Abstract
In response to exogenous rhythms of light and temperature, most organisms exhibit endogenous circadian rhythms (i.e. cycles of behavior and gene expression with a periodicity of approximately 24 h). One of the defining characteristics of the circadian clock is its ability to synchronize (entrain) to an environmental rhythm. Entrainment is arguably the most salient feature of the clock in evolutionary terms. Previous quantitative trait studies of circadian characteristics in Arabidopsis (Arabidopsis thaliana) considered leaf movement under constant (free-running) conditions. This study, however, addressed the important circadian parameter of phase, which reflects the entrained relationship between the clock and the external cycle. Here it is shown that, when exposed to the same photoperiod, Arabidopsis accessions differ dramatically in phase. Variation in the timing of circadian LUCIFERASE expression was used to map loci affecting the entrained phase of the clock in a recombinant population derived from two geographically distant accessions, Landsberg erecta and Cape Verde Islands. Four quantitative trait loci (QTL) were found with major effects on circadian phase. A QTL on chromosome 5 contained SIGNALING IN RED LIGHT REDUCED 1 and PSEUDORESPONSE REGULATOR 3, both genes known to affect the circadian clock. Previously unknown polymorphisms were found in both genes, making them candidates for the effect on phase. Fine mapping of two other QTL highlighted genomic regions not previously identified in any circadian screens, indicating their effects are likely due to genes not hitherto considered part of the circadian system.
Circadian phase is a measure of the temporal relationship between the circadian oscillator and the environmental cycle of night and day. It can be defined as the timing of a point in a circadian rhythm relative to a marker such as dawn or dusk. The circadian clock is synchronized with the external cycle so that a specific point in each output rhythm always occurs at the same time relative to dawn or dusk (Pittendrigh, 1981). This synchronization is termed entrainment. The significance of entrainment should be easily appreciated by anyone who has suffered from jet lag, the result of sudden desynchronization between the internal clock and the surrounding environment. Many metabolic processes in Arabidopsis (Arabidopsis thaliana) are under circadian control (Harmer et al., 2000); the clock coordinates the temporal synchronization of a range of interconnected metabolic pathways with the light-dark (LD) cycle and each other, a further indication of how important entrainment is to this organism. Natural selection is predicted to favor a circadian period close to 24 h because this eases the process of entrainment to the daily solar cycle (Pittendrigh, 1960, 1981; Dunlap, 1999). As a result of entrainment, the period length of a rhythm becomes that of the environmental cycle. Therefore, the only properties of the circadian clock expressed in natural conditions are phase and amplitude; these are the phenotypes directly subjected to natural selection. Variation in phase of an entrained circadian rhythm between populations is likely to reflect differences in selection on circadian parameters in their progenitors.
Naturally existing variation in the model plant Arabidopsis was used as the basis for a study of entrained phase. This plant has a very wide geographical range across Eurasia but has been introduced to other continents and occupies a variety of habitats; natural variation between accessions is extensive and usually multigenic (Koornneef et al., 2004). Significant variation in the free-running period length has been observed previously between Arabidopsis accessions (e.g. Swarup et al., 1999; Michael et al., 2003; Edwards et al., 2005). These earlier experiments considered only free-running systems in continuous light. In contrast, here we consider whether Arabidopsis populations differ in the parameters of their circadian rhythms when entrained, an experimental approach that better reflects the conditions under which the circadian system evolved and usually functions.
The advantages of working with a model organism such as Arabidopsis include the availability of controlled sources of natural variation, such as accessions obtained by single-seed descent and recombinant inbred lines (RILs). RILs are immortal recombinant populations derived from an initial cross between two accessions, each individual line being a homozygous mosaic of the original parental genotypes. This allows many phenotypes to be compared to a single genetic map. Thus, naturally occurring differences between wild-type populations can be associated with specific genomic regions. Naturally existing variation between two geographically distant accessions, Landsberg erecta (Ler) and Cape Verde Islands (Cvi), was used to determine loci controlling circadian phase in these populations. As circadian phase varied continuously between the different genotypes of a Ler × Cvi RIL population, we could analyze it as a quantitative trait. Using the combination of phenotypic and genotypic data, we were able to identify, and then dissect by fine mapping, quantitative trait loci (QTL) controlling circadian phase. This indicated loci that were responsible for advancing or delaying the clock.
There is a positive correlation between period and phase in circadian mutants such as zeitlupe (ztl) and timing of cab 1 (toc1), where the endogenous period is very long or short and the phase of a rhythm is delayed or advanced, respectively, during entrainment (Somers et al., 2000; Strayer et al., 2000). Michael et al. (2003) showed no correlation in period and phase in accessions and a negative correlation in RILs. Therefore, in wild-type plants, the relationship of period and phase is ambiguous. By measuring natural variation between populations, QTL affecting circadian phase were identified without requiring mutants, which may alter the working of the clock very dramatically. In addition, factors other than period that affect phase could be investigated.
The power of our analysis was increased by the use of a well-characterized hand of the circadian clock, the rhythm of bioluminescence from a modified firefly LUCIFERASE (LUC) gene coupled to the CAB2 promoter of Arabidopsis (described in Millar et al., 1992, 1995) to obtain highly accurate measurements of circadian phase after entrainment. Previous studies identifying circadian QTL in Arabidopsis have only considered the rhythm of leaf movement during constant light (Swarup et al., 1999; Michael et al., 2003; Edwards et al., 2005). Two of these studies (Swarup et al., 1999; Edwards et al., 2005) used fast Fourier transform nonlinear least squares (FFT-NLLS; Plautz et al., 1997) to analyze their leaf movement data, obtaining measures of free-running period and amplitude of rhythms, but were unable to measure phase.
Although Michael et al. (2003) used FFT-NLLS to estimate phase of the Arabidopsis leaf movement rhythm, we did not consider that FFT-NLLS produces an accurate estimate of phase. This method fits a period to the waveform over several cycles during constant conditions and then extrapolates back to the end of the LD cycle to estimate phase. When a plant is moved from LD cycles (entraining conditions) to constant conditions (free-running conditions), the period alters, becoming longer in low light or darkness and shorter in bright light (Devlin and Kay, 2000). Attempts to measure phase using such data are confounded by the change in period because each successive peak (or trough or midpoint) will be earlier or later depending on whether the period is shortening or lengthening. In contrast, the phase assay presented here, a single entrained peak of the CAB2∷LUC+ rhythm, is independent of the subsequent behavior of the system during a free run.
Gene expression assayed with a promoter∷reporter construct has not previously been used as a quantitative trait in the context of QTL analysis. This study shows that reporter gene expression can be used to map a circadian parameter in Arabidopsis; the data presented here show that this method allows identification of the QTL controlling phase.
RESULTS
Analysis of Phase Detected Four QTL
The measurement of the entrained phase of CAB2∷LUC+ expression in 26 Arabidopsis accessions revealed a wide range of responses to a LD cycle of 12 h light and 12 h dark (LD 12:12 photoperiod; Table I; Supplemental Tables I and II). Under these conditions, the timing of peak reporter gene expression was found to vary significantly (P < 0.001; Supplemental Table III) between accessions from less than 1 h after subjective dawn to nearly 5 h after subjective dawn.
Table I.
