Abstract
In Gouda and Cheddar type cheeses the amino acid conversion to aroma compounds, which is a major process for aroma formation, is essentially due to lactic acid bacteria (LAB). In order to evaluate the respective role of starter and nonstarter LAB and their interactions in cheese flavor formation, we compared the catabolism of phenylalanine, leucine, and methionine by single strains and strain mixtures of Lactococcus lactis subsp. cremoris NCDO763 and three mesophilic lactobacilli. Amino acid catabolism was studied in vitro at pH 5.5, by using radiolabeled amino acids as tracers. In the presence of α-ketoglutarate, which is essential for amino acid transamination, the lactobacillus strains degraded less amino acids than L. lactis subsp. cremoris NCDO763, and produced mainly nonaromatic metabolites. L. lactis subsp. cremoris NCDO763 produced mainly the carboxylic acids, which are important compounds for cheese aroma. However, in the reaction mixture containing glutamate, only two lactobacillus strains degraded amino acids significantly. This was due to their glutamate dehydrogenase (GDH) activity, which produced α-ketoglutarate from glutamate. The combination of each of the GDH-positive lactobacilli with L. lactis subsp. cremoris NCDO763 had a beneficial effect on the aroma formation. Lactobacilli initiated the conversion of amino acids by transforming them mainly to keto and hydroxy acids, which subsequently were converted to carboxylic acids by the Lactococcus strain. Therefore, we think that such cooperation between starter L. lactis and GDH-positive lactobacilli can stimulate flavor development in cheese.
In Gouda and Cheddar type cheeses, the microbial enzymes are of main importance for development of flavor. The starter culture of those cheeses basically consists of Lactococcus lactis subsp. lactis and L. lactis subsp. cremoris. These microorganisms, in addition to their essential role in milk acidification, contribute considerably to the formation of cheese flavor by production of peptides and amino acids, which contribute to basic taste of cheese, and subsequent conversion of amino acids to aroma compounds.
Nonstarter lactobacilli (NSLAB) present in cheese are adventitious microflora, which originate either from the factory environment or from the milk. Lactobacilli are found in cheese in low numbers (<50 CFU g−1) at the day of manufacture and become the dominant viable microflora (107 to 108 CFU g of cheese−1) in the mature cheese (10). Many authors have postulated that NSLAB play a significant role in the development of cheese flavor since Cheddar cheese made in aseptic vats lack the full mature flavor (18, 29). In the last decade, many studies have dealt with the effect of adjunct lactobacilli on cheese flavor development (6, 14, 21, 26). Most authors have reported that cheese made with adjunct is characterized by higher concentrations of free amino acids and enhanced flavor intensity (14, 22). However, the selection of adjunct strains seems to be a crucial step since deliberate addition of NSLAB to cheese milk has had a varying success (6). Indeed, some strains do not have any effect on cheese flavor, while others can significantly accelerate flavor development (6).
Both starter lactococci and NSLAB have the potential to produce aroma compounds from amino acids (7, 13, 23, 33). The conversion of amino acids to aroma compounds is mainly initiated by amino acid transamination, which requires an α-keto acid as an amino group acceptor for the aminotransferases. Keto acids produced by transamination can further be transformed to various aroma compounds (33). Aminotransferase activity is widespread among NSLAB as well as in lactococci (30, 34). However, the presence of the α-keto acid is the limiting factor for amino acid transamination by most LAB. Therefore, addition of α-ketoglutarate (α-KG) to the cheese curd enhanced amino acid catabolism and consequently increased cheese flavor production (3, 31). It has also been shown that a genetically modified L. lactis strain, which overproduced a glutamate dehydrogenase (GDH), was capable of producing α-KG from glutamate (Glu) and produced aroma compounds from amino acids in a cheese model (20). Natural GDH activity has been reported in some NSLAB such as Lactobacillus plantarum and Lactobacillus fermentum (16). This strain-specific activity could possibly explain the beneficial effect of some adjunct lactobacilli on cheese aroma formation.
In Gouda and Cheddar type cheese, many interactions may occur between starter and NSLAB. The reports on microbial interactions in cheese deal mostly with the growth inhibition or stimulation of some bacterial strains by others, however, mechanisms of such actions still remain unclear (12). Nevertheless, it has been suggested that lactic starters may influence the development of adventitious NSLAB, which may affect, favorably or unfavorably, the development of cheese flavor (2, 15, 17, 19, 25). Several reports show that these two populations can be present together in high numbers (107 to 108 CFU g−1 of cheese) for at least 2 months of cheese ripening, and especially when adjunct strain was used in cheese production (8, 14). Therefore, it is likely that cooperation between L. lactis and NSLAB in the formation of aroma compounds take place in cheese.
