciences and Engineering Research Council of Canada Strategic Projects Grant and operating grant 3100AO–107631/1 from the Swiss National Science Foundation.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Christian Hardtke (christian.hardtke@unil.ch).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.075382.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH (1990. a) Combinatorial and synergistic properties of CaMV 35S enhancer subdomains. EMBO J 9: 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benfey PN, Ren L, Chua NH (1990. b) Tissue-specific expression from CaMV 35S enhancer subdomains in early stages of plant development. EMBO J 9: 1677–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Hokamp K, Wolfe KH (2003) A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res 13: 137–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche N, Bouchez D (2001) Arabidopsis gene knockout: phenotypes wanted. Curr Opin Plant Biol 4: 111–117 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Patel SR, Whitelam GC (1998) Phytochrome E influences internode elongation and flowering time in Arabidopsis. Plant Cell 10: 1479–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drobak BK, Heras B (2002) Nuclear phosphoinositides could bring FYVE alive. Trends Plant Sci 7: 132–138 [DOI] [PubMed] [Google Scholar]
- Fan HY, Hu Y, Tudor M, Ma H (1997) Specific interactions between the K domains of AG and AGLs, members of the MADS domain family of DNA binding proteins. Plant J 12: 999–1010 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Ckurshumova W, Vidaurre DP, Singh SA, Stamatiou G, Tiwari SB, Hagen G, Guilfoyle TJ, Berleth T (2004) Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 131: 1089–1100 [DOI] [PubMed] [Google Scholar]
- Hardtke CS, Gohda K, Osterlund MT, Oyama T, Okada K, Deng XW (2000) HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J 19: 4997–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayakawa A, Hayes SJ, Lawe DC, Sudharshan E, Tuft R, Fogarty K, Lambright D, Corvera S (2004) Structural basis for endosomal targeting by FYVE domains. J Biol Chem 279: 5958–5966 [DOI] [PubMed] [Google Scholar]
- Hirschi KD (2003) Insertional mutants: a foundation for assessing gene function. Trends Plant Sci 8: 205–207 [DOI] [PubMed] [Google Scholar]
- Holm M, Ma LG, Qu LJ, Deng XW (2002) Two interacting bZIP proteins are direct targets of COP1-mediated control of light-dependent gene expression in Arabidopsis. Genes Dev 16: 1247–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen RB, La Cour T, Albrethsen J, Nielsen M, Skriver K (2001) FYVE zinc-finger proteins in the plant model Arabidopsis thaliana: identification of PtdIns3P-binding residues by comparison of classic and variant FYVE domains. Biochem J 359: 165–173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kempin SA, Savidge B, Yanofsky MF (1995) Molecular basis of the cauliflower phenotype in Arabidopsis. Science 267: 522–525 [DOI] [PubMed] [Google Scholar]
- Lemmon MA (2004) Pleckstrin homology domains: not just for phosphoinositides. Biochem Soc Trans 32: 707–711 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Lee DK, Vu TH, Tej SS, Edberg SB, Matvienko M, Tindell LD (2004) Arabidopsis MPSS: an online resource for quantitative expression analysis. Plant Physiol 135: 801–813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS (2004) Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18: 700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett FB, Meeks-Wagner DR (1995) Seeing double: appreciating genetic redundancy. Plant Cell 7: 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault L, Nassar N, Vetter I, Becker J, Klebe C, Roth M, Wittinghofer A (1998) The 1.7 A crystal structure of the regulator of chromosome condensation (RCC1) reveals a seven-bladed propeller. Nature 392: 97–101 [DOI] [PubMed] [Google Scholar]
- Rutledge RG, Cote C (2003) Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res 31: e93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon S, Meeks-Wagner DR (1993) Genetic interactions that regulate inflorescence development in Arabidopsis. Plant Cell 5: 639–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y (2002) The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA 99: 13627–13632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somerville C, Dangl J (2000) Genomics: plant biology in 2010. Science 290: 2077–2078 [DOI] [PubMed] [Google Scholar]
- Strader LC, Ritchie S, Soule JD, McGinnis KM, Steber CM (2004) Recessive-interfering mutations in the gibberellin signaling gene SLEEPY1 are rescued by overexpression of its homologue, SNEEZY. Proc Natl Acad Sci USA 101: 12771–12776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D (1992) Redundancies, development and the flow of information. Bioessays 14: 263–266 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ (1999) Dimerization and DNA binding of auxin response factors. Plant J 19: 309–319 [DOI] [PubMed] [Google Scholar]
- van Leeuwen W, Okresz L, Bogre L, Munnik T (2004) Learning the lipid language of plant signalling. Trends Plant Sci 9: 378–384 [DOI] [PubMed] [Google Scholar]
- Vision TJ, Brown DG, Tanksley SD (2000) The origins of genomic duplications in Arabidopsis. Science 290: 2114–2117 [DOI] [PubMed] [Google Scholar]
- Weigel D, Alvarez J, Smyth DR, Yanofsky MF, Meyerowitz EM (1992) LEAFY controls floral meristem identity in Arabidopsis. Cell 69: 843–859 [DOI] [PubMed] [Google Scholar]
- Weintraub H (1993) The MyoD family and myogenesis: redundancy, networks, and thresholds. Cell 75: 1241–1244 [DOI] [PubMed] [Google Scholar]