Entrained phase of Arabidopsis accessions from diverse geographic origins
Phase was measured as the timing of peak CAB2∷LUC+ reporter activity relative to subjective dawn after an LD 12:12 cycle. sem, se of the mean. Number of ecotypes = 26. Information on the origin of each accession was obtained from the NASC (http://Arabidopsis.info).
| Accession No. | Country of Origin | NASC No. | Latitude | Altitude | Phase | sem |
|---|---|---|---|---|---|---|
| °N | m | h | ||||
| Cvi-0 | Cape Verde | N902 | 16 | 1,200 | 3.49 | 0.18 |
| Mt-0 | Libya | N1380 | 32 | 312 | 4.19 | 0.22 |
| Tsu-1 | Japan | N1640 | 35 | 100 | 2.23 | 0.21 |
| CT-1 | Italy | N1094 | 37 | 100 | 2.06 | 0.27 |
| Shah | Tajikistan | N929 | 39 | 3,400 | 2.35 | 0.17 |
| Kondara | Tajikistan | N916 | 39 | 1,100 | 3.35 | 0.18 |
| Col-0 | US | N1092 | 39 | 150 | 2.22 | 0.17 |
| Fei-0 | Portugal | N22645 | 41 | 100 | 2.45 | 0.29 |
| Ts-1 | Spain | N1552 | 42 | 0 | 0.79 | 0.29 |
| Kin-0 | US | N1273 | 42 | 258 | 4.00 | 0.30 |
| Ll-0 | Spain | N1338 | 42 | 50 | 1.54 | 0.26 |
| Mr-0 | Italy | N1372 | 45 | 1,500 | 1.92 | 0.18 |
| Ag-0 | France | N901 | 45 | 150 | 2.14 | 0.27 |
| Ga-0 | Germany | N1180 | 50 | 150 | 2.22 | 0.17 |
| Van-0 | Canada | N1584 | 50 | 100 | 2.24 | 0.27 |
| Gy-0 | Germany | N1216 | 51 | 200 | 2.01 | 0.24 |
| No-0 | Germany | N1394 | 51 | 250 | 4.80 | 0.14 |
| An-1 | Belgium | N944 | 52 | 50 | 1.55 | 0.37 |
| NOK-3 | Netherlands | N1404 | 52 | 50 | 2.94 | 0.27 |
| Wt-5 | Germany | N1612 | 52 | 100 | 3.12 | 0.34 |
| Ws | Belarus | N915 | 53 | 100 | 2.48 | 0.17 |
| Ler | Poland | NW20 | 53 | 100 | 2.37 | 0.16 |
| La-0 | Poland | N1298 | 53 | 100 | 3.76 | 0.19 |
| Kil-0 | UK | N1270 | 56 | 450 | 3.88 | 0.63 |
| RLD | Russia | N1641 | 57 | 100 | 3.70 | 0.17 |
| Est-0 | Estonia | N1148 | 59 | 150 | 3.81 | 0.18 |
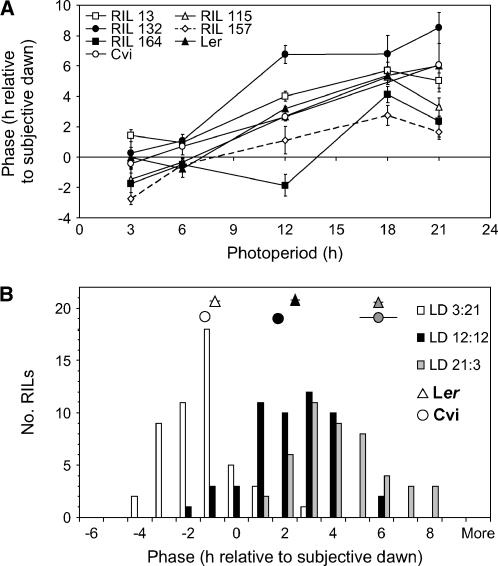
A population of RILs derived from Ler and Cvi accessions was used as a controlled source of phenotypic and genetic variation. Initially, the entrained phase after different photoperiods in a subset of RILs and the two parental lines was measured (representative data are shown in Fig.1A). This showed very marked differences between RILs (Supplemental Table III). Although there was a trend of earlier phase in shorter photoperiods, the shape of the response to photoperiod differed between RILs. For any given photoperiod, the phase of the rhythm varied by several hours between RILs (Fig. 1A).
Figure 1.
Phase in Cvi × Ler RILs and parental lines. A, Phase in response to different photoperiods. Means ± 95% confidence interval. B, Distribution of phases in RIL population after three photoperiods; parental mean phase ± 95% confidence interval.
The effect of photoperiod on phase was tested systematically by measuring the responses of all transformed RILs to three photoperiods (very short days [LD 3:21], intermediate days [LD 12:12], and very long days [LD 21:3]; summarized in Fig. 1B; detailed information on numbers of seedlings of each genotype used in phase estimation in Supplemental Tables II–VI). Variation between RILs was statistically significant, and there was also a significant interaction between genotype and photoperiod (Supplemental Table III; Supplemental Fig. 1). Phase was slightly, but significantly, correlated between all photoperiods (Supplemental Table VIII), indicating that some of its genetic control is shared between environments. Figure 1B shows the mean phases in response to the three photoperiods. At each daylength, there was a wider variation in phase in the RILs than between the parental ecotypes. This transgressive variation suggests that the parental phenotypes result from the balancing effects of alleles advancing and delaying phase.
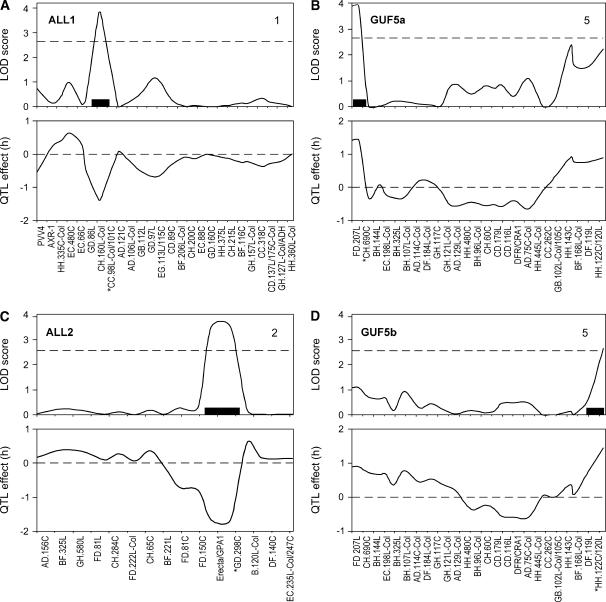
The recombinant population was used to map phase advances or delays in the CAB2∷LUC+ rhythm to specific genomic regions, indicating that these responses were caused by variation at several loci. The mean phase values for each RIL were used in a QTL analysis to identify the number, effect, and location of QTL controlling phase in each set of photoperiodic conditions. Using the multiple QTL model mapping (MQM) method (van Ooijen, 1999; van Ooijen and Maliepaard, 2000) of the MapQTL program, four QTL affecting phase of the CAB2∷LUC+ rhythm were identified (Fig. 2; Table II). QTL were named ALLODOLA (ALL; Italian for lark) if Cvi alleles advanced the phase of the clock and GUFO (GUF; Italian for owl) if Cvi alleles delayed the phase relative to subjective dawn. They were numbered by chromosome with their order alongside it.
Figure 2.