In the present study, we compared the ability of L. lactis subsp. cremoris NCDO763 and GDH-positive or -negative strains of nonstarter lactobacilli, to produce aroma compounds from phenylalanine (Phe), leucine (Leu), and methionine (Met) in vitro. We evaluated, in addition, the impact of combination of strains on the amino acid catabolism. Finally, we evaluated the effect of salt on amino acid catabolism by selected strains.
MATERIALS AND METHODS
Bacterial strains.
Lactococcus lactis subsp. cremoris NCDO763 was obtained from the National Collection of Food Bacteria (Shinfield, Reading, England). Lactobacillus paracasei 1244 was obtained from CNRZ culture collection (INRA, Jouy-en-Josas, France). L. paracasei subsp. paracasei INF15D, isolated from Norvegia cheese was obtained from the culture collection of the Department of Food Science (Agricultural University of Norway, Aas, Norway). Lactobacillus casei 2756, isolated from Cheddar cheese was obtained from Department of Food Chemistry (University College, Cork, Ireland).
Preparation of cells for amino acid degradation.
L. lactis subsp. cremoris NCDO763 was grown (2% inoculum) in 10% NILAC milk (NIZO, Ede, The Netherlands) buffered with 75 mM sodium β-glycerophosphate. Lactobacillus strains were grown (1% inoculum) in MRS broth (Difco Laboratories, Detroit, Mich.). Cells, in the late exponential growth phase, were harvested by centrifugation (4,100 × g for 15 min at 4°C), washed twice with 50 mM sodium glycerophosphate buffer (pH 7.5), and suspended in the same buffer to an optical density at 480 nm (OD480) of 200 (OD480 = 2 in dilution 1/100). The cell suspensions (200 μl) were stored at −80°C until use.
Amino acid catabolism.
The catabolism of Phe, Leu, and Met by single strains and mixed strains were investigated in four different reaction mixtures. The basic reaction mixture (reaction mixture I) consisted of 70 mM potassium phosphate buffer (pH 5.5), 3 mM unlabeled amino acid, 0.05 μM of radiolabeled amino acid (l-[2,3,4,5,6-3H]phenylalanine, l-[4,5-3H]leucine or l-[methyl-3H]methionine), and 0.05 mM pyridoxal phosphate. The second reaction mixture (reaction mixture II) contained in addition 10 mM α-KG, while the third (reaction mixture III) contained both 10 mM α-KG and 1.5% NaCl in order to evaluate the effect of salt on amino acid catabolism. In the fourth reaction mixture (reaction mixture IV), 10 mM Glu was added to the basic reaction mixture (reaction mixture I).
For incubations with single strains, 50 μl of cell suspension was added to the different reaction mixtures, whereas 25 μl of each of the cell suspension of Lactococcus and Lactobacillus were used for the incubations with mixed strains. The final volume of the reaction mixture was 0.5 ml. Incubations were performed for 40 h at 37°C. Aliquots (100 μl) of the reaction mixture were taken after 0, 10, 20, and 40 h of incubation and cells were removed by centrifugation (8,000 × g for 5 min). The metabolites were then analyzed by reverse-phase and ion-exclusion high-performance liquid chromatography (IE-HPLC), with both UV and radioactivity detection as previously described (32, 34). Samples containing Phe were analyzed by reverse-phase HPLC (RP-HPLC) only. The quantitative analysis of Phe degradation was made by calculating the percentage of radioactivity associated with each peak (amino acid and metabolites). Samples containing Leu or Met were initially analyzed by RP-HPLC, which allowed the separation of amino acid from metabolites (but the different metabolites were not well separated) and followed by IE-HPLC which allowed the separation of different metabolites (but the amino acid was not eluted from the column). The quantitative analysis of Leu or Met degradation was made by calculating the percentage of total radioactivity of the sample associated with the amino acid peak in RP-HPLC and with each metabolite peak in IE-HPLC. Identification was made by comparison of the retention times with those of appropriate standard compounds as previously described (32, 34). The standard components were obtained from Sigma Chemical Co. (St. Louis, Mo.). The 3-methylthiopropionic acid was prepared by acidic hydrolysis of the methyl ester of 3-methylthiopropionate obtained from Sigma (9).
Preparation of cell extract for GDH determination.