Positions (top graph) and effects (bottom graph) of phase QTL. A, ALL1, chromosome 1: Cvi alleles give early phase in LD 3:21. B, GUF5a, chromosome 5: Cvi alleles give late phase in LD 3:21. C, ALL2, chromosome 2: Cvi alleles give early phase in LD 12:12. D, GUF5b, chromosome 5: Cvi alleles give late phase in LD 12:12. Genotyping markers shown on horizontal axis. Markers asterisked where cofactors fitted. Black bar, 95% confidence interval for a QTL. Genome-wide 95% confidence levels: LD 3:21 LOD score ≥ 2.65; LD 12:12 LOD score ≥ 2.55.
Table II.
Summary of QTL, positions, and their effects
Environment describes the 7-d entrainment protocol used to discover the QTL. Location is the 95% confidence interval for the location of the QTL. Effect is the double additive effect on phase associated with Cvi alleles relative to Ler at the QTL. Var is the percentage of the total phenotypic variation explained by the QTL.
| QTL Name | Environment | Location | P | Effect | Var |
|---|---|---|---|---|---|
| cM | h | % | |||
| ALL1 | LD 3:21 | 24.3–40.5 | <0.01 | −1.27 | 24.1 |
| ALL2 | LD 12:12 | 44.6–58.2 | <0.01 | −1.67 | 22.7 |
| GUF5a | LD 3:21 | 0–7.6 | <0.01 | +1.25 | 24.6 |
| GUF5b | LD 12:12 | 103.5–117 | <0.05 | +1.46 | 16.5 |
Two QTL were found after entrainment to LD 3:21: ALL1 (chromosome 1) and GUF5a (chromosome 5, top; Fig. 2, A and B). Two additional QTL were found after entrainment to LD 12:12: ALL2 (chromosome 2) and GUF5b (chromosome 5, bottom; Fig. 2, C and D). Each QTL explained between 16.5% and 24.1% of the variation in phase between the RIL lines (Table II). The total variation explained by the QTL in each environment is an estimate of broad sense heritability of the trait; the estimates are 39.2% and 48.7% for phase after entrainment to LD 12:12 and LD 3:21, respectively (Table II). There was a very strong effect of the environment as no QTL was found in more than one condition. Indeed, the environment (i.e. photoperiod × genotype interaction) was highly significant (P < 0.001; Supplemental Table III). It is noticeable that we found both a phase-advancing and a phase-delaying QTL in each photoperiod; this, and the similar amount of variation explained by the QTL, suggests that the closeness of the parental phenotypes can be explained by a simple model of stabilizing selection. Stabilizing selection is also indicated by the narrower range of variation in phase in all accessions compared to RILs. No significant epistatic interactions were found between the QTL (P > 0.05; Chase et al., 1997).
No QTL were detected after entrainment to LD 21:3. Despite considerable phenotypic variation (Fig. 1B), Ler × Cvi RILs did not differ systematically at loci regulating phase after very-long-day entrainment. This may be a reflection of the smaller proportion of variation attributable to the RIL compared with the transformant in this particular environment. This can be quantified by the F ratio (F), which shows the variation explained by each factor relative to the error variation: F(RIL) = 1.18, F(transformant) = 10.29 (Supplemental Table III). Without knowledge of the genes, pathways, and mechanisms involved, it is impossible to be certain why so much more variation is explained by transformants after entrainment to LD 21:3. However, it may be because different genes affect phase in different photoperiods.
QTL Were Confirmed in Near-Isogenic Lines
Near-isogenic lines (NILs) containing small areas of Cvi chromosomes introgressed in an otherwise entirely Ler genomic background were used to test for our QTL effects (Swarup et al., 1999; Edwards et al., 2005); each result was confirmed in at least two independent experiments (Table III; Supplemental Table VII).
Table III.
Confirmation of QTL effects in NIL lines
Environment describes the 7-d entrainment protocol after which phase was measured as appropriate to the QTL being confirmed. Phase in hours relative to subjective dawn. sem, se of the mean; t tests are heteroscedastic one-tailed tests comparing the phase of each NIL to the Ler control included in the same experiment.
| NIL | Environment | Mean Phase | sem | Total No. Plants | t | P | Effect Relative to Ler | Expected QTL Effect |
|---|---|---|---|---|---|---|---|---|
| h | ||||||||
| Ler | LD 3:21 | −1.95 | 0.35 | 41 | ||||
| 18 | LD 3:21 | −1.51 | 0.35 | 111 | −0.89 | >0.05 | None | Early (ALL1) |
| 18.32 | LD 3:21 | −0.81 | 0.49 | 61 | −0.50 | >0.05 | None | Early (ALL1) |
| 42 | LD 3:21 | −3.15 | 0.24 | 113 | 2.84 | <0.01 | Early | Early (ALL1) |
| 45 | LD 3:21 | −1.00 | 0.27 | 148 | −2.14 | <0.05 | Late | Early (ALL1) |
| 251 | LD 3:21 | −0.81 | 0.37 | 78 | −2.24 | <0.05 | Late | Early (ALL1) |
| Ler | LD 3:21 | −0.83 | 0.43 | 39 | ||||
| 6 | LD 3:21 | 0.42 | 0.27 | 149 | −2.46 | <0.01 | Late | Late (GUF5a) |
| 55 | LD 3:21 | 0.83 | 0.31 | 105 | −3.11 | <0.01 | Late | Late (GUF5a) |
| 46 | LD 3:21 | 0.38 | 0.31 | 89 | −2.28 | <0.05 | Late | Late (GUF5a) |
| 85 | LD 3:21 | 0.33 | 0.33 | 76 | −2.16 | <0.05 | Late | Late (GUF5a) |
| 45a | LD 3:21 | −0.45 | 0.59 | 25 | −0.52 | >0.05 | None | Late (GUF5a) |
| 106 | LD 3:21 | 0.12 | 0.32 | 48 | −1.77 | <0.05 | Late | Late (GUF5a) |
| Ler | LD 12:12 | 4.84 | 0.43 | 27 | ||||
| 19.2 | LD 12:12 | 5.94 | 0.26 | 81 | −2.22 | <0.05 | Late | Late (GUF5b) |
| 26.4 | LD 12:12 | 6.13 | 0.32 | 66 | −2.43 | <0.01 | Late | Late (GUF5b) |
| 30.2 | LD 12:12 | 6.01 | 0.20 | 132 | −2.49 | <0.01 | Late | Late (GUF5b) |
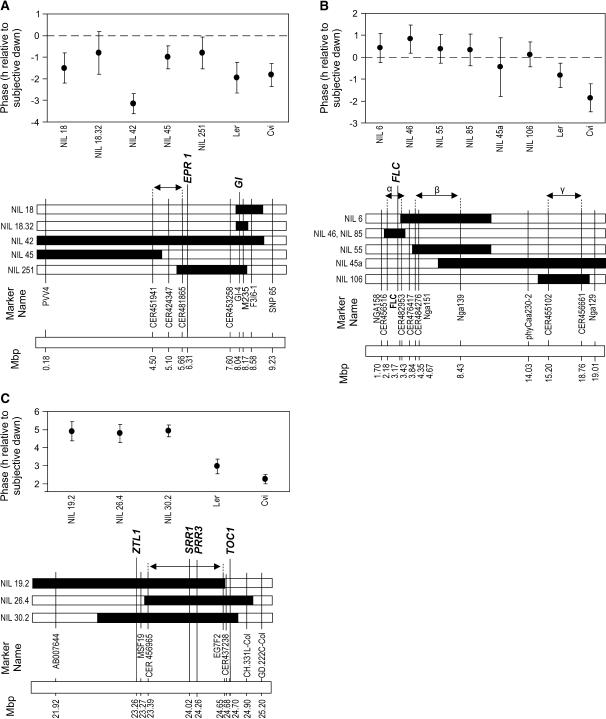
NILs confirmed the existence of the ALL1 QTL; Cvi alleles at this locus caused early phase in LD 3:21. NIL 42 had an earlier phase than Ler by 1.2 h (Fig. 3A) in LD 3:21. However, NILs 18, 45, and 251, which have smaller Cvi introgressions, did not show an earlier phase of CAB2∷LUC+ relative to Ler (Fig. 3A); NIL 18.32, derived from NIL 18, also did not show the ALL1 effect. This mapped the ALL1 QTL to a small region between CER451941 and CER481865 (approximately 4.50–5.66 Mb; Fig. 3A). There are approximately 350 predicted genes in this region.
Figure 3.
Confirmation of phase QTL effects (relative to Ler) and position of markers used to map breakpoints of NILs. Top, x axis depicts NIL and parental genotypes. Double-headed arrows show QTL interval predicted from NIL phase data. A, ALL1 QTL in LD 3:21: only NIL 42 shows early phase. B, GUF5a QTL in LD 3:21: all NILs except 45a show late phase. C, GUF5b QTL in LD 12:12: all NILs show late phase. Means ± 95% confidence interval. Black, Cvi introgression; white, Ler background.
The effect of the ALL2 QTL (where Cvi alleles are associated with late phase in LD 12:12) could not be confirmed using NILs spanning its interval on chromosome 2. None of the NILs containing Cvi introgressions in this region showed the early phase effect predicted by the original analysis (data not shown). Landsberg ERECTA (wild type at the ERECTA locus) was also phenotyped; this line also was not early with respect to Ler, indicating that the erecta mutation was not responsible for the early phase. The ALL2 QTL might be a false positive, the association between phase and genotype arising by chance. However, this is unlikely given that the probability of the QTL occurring by chance is so small (P = 0.004 for a log of the odds [LOD] score of 3.5). An alternative explanation is that epistatic interactions within the area suppress the expected QTL effect.
Mapping of NIL breakpoints within the GUF5a QTL region (where Cvi alleles cause late phase in LD 3:21; Fig. 3B) suggested that this QTL was made up of at least three distinct loci (Fig. 3B; marked α, β, and γ), each of which caused late phase. We estimate that these regions contain approximately 570, 1,220, and 1,253 predicted genes, respectively. A closely related pair of NILs (46 and 85) showed late phase of CAB2∷LUC+ relative to Ler. This is consistent with a late-phase QTL on NILs 46 and 85 in a region (α) containing the floral regulator FLOWERING LOCUS C (FLC), which regulates photoperiodic control of flowering. NILs 6 and 55, also from the GUF5a region of chromosome 5, showed later phase of CAB2∷LUC+ expression than Ler (Fig. 3B). Because NILs 46 and 85 share no region of introgression with NIL 55, the late phase of NIL 55 must have an independent cause (β). Although NIL 6 shares a small region of Cvi introgression with NILs 46 and 85, it has a much larger region of overlap with NIL 55 (Fig. 3B). The possibility that NIL 6 contains both a unique QTL and the one responsible for the behavior of NIL 55 is unlikely given the absence of additive effects in NIL 6. Late phase was also observed in NIL 106, where the Cvi introgression does not overlap with the other NILs, implying the existence of yet another late-phase QTL (γ) further down chromosome 5. A late-phase effect was not detected in NIL 45a despite its introgression overlapping that of NIL 106. It is possible that epistasis between loci within NIL 45a is responsible for suppressing this effect.
The GUF5b QTL (where Cvi alleles cause phase delays in LD 12:12) was confirmed in three NIL lines (Fig. 3C). Breakpoint mapping of NILs ruled out the core clock component TOC1 and the clock-associated gene ZTL as causes of late phase of the GUF5b QTL. The QTL was placed in a small region bound by the markers MSF19 and CER437238 (between approximately 23.27 and 24.68 Mb) and estimated to include approximately 422 predicted genes. This region contains SIGNALING IN RED LIGHT REDUCED 1 (SRR1; Staiger et al., 2003) and PSEUDORESPONSE REGULATOR 3 (PRR3; Matsushika et al., 2000). As altered expression of these genes has been shown to affect the clock (Staiger et al., 2003; Murakami et al., 2004), they were considered to be candidates for the GUF5b QTL and sequenced from Ler and Cvi.
Sequencing and Analysis of GUF5b Candidate Genes
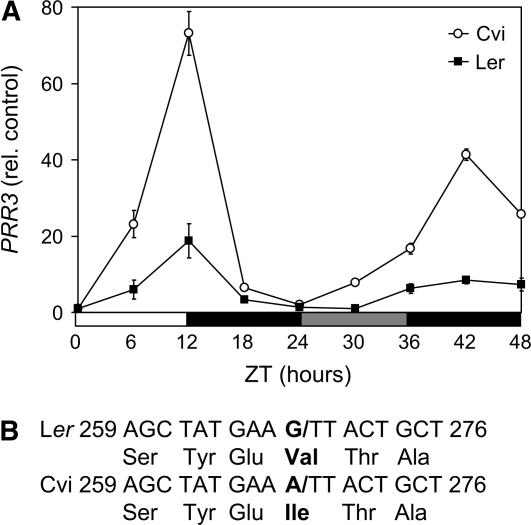
Nucleotide sequencing of PRR3 predicted changes in both Ler and Cvi from Columbia (Col) at amino acid positions 210 (Gln to Glu), 267 (Asn to Glu), 268 (Ala to Ser), and 330 (Asn to Lys). These changes were common to both ecotypes so they could not be responsible for the GUF5b QTL. However, a nonsynonymous polymorphism (G to A) was detected at the end of the first exon of PRR3 in Cvi, but not in Ler. This nucleotide is the first in the triplet coding for a Val residue, which is conserved throughout all five members of the TOC1/PRR family (Matsushika et al., 2000); in the toc1-2 mutant, an identical mutation leads to missplicing and a dramatic reduction in the amount of correctly spliced TOC1 mRNA (Strayer et al., 2000).
The transcript levels of PRR3 in Cvi and Ler were therefore determined. Transcripts amplified over the first exon boundary showed rhythmic gene expression in both Cvi and Ler (Fig. 4A); transcripts spanning the splice site of the third intron gave the same rhythm (data not shown). cDNA sequencing revealed that the first intron was spliced out correctly in both Cvi and Ler transcripts, indicating that the nucleotide change at the end of the first exon in Cvi did not alter the splice point. It is therefore predicted that the conserved Val is replaced in Cvi with Ile (Fig. 4B), the remainder of the transcript sequence being identical between accessions.
Figure 4.
A, Real-time RT-PCR transcript levels of PRR3 cDNA, normalized to β-TUBULIN4 level and ZT 0 in Ler and Cvi ecotypes. Primers span first exon boundary. LD 12:12 ZT 0 to 24, then constant darkness. White bar, Day; black bar, night; gray bar, subjective night. The mean of two repeats is plotted for each ecotype; error bars show 95% confidence interval. PRR3 levels were normalized against those of β-TUBULIN4 for each time point before being normalized to ZT 0. B, Nucleotide sequence and predicted translation of PRR3 first and second exon boundary (/) from Ler and Cvi; numbering relative to coding region start site.