Five hundred microliters of the cell suspension (OD480 of 200) was added to 500 μl of 50 mM sodium glycerophosphate buffer (pH 7.5) containing 0.6 g of glass beads (106 μm). The cells were disrupted in a Mini-beadbeater 8TM cell disrupter (Biospec Products, Bartlesville, Ill.) three times for 1 min each, with 1 min of cooling on ice after each treatment. After centrifugation (14 000 × g for 20 min at 4°C) the supernatant was filtered through a 0.45-μm-pore-size filter (hydrophilic polyvinylidene difluoride; Millipore Co., Bedford, Mass.) and was thereafter used as cell extract (CFE). The protein concentration in the CFE was determined by the Bradford assay, by using bovine serum albumin as the standard (5).
Determination of GDH activity.
The GDH activities of the CFE were determined by using the test based on the colorimetric glutamic acid assay of Boehringer (4). In this test, the reduced cofactor (NADH or NADPH) produced by oxidative deamination of Glu reacts with iodonitrotetrazolium in the presence of diaphorase to produce a colorimetric product. The test was performed by using enzyme-linked immunosorbent assay plates. The reaction mixture consisted of 40 μl of 50 mM potassium phosphate-50 mM TEA buffer (pH 9.0) containing 1% of Triton X-100 (Promega Corporation, Madison, Wis.), 20 μl of 100 mM Glu, 20 μl of 2 mM iodonitrotetrazolium, 20 μl of diaphorase (1.76 U/ml), 20 μl of NADP+ (13.8 mM) or NAD+ (17.33 mM), and 180 μl of water (300 μl in total). The colorimetric reaction started immediately by the addition of 30 μl of CFE to the reaction mixture. The changes in concentration of NAD(P)H were measured after incubation for 1 h at 37°C, by measuring the absorbance at 492 nm. In order to subtract the nonspecific reactions, which could produce the reduced cofactors, a control test was prepared, for each strain, without addition of Glu. The GDH activity was expressed by the increase in Abs at 492 nm, per mg of protein of CFE and per min of reaction.
Statistical analysis.
The results are the mean of two or three replicates. The effect of α-KG, Glu and salt on amino acid degradation by single strain was evaluated by one-way analysis of variance with α-KG, Glu and salt as experimental factors, respectively. Two-way analysis of variance, with the Lactococcus strain and each of lactobacilli as experimental factors, respectively, was performed to define the significance of interactions between strains in formation of carboxylic acids from Phe, Leu, and Met. The analyses were performed using Minitab software (Minitab Inc., State College, Penn.).
RESULTS
Effect of α-KG and salt on amino acid catabolism by single strains.
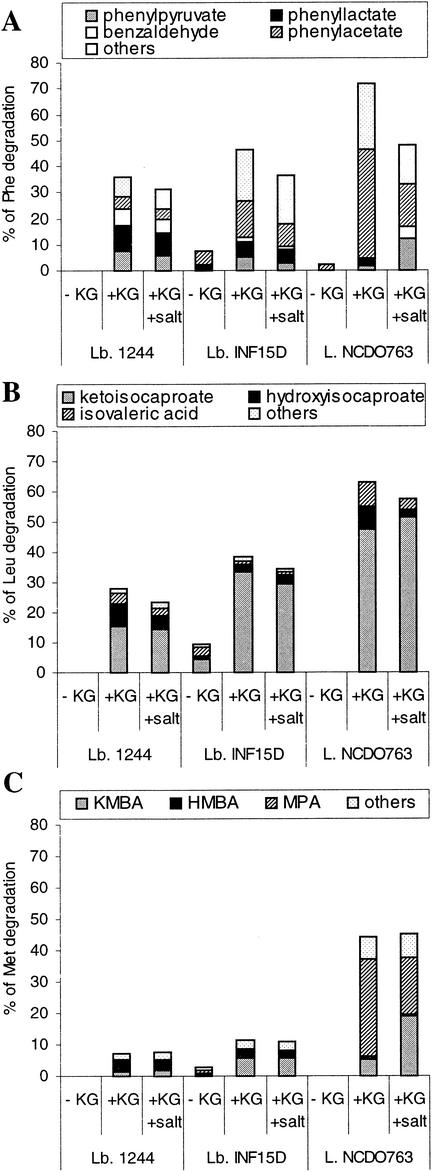
The amino acid catabolism by L. lactis subsp. cremoris NCDO763 and three lactobacillus strains was studied in vitro using radiolabeled Phe, Leu, and Met as tracers. Figure 1 shows the effect of α-KG and salt addition on the amino acid degradation and metabolites production by single strains.