SRR1 was predicted to encode a protein of 275 amino acids in Col (Staiger et al., 2003); sequencing from the parental accessions (Fig. 5) suggested a protein of the same length in Ler, but a protein of 277 amino acids in Cvi. Analysis of the predicted proteins revealed one polymorphism between Ler and Col at position 77 (Ala to Thr) with respect to the first Met. The Cvi sequence differed from Ler more greatly, with substitutions at positions 73 (Ser to Asn), 77 (Thr to Val), 90 (Gln to His), and 235 (Arg to Ser) and conversion of the stop codon (position 276 in Ler and Col) to Glu plus the insertion of a Phe at position 277 before a novel stop codon (Fig. 5).
Figure 5.
Predicted amino acid sequence of SRR1 from Col (At5g59560), Ler, and Cvi ecotypes. Polymorphic residues (bold) are indicated by asterisk; amino acids conserved across multiple species (Staiger et al., 2003) are highlighted in gray.
DISCUSSION
The circadian clock exists to set phase, allowing coordination of physiological rhythms with environmental ones. This study was designed to identify novel QTL for phase. The first peak of expression of CAB2∷LUC+ relative to subjective dawn was used as a phase marker.
Previous studies have determined geographic clines in some circadian-associated traits in Arabidopsis (e.g. flowering time [Stinchcombe et al., 2004], hypocotyl length [Maloof et al., 2001], and period length [Michael et al., 2003]). The presence of clines in circadian parameters is observable in other species (Drosophila melanogaster in Europe—temperature compensation [Sawyer et al., 1997]; Drosophila auraria in Japan—phase, period, amplitude [Pittendrigh and Takamura, 1989]). This study did not find a geographic cline in phase despite examining accessions from a wide range of latitudes (14°N–55°N). Because previous clines in circadian traits reported from this species are relatively weak, we conjecture that the high self-fertilization rate in Arabidopsis populations, which reduces the intrapopulation variation (Bergelson et al., 1998), may in some part act to counter the establishment of a cline in the absence of strong directional selection for a trait. Given this, a larger number of accessions may be needed to determine whether a cline is present.
Populations showed considerable differences in their response to a set photoperiod, but the true extent of the pool of potential variation was hidden, being revealed only in a recombinant population. It is noticeable that, after entrainment to LD 12:12, phases of Ler × Cvi RILs ranged from 1 h before subjective dawn to 7 h after it. This range is approximately double that seen across the wild-type accessions (Table I), suggesting stabilizing selection in wild-type populations acts against the most extreme phase phenotypes. The much wider range of phase phenotypes found in the RIL population than might have been predicted from merely observing the behavior of Ler and Cvi (Table I; Supplemental Table II; Fig. 1B) implies that the similar phase phenotypes of the parental accessions were due to balancing effects of early and late alleles.
Different photoperiods invoked different responses from plants. The results presented imply that phase is affected by both the environment and genotype. Earlier phase was observed after short photoperiods in both Ler and Cvi and a derived RIL population; however, observations of individual RILs showed that this was not a simple linear relationship (Fig. 1A; Supplemental Fig. 1), indicating a strong interaction with the environment that took the form of changes in both rank and scale. In very short daylengths, early phase of CAB2∷LUC+ expression may reflect earlier timing of all aspects of the metabolic machinery, thus maximizing the ability of the plant to harvest light for photosynthesis (Harmer et al., 2000) and thus increasing the efficiency of use of available resources. In longer days, when light was not a limited resource, the plants switched to later phase.
Analysis of circadian reporter gene expression in a recombinant population showed this technique to be an effective method for finding QTL affecting parameters of the circadian clock. The results revealed regions containing novel candidates for pathways controlling circadian entrainment. Phenotypic and mapping analysis of RILs and NILs confirmed three QTL, ALL1, GUF5a, and GUF5b, which changed the phase of the CAB2∷LUC+ rhythm. A previous study of leaf movement (Michael et al., 2003) suggested period and phase QTL in the Ler×Col RIL population in similar positions to our phase QTL. However, of the five phase QTL named, only one (PHI5b; mapping in a similar position to GUF5b) was significant at the 0.05% level. In addition, not all of the period QTL named were significant at this level. As the QTL were not confirmed in NILs, it is difficult to compare results with this study.
Another leaf movement QTL study (Edwards et al., 2005) suggested that GIGANTEA (GI) was responsible for the effects of PerCV1b/ESP, a short-period QTL on chromosome 1 overlapping the ALL1 reported here. In this study, the phenotypes of the NILs and breakpoint mapping in this region ruled out GI as the cause of the ALL1 QTL (Fig. 3A). Breakpoint mapping of the ALL1 NILs also eliminated EPR1 (a MYB transcription factor identified through its homology to CCA1 and LHY as a slave oscillator involved in controlling CAB2 output [Kuno et al., 2003]) as a cause of the QTL. In fact, mapping of the ALL1 QTL suggested it was a novel circadian locus controlling phase on chromosome 1. Within this region, four genes showed circadian regulation based on analysis of a previous microarray experiment (Harmer et al., 2000): the transcription factor RAV1 (At1g13260), Lhcb6 (At1g15820), an auxin-induced gene (At1g16510), and an expressed protein (At1g13930). Future work needs to determine whether any of these are responsible for the early phase of ALL1.
It is likely that the α-region of GUF5a, linked to the FLC marker, overlaps with PerCV5b, a long-period QTL probably caused by FLC (K. Edwards, personal communication). FLC is currently the strongest candidate for the late phase of NILs 46 and 85; this is supported by a previously cataloged polymorphism between Ler and Cvi FLC sequences (Caicedo et al., 2004). The Ler allele of FLC has been shown to be only weakly expressed due to the insertion of a transposable element (Gazzani et al., 2003). FLC was first identified as a floral repressor (Koornneef et al., 1994), but has also been suggested as the cause of a period QTL (Swarup et al., 1999); the effect on phase seen here may be related to a change in the period length.
It is important to note that the late phase of the GUF5a QTL cannot be accounted for solely by a single locus. Two regions subtending the QTL (β and γ; Fig. 3B) are likely to include novel loci controlling phase of the clock. Analysis of existing microarray data (Harmer et al., 2000) found two genes in the GUF5a region showing circadian regulation: KNOTTED1-LIKE 4 (KNAT4; At5g11060), a homeobox gene previously reported as being involved in the response to light (Serikawa et al., 1996), and GUN5 (At5g13630), a magnesium chelatase subunit involved in plastid-to-nucleus signal transduction.
Two genes were considered to be particularly strong candidates for the GUF5b effect on circadian phase (Fig. 3C): SRR1, required for normal circadian gene expression and other clock outputs in a variety of photic environments, and PRR3, a member of the TOC1/PRR gene family. SRR1 acts through modifying the signal from the phytochrome B (phyB) photoreceptor; in this context, it is noteworthy that the out of phase 1 (oop1) mutant of PHYB causes early phase of circadian rhythms (Salome et al., 2002). In mice, the SRR1 homolog maps close to a QTL affecting free-running period length (Shimomura et al., 2001). PRR3 is one of the TOC1/PRR gene family whose members all have clock-associated phenotypes (Matsushika et al., 2000; Makino et al., 2001, 2002).
Sequencing these genes found polymorphisms between the parental accessions. Transcript analysis of PRR3 found neither missplicing nor change in expression between ecotypes; however, a highly conserved amino acid was substituted in Cvi. Previous reports have shown that T-DNA insertions in the untranslated region or coding regions of PRR3 (Michael et al., 2003) or overexpression of the gene (Murakami et al., 2004) have relatively small effects upon the circadian rhythmicity of Arabidopsis; this is consistent with the relatively small effect of the GUF5b QTL. A number of dissimilar amino acid substitutions plus the insertion of two extra residues were found in the predicted coding sequence of SRR1 in Cvi relative to those found in the Ler/Col versions of the protein, although none occurred in conserved regions of the protein (Staiger et al., 2003). Both SRR1 and PRR3 are known to affect multiple circadian outputs in Arabidopsis (Makino et al., 2001, 2002; Murakami-Kojima et al., 2002; Michael et al., 2003; Staiger et al., 2003); our results suggest that natural allelic variation within them also affects the circadian system.
SRR1 and PRR3 are therefore both possible causes for the GUF5b QTL effect and, as the two loci are closely linked (<1 cM apart), their effects on phase cannot be distinguished at present. However, we consider SRR1 to be a stronger candidate for the GUF5b QTL than PRR3 because of the greater differences between the Ler and Cvi predicted proteins. If SRR1 is the cause of the GUF5b phase QTL (either singly or with PRR3), then it may shed light on the early phase phenotype of the PHYB oop1 mutant (Salome et al., 2002), which produces a truncated version of the PHYB protein lacking the C-terminal kinase domain. We suggest this truncation affects the interaction with SRR1, leading to early phase. A putative nuclear localization domain has been identified in SRR1 (Staiger et al., 2003); if it enters the nucleus, it is in the correct place to interact with PHYB. In Cvi, SRR1 structure may be altered sufficiently to change an interaction in the PHYB signaling pathway and, hence, cause later phase.
Detailed analysis of the ALL1 and GUF5a QTL intervals shows that the causes of their effects on phase can be attributed to novel loci. Although the part of GUF5a that contains FLC has previously been linked with period length, fine mapping of the remainder of GUF5a and ALL1 placed them at loci not previously associated with period (Swarup et al., 1999; Edwards et al., 2005). This means they must be caused by genes not previously considered part of the circadian system or its output. That different loci are identified in screens for phase and period supports the idea that the entrained phase of a rhythm does not depend solely on the period length. Future work will focus on identifying candidate genes for ALL1 and the β- and γ-regions of GUF5a, and determining whether SRR1 and/or PRR3 are responsible for the phase effects of GUF5b.
MATERIALS AND METHODS
Plant Materials and Growth
A selection of Arabidopsis (Arabidopsis thaliana) accessions reflecting a wide geographical range (see Table I) and the core set (N22477) of 50 Cvi × Ler RILs (Alonso-Blanco et al., 1998a, 1998b) plus additional lines (RILs 6 [N22005], 8 [N22007], 13 [N22012], and 191 [N22160]) from the same population were obtained from the Nottingham Arabidopsis Stock Centre (NASC). NILs, consisting of an almost complete Ler genome with a single small Cvi introgression in the region of interest, were generated by backcrossing appropriate RILs with Ler. The chromosome 5 NILs 45a, 106, and 187 were provided by Professor Maarten Koornneef; NILs 6, 46, 55, and 85 were produced by backcrossing NIL 187 to Ler. These and the other NILs used in this project have been described previously (Edwards et al., 2005).
Accessions, RILs, and NILs were transformed by floral dip with the CAB2∷LUC+ reporter (Hall et al., 2002). T1 transformants were selected on antibiotic plates and selfed. T2 plants expressing LUC activity were assayed. Multiple independently transformed lines (mean no. transformants = 3.81) were generated for each RIL or NIL to control for possible positional effects caused by the insertion of the transgene.
For measurements of phase or collection of a tissue time course for real-time reverse transcription (RT)-PCR analysis, seeds were surface sterilized, then placed on Murashige and Skoog medium (M5524; Sigma; +3% Suc, 1% agar), and stratified for 48 h at 4°C before being transferred to a LD cycle in a growth chamber (55 μmol m−1 s−1) at 22°C.
Measurement of Phase
After 7 d of growth, seedlings were placed in 96-well plates and 5 μm luciferin solution added to each well, as described previously (McWatters et al., 2000). Seedlings were transferred to constant darkness at dusk (i.e. Zeitgeber [ZT] 3 following entrainment to LD 3:21, ZT 12 following entrainment to LD 12:12, or ZT 21 following entrainment to LD 21:3), and bioluminescence levels were recorded from individual seedlings for 36 to 48 h in a Packard Topcount luminometer. Although CAB2∷LUC+ activity damps in darkness after 36 h in some ecotypes, including Columbia, Cvi, and Ler, recording for this length of time is sufficient to measure this peak (Supplemental Fig. 2). All experiments were conducted at a constant 22°C throughout.
Luminescence rhythms of individual seedlings (see Supplemental Tables II, IV, and V–VII for the numbers of seedlings and transformants) were plotted as a three-point moving average and the time of the first peak of each seedling's CAB2∷LUC+ rhythm recorded using Biological Rhythms Analysis Software System (BRASS version 1.3.1; Johnson and Frasier, 1985; Straume et al., 1991; Plautz et al., 1997; Brown, 2004). Circadian phase was expressed as the time of this peak in hours relative to the first subjective dawn. Hence, if the rhythm peaked before subjective dawn, the phase is negative; peaks after dawn give positive phase values.
We decided to measure the first peak after a LD cycle because the acute response of CAB2∷LUC+ to light (McWatters et al., 2000; Hall et al., 2003) makes it difficult in constant light to determine accurately the timing of the circadian peak of activity if this falls before or around dawn. The time of the first CAB2∷LUC+ peak after discontinuation of a LD cycle is well established in circadian studies of Arabidopsis as an accurate representation of the clock (e.g. McWatters et al., 2000; Hall et al., 2003). In addition, real-time RT-PCR on central clock components and a variety of input and output genes confirmed that gene expression rhythms did not change in the first subjective day after entrainment to LD 12:12 (data not shown). It is considered that the first peak after the discontinuation of a LD cycle is a measure of the entrained phase of a rhythm (Love et al., 2004).
QTL Mapping
We used the base set of 50 Ler × Cvi RILs, which were originally selected as a starting set for molecular mapping because they have higher recombination and lower segregation distortion than average, and supplemented it with selected lines. The total number of RILs transformed and used in this analysis (50; see Supplemental Table II) is comparable to other circadian QTL studies (e.g. 48 Ler × Cvi [Swarup et al., 1999; Edwards et al., 2005] and 76 Ler × Col [Michael et al., 2003]).
The positions of QTL for phase were mapped using interval mapping (IM) and approximate MQM procedures in MapQTL 4.0 (van Ooijen, 1999; van Ooijen and Maliepaard, 2000). MQM uses selected markers as cofactors to define the position of QTL detected by IM. The effects of these QTL are removed from the subsequent IM of all other loci, which reduces the residual variance and thus increases the power of the analysis. MapQTL 4.0 was used to carry out permutation tests to set 95% genome-wide significance levels for the LOD score; 95% confidence intervals for the QTL were obtained by constructing 2-LOD support intervals around them (Lander and Botstein, 1989; van Ooijen, 1999; van Ooijen and Maliepaard, 2000). EPISTAT (Chase et al., 1997) was used to test for epistatic interactions between the QTL. Total genomic DNA was extracted using the Edwards protocol (Edwards et al., 1991); NIL breakpoints were finely mapped using PCR-based markers (single-nucleotide polymorphisms and simple sequence-length polymorphisms) designed using the CEREON polymorphism database. All novel markers have been submitted to The Arabidopsis Information Resource (TAIR) database (www.arabidopsis.org), which was used to predict the approximate number of genes in a QTL.