FIG. 1.
Effect of α-KG and salt on amino acid catabolism by single strains of L. paracasei 1244, L. paracasei subsp. paracasei INF15D, and L. lactis subsp. cremoris NCDO763. (A) Metabolites produced from Phe; (B) metabolites produced from Leu; (C) metabolites produced from Met (KMBA, HMBA, and MPA). One hundred percent of degraded amino acids corresponds to a concentration of 3 mM.
L. paracasei subsp. paracasei INF15D as well as L. casei 2756 (results not shown) degraded, to a certain extent, Phe, Leu, and Met in reaction mixture without α-KG (I), while only little or no amino acid degradation was observed with L. lactis subsp. cremoris NCDO763 and L. paracasei 1244. In general, the results obtained for L. casei 2756 in different reaction mixtures were similar to those obtained for L. paracasei subsp. paracasei INF15D and are not shown. Addition of α-KG to the reaction medium increased the degradation of amino acids significantly (P ≤ 0.05) by both L. lactis subsp. cremoris NCDO763 and the lactobacillus strains. In the reaction mixture with α-KG, L. lactis subsp. cremoris NCDO763 degraded more Phe, Leu, and Met than the tested lactobacilli and produced more carboxylic acids, which are potent aroma compounds. Indeed, high amounts of phenyl acetate and methylthiopropionate (MPA) were formed as a result of the degradation of Phe and Met, respectively. The lactobacillus strains produced mainly keto acids (phenylpyruvate, ketoisocaproate, and ketomethylthiobutyrate [KMBA]) and hydroxy acids (phenyllactate, hydroxyisocaproate, and hydroxymethylthiobutyrate [HMBA]), which are not aroma compounds, and smaller amounts of carboxylic acids.
L. lactis subsp. cremoris NCDO763 and the lactobacillus strains produced in addition some other metabolites, especially from Phe and Met. Most of these metabolites are not yet identified, however, a few were aldehydes and alcohols derived from the corresponding amino acids (Fig. 1 to 3). Salt at concentration of 1.5% NaCl slightly inhibited the degradation of Phe and Leu by all tested strains and significantly (P ≤ 0.05) reduced the formation of carboxylic acids, especially from Phe and Met, by L. lactis subsp. cremoris NCDO763.
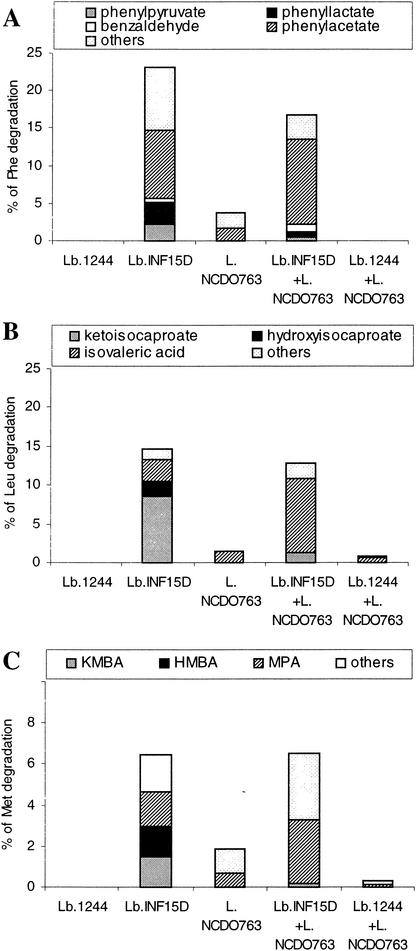
FIG. 3.
Percent amino acid degradation in reaction medium added Glu, by single strains of L. paracasei 1244, L. paracasei subsp. paracasei INF15D, and L. lactis subsp. cremoris NCDO763 and strain mixtures (L. paracasei subsp. paracasei INF15D with L. lactis subsp. cremoris NCDO763 and L. paracasei 1244 with L. lactis subsp. cremoris NCDO763). (A) Metabolites produced from Phe; (B) metabolites produced from Leu; (C) metabolites produced from Met (MBA, HMBA, and MPA). Significant interactions (P ≤ 0.05) in formation of isovalerate and MPA were found for mixtures of L. lactis subsp. cremoris NCDO763 and L. paracasei subsp. paracasei INF15D. One hundred percent of degraded amino acids corresponds to a concentration of 3 mM.