Sequencing of Candidate Genes
Genomic DNA was extracted from 10-d-old Ler and Cvi seedlings (Dellaporta et al., 1983) and was used as a template to amplify regions of SRR1 (At5G59560) and PRR3 (At5G60100) by PCR, using gene-specific primers: SRR1 5′-TTGGGCCGTATCTAGACCCA-3′, SRR1 5′-CAAGACTACTATCTGTTTTTGGAAATGG-3′, PRR3 5′-TTTGGTGAAGGGATTAGAATAAGTTTG-3′, and PRR3 5′-CAAGACTACTATCTGTTTTTGGAAATGG-3′.
Amplified products were ligated into the pCR-BLUNT vector (44–0302; Invitrogen) in accordance with the manufacturer's instructions. SRR1 and PRR3 were sequenced (Department of Biochemistry, University of Oxford) using M13 forward and reverse primers and gene-specific primers: SRR1 5′-ACACAGCATTCTTGGAGCAGC-3′, SRR1 5′-AGACCCTAACACGAGGCGAA-3′, PRR3 5′-ATGTGTTTTAATAACATTGAAACTGGTGATG-3′, PRR3 5′-CCAGCTTCAATATGCCATGCT-3′, PRR3 5′-TAGGCACGGGATCACAGACA-3′, PRR3 5′-TTGCAAAACTGTTGGGTTCG-3′, PRR3 5′-CAAGGACATCCGGAACAGCAGTAA-3′, PRR3 5′-CAGCAGACCGGTTCCTGAAT-3′, PRR3 5′-TTTGAAGGCGAGGTGCTCTT-3′, and PRR3 5′-TCGAACCCAACAGTTTTGCA-3′.
Nucleotide sequences have been deposited at GenBank (accession nos. DQ060152, DQ060153, DQ060154, and DQ060155). Nucleotide sequences were translated using Proseq software (Filatov, 2002) and alignments carried out using T-COFFEE at EMBnet (Notredame et al., 2000).
Real-Time PCR and Transcript Sequencing
Replicated samples of Ler and Cvi seedlings were collected and immediately frozen in liquid nitrogen, starting at dawn on day 8 of a LD 12:12 cycle; this cycle was discontinued at dawn on day 9, after which time seedlings were kept in continuous darkness. RNA was extracted (RNeasy kit 74904; Qiagen) in accordance with the manufacturer's instructions and cDNA synthesized (TaqMan N808–0234; Applied Biosystems). Real-time PCR was carried out in an ABI Prism 3700 using SYBR Green PCR master mix (4309155; Applied Biosystems) and gene-specific primers in accordance with the manufacturer's instructions. Levels of PRR3 were calculated using the standard curve method and normalized using products amplified from the constitutively expressed β-TUBULIN4 gene as a control before normalizing to PRR3 levels at ZT 0. All real-time PCR was performed on two cDNA time courses (biological replicates); each containing two technical repeats. Data shown use β-TUBULIN4 as a control (Fig. 4). Similar experiments using UBIQUITIN10 and EF1α as controls gave comparable data (data not shown). UBIQUITIN10, EF1α, and PRR3 third exon primer sequences were as previously published (Czechowski et al., 2004); PRR3 first exon and β-TUBULIN4 primers were designed using Primer Express (Applied Biosystems): β-TUBULIN4 F 5′-AGATCTGGTCCGTTCGGTCAG-3′, β-TUBULIN4 R 5′-CGGCACCAGATTGACCAAAG-3′, PRR3 exon 1F 5′-ACGCCATATTGTTACTGCCCTT-3′, PRR3 exon 1R (Ler) 5′-GACATCCGGAACAGCAGTAACTT-3′, and PRR3 exon 1R (Cvi) 5′-GACATCCGGAACAGCAGTAATTT-3′.
To sequence the boundary between the PRR3 first and second exons, RNA extracted from 10-d-old Ler and Cvi seedlings was reverse transcribed; cDNA was cloned into pCR-BLUNT and used as a template to amplify a PRR3 fragment using gene-specific primers: PRR3 5′-TTTGGTGAAGGGATTAGAATAAGTTTG-3′ and PRR3 5′-CAGCAGACCGGTTCCTGAAT-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers DQ060152, DQ060153, DQ060154, and DQ060155.
Supplementary Material
Acknowledgments
We are grateful to Maarten Koornneef, Joost Keurentjes, and Julin Maloof for sharing NILs and mapping data with us ahead of publication. We would like to thank Caroline O'Brien for technical assistance and Marc Knight for critical reading of the manuscript.
This work was supported by the Biotechnology and Biological Sciences Research Council (BBSRC; grant no. 43/G17845 to H.G.M.) and a BBSRC studentship (to C.D.). H.G.M. is a Royal Society University Research Fellow.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Harriet G. McWatters (harriet.mcwatters@plants.ox.ac.uk).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.074518.
References
- Alonso-Blanco C, El-Assal SED, Coupland G, Koornneef M (1998. a) Analysis of natural allelic variation at flowering time loci in the Landsberg erecta and Cape Verde Islands ecotypes of Arabidopsis thaliana. Genetics 149: 749–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJM, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MTR (1998. b) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Bergelson J, Stahl E, Dudek S, Kreitman M (1998) Genetic variation within and among populations of Arabidopsis thaliana. Genetics 148: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PE (2004) BRASS—Biological Rhythms Analysis Software System Version 1.3.1. http://www.amillar.org/Downloads.html (February 11, 2004)
- Caicedo AL, Stinchcombe JR, Olsen KM, Schmitt J, Purugganan MD (2004) Epistatic interaction between Arabidopsis FRI and FLC flowering time genes generates a latitudinal cline in a life history trait. Proc Natl Acad Sci USA 101: 15670–15675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase K, Adler FR, Lark KG (1997) Epistat: a computer program for identifying and testing interactions between pairs of quantitative trait loci. Theor Appl Genet 94: 724–730 [Google Scholar]
- Czechowski T, Bari RP, Stitt M, Scheible WR, Udvardi MK (2004) Real-time RT-PCR profiling of over 1400 Arabidopsis transcription factors: unprecedented sensitivity reveals novel root- and shoot-specific genes. Plant J 38: 366–379 [DOI] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation version II. Plant Mol Biol Rep 1: 19–21 [Google Scholar]
- Devlin PF, Kay SA (2000) Cryptochromes are required for phytochrome signaling to the circadian clock but not for rhythmicity. Plant Cell 12: 2499–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap JC (1999) Molecular bases for circadian clocks. Cell 96: 271–290 [DOI] [PubMed] [Google Scholar]
- Edwards K, Johnstone C, Thompson C (1991) A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res 19: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA (2002) Proseq. A software for preparation and evolutionary analysis of DNA sequence datasets. Mol Ecol Notes 2: 621–624 [Google Scholar]
- Gazzani S, Gendall AR, Lister C, Dean C (2003) Analysis of the molecular basis of flowering time variation in Arabidopsis accessions. Plant Physiol 132: 1107–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Bastow RM, Davis SJ, Hanano S, McWatters HG, Hibberd V, Doyle MR, Sung S, Halliday KJ, Amasino RM, et al (2003) The TIME FOR COFFEE gene maintains the amplitude and timing of Arabidopsis circadian clocks. Plant Cell 15: 2719–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A, Kozma-Bognar L, Bastow RM, Nagy F, Millar AJ (2002) Distinct regulation of CAB and PHYB gene expression by similar circadian clocks. Plant J 32: 529–537 [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T, Wang X, Kreps JA, Kay SA (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113 [DOI] [PubMed] [Google Scholar]
- Johnson ML, Frasier SG (1985) Nonlinear least squares analysis. Methods Enzymol 117: 301–342 [Google Scholar]
- Koornneef M, Alonso-Blanco C, Vreugdenhil D (2004) Naturally occurring genetic variation in Arabidopsis thaliana. Annu Rev Plant Physiol Plant Mol Biol 55: 141–172 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Blankestijn-de Vries H, Hanhart C, Soppe W, Peeters T (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6: 911–919 [Google Scholar]
- Kuno N, Moller SG, Shinomura T, Xu X, Chua NH, Furuya M (2003) The novel MYB protein EARLY-PHYTOCHROME-RESPONSIVE1 is a component of a slave circadian oscillator in Arabidopsis. Plant Cell 15: 2476–2488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: 185–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love J, Dodd AN, Webb AA (2004) Circadian and diurnal calcium oscillations encode photoperiodic information in Arabidopsis. Plant Cell 16: 956–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Oda Y, Mizuno T (2001) Light response of the circadian waves of the APRR1/TOC1 quintet: When does the quintet start singing rhythmically in Arabidopsis? Plant Cell Physiol 42: 334–339 [DOI] [PubMed] [Google Scholar]
- Makino S, Matsushika A, Kojima M, Yamashino T, Mizuno T (2002) The APRR1/TOC1 quintet implicated in circadian rhythms of Arabidopsis thaliana: I. Characterization with APRR1-overexpressing plants. Plant Cell Physiol 43: 58–69 [DOI] [PubMed] [Google Scholar]
- Maloof JN, Borevitz JO, Dabi T, Lutes J, Nehring RB, Redfern JL, Trainer GT, Wilson JM, Asami T, Berry CC, et al (2001) Natural variation in light sensitivity of Arabidopsis. Nat Genet 29: 441–446 [DOI] [PubMed] [Google Scholar]
- Matsushika A, Makino S, Kojima M, Mizuno T (2000) Circadian waves of expression of the APRR1/TOC1 family of pseudo-response regulators in Arabidopsis thaliana: insight into the plant circadian clock. Plant Cell Physiol 41: 1002–1012 [DOI] [PubMed] [Google Scholar]
- McWatters HG, Bastow RM, Hall A, Millar AJ (2000) The ELF3 zeitnehmer regulates light signalling to the circadian clock. Nature 408: 716–720 [DOI] [PubMed] [Google Scholar]
- Michael TP, Salome PA, Yu HJ, Spencer TR, Sharp EL, McPeek MA, Alonso JM, Ecker JR, McClung CR (2003) Enhanced fitness conferred by naturally occurring variation in the circadian clock. Science 302: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Carre IA, Strayer CA, Chua NH, Kay SA (1995) Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163 [DOI] [PubMed] [Google Scholar]
- Millar AJ, Short SR, Chua NH, Kay SA (1992) A novel circadian phenotype based on firefly luciferase expression in transgenic plants. Plant Cell 4: 1075–1087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami M, Yamashino T, Mizuno T (2004) Characterization of circadian-associated APRR3 pseudo-response regulator belonging to the APRR1/TOC1 quintet in Arabidopsis thaliana. Plant Cell Physiol 45: 645–650 [DOI] [PubMed] [Google Scholar]
- Murakami-Kojima M, Nakamichi N, Yamashino T, Mizuno T (2002) The APRR3 component of the clock-associated APRR1/TOC1 quintet is phosphorylated by a novel protein kinase belonging to the WNK family, the gene for which is also transcribed rhythmically in Arabidopsis thaliana. Plant Cell Physiol 43: 675–683 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins D, Heringa J (2000) T-COFFEE: a novel method for multiple sequence alignments. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1960) Circadian rhythms and the circadian organisation of living systems. Cold Spring Harb Symp Quant Biol 25: 159–184 [DOI] [PubMed] [Google Scholar]
- Pittendrigh CS (1981) Circadian systems: entrainment. In J Aschoff, ed, Handbook of Behavioral Neurobiology, Vol 4. Plenum, New York, pp 95–124
- Pittendrigh CS, Takamura T (1989) Latitudinal clines in the properties of a circadian pacemaker. J Biol Rhythms 4: 217–235 [PubMed] [Google Scholar]
- Plautz JD, Straume M, Stanewsky R, Jamison CF, Brandes C, Dowse HB, Hall JC, Kay SA (1997) Quantitative analysis of Drosophila period gene transcription in living animals. J Biol Rhythms 12: 204–217 [DOI] [PubMed] [Google Scholar]
- Salome PA, Michael TP, Kearns EV, Fett-Neto AG, Sharrock RA, McClung CR (2002) The out of phase 1 mutant defines a role for PHYB in circadian phase control in Arabidopsis. Plant Physiol 129: 1674–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer LA, Hennessy JM, Peixoto AA, Rosato E, Parkinson H, Costa R, Kyriacou CP (1997) Natural variation in a Drosophila clock gene and temperature compensation. Science 278: 2117–2120 [DOI] [PubMed] [Google Scholar]
- Serikawa KA, Martinez-Laborda A, Zambryski P (1996) Three knotted1-like homeobox genes in Arabidopsis. Plant Mol Biol 32: 673–683 [DOI] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, et al (2001) Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res 11: 959–980 [DOI] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101: 319–329 [DOI] [PubMed] [Google Scholar]
- Staiger D, Allenbach L, Salathia N, Fiechter V, Davis SJ, Millar AJ, Chory J, Fankhauser C (2003) The Arabidopsis SRR1 gene mediates phyB signaling and is required for normal circadian clock function. Genes Dev 17: 256–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stinchcombe JR, Weinig C, Ungerer M, Olsen KM, Mays C, Halldorsdottir SS, Purugganan MD, Schmitt J (2004) A latitudinal cline in flowering time in Arabidopsis thaliana modulated by the flowering time gene FRIGIDA. Proc Natl Acad Sci USA 101: 4712–4717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straume M, Frasier-Cadoret SG, Johnson ML (1991) Least squares analysis of fluorescence data. In JR Lakowicz, ed, Topics in Fluorescence Spectroscopy, Vol 2: Principles. Plenum, New York, pp 117–240
- Strayer C, Oyama T, Schultz TF, Raman R, Somers DE, Mas P, Panda S, Kreps JA, Kay SA (2000) Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289: 768–771 [DOI] [PubMed] [Google Scholar]
- Swarup K, Alonso-Blanco C, Lynn JR, Michaels SD, Amasino RM, Koornneef M, Millar AJ (1999) Natural allelic variation identifies new genes in the Arabidopsis circadian system. Plant J 20: 67–77 [DOI] [PubMed] [Google Scholar]
- van Ooijen JW (1999) LOD significance thresholds for QTL analysis in experimental populations of diploid species. Heredity 83: 613–624 [DOI] [PubMed] [Google Scholar]
- van Ooijen JW, Maliepaard C (2000) MapQTL Version 4.0: Software for the Calculation of QTL Positions on Genetic Maps. CPRO-DLO, Wageningen, The Netherlands
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.