Amino acid degradation by strain mixtures in the presence of α-KG and salt.
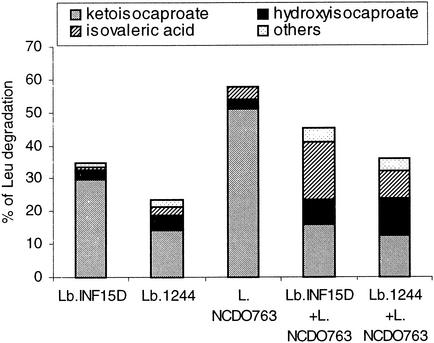
Degradation of amino acids by the combination of each strain of Lactobacillus with L. lactis subsp. cremoris NCDO763 was studied in reaction mixtures supplemented with α-KG (reaction mixture II) and both α-KG and NaCl (reaction mixture III). In general, the percentage of amino acid degradation obtained in incubations with mixed strains was the average of the degradations obtained with each single strain. Also the quantity of carboxylic acids produced by strain mixtures was the average of the amount produced by each single strain. This was, however, different for Leu degradation in presence of α-KG and NaCl, where a significant interaction effect (P ≤ 0.05) was found, and mixture of strains formed higher levels of isovalerate than each single strain (Fig. 2)
FIG. 2.
Leu degradation (percent) in the presence of α-KG and salt, by single strains of L. paracasei 1244, L. paracasei subsp. paracasei INF15D, and L. lactis subsp. cremoris NCDO763 and strain mixtures (L. paracasei INF15D with L. lactis subsp. cremoris NCDO763 and L. paracasei 1244 with L. lactis subsp. cremoris NCDO763). One hundred percent of degraded Leu corresponds to a concentration of 3 mM.
GDH activity and amino acid degradation in the presence of Glu.
The GDH activities of the experimental strains are presented in Table 1. Both L. paracasei subsp. paracasei INF15D and L. casei 2756 exhibited a high NADP-specific GDH activity, while no GDH activity was detected for L. paracasei 1244. Only very low NAD-specific GDH activity was found for L. lactis subsp. cremoris NCDO763.
TABLE 1.
Occurrence of NAD and NADP-specific GDH activitya in strains
| Species | Strain | GDH
|
|
|---|---|---|---|
| NADP+ | NAD+ | ||
| Lactobacillus | INF15D | 0.18 | 0.00 |
| Lactobacillus | 2756 | 0.10 | 0.00 |
| Lactobacillus | 1244 | 0.00 | 0.00 |
| Lactococcus | NCDO763 | 0.00 | 0.02 |
Specific activities of GDH are expressed as increase in A492 per milligram of protein of CFE and per minute of reaction.
In order to verify the role of GDH in the amino acid catabolism, degradation of Phe, Leu, and Met was studied in reaction mixtures I (without Glu) and IV (with Glu) (Table 2). The apparent but low amino acid degradation observed in reaction mixture I was probably due to utilization of intracellular pool of Glu by GDH-possessing lactobacilli. Addition of Glu significantly (P ≤ 0.05) increased the degradation of Phe, Leu, and Met by L. paracasei subsp. paracasei INF15D and Leu and Phe by L. casei 2756, both GDH-positive strains. A minor increase in amino acid degradation was observed in the presence of Glu with L. lactis subsp. cremoris NCDO763, which exhibited only very low NAD-dependent GDH activity. No amino acid degradation was found with L. paracasei 1244, the GDH-negative strain.
TABLE 2.
Amino acid degradation (%) by experimental strains in reaction mixture without glutamate (−Glu) or with added glutamate (+Glu)
| Species | Strain | Phe
|
Leu
|
Met
|
|||
|---|---|---|---|---|---|---|---|
| −Glu | +Glu | −Glu | +Glu | −Glu | +Glu | ||
| Lactobacillus | INF15D | 7.5 | 23.1a | 9.7 | 14.6a | 2.6 | 6.5a |
| Lactobacillus | 2756 | 6.3 | 18.2a | 6.4 | 13.3a | 3.8 | 6.0 |
| Lactobacillus | 1244 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Lactococcus | NCDO763 | 2.7 | 3.8 | 0.0 | 1.5 | 0.0 | 1.8 |
Significant effect of Glu addition (P ≤ 0.05).
Effect of the combination of L. lactis subsp. cremoris NCDO763 with the lactobacilli on the amino acid conversion to aroma compounds.
Amino acid catabolism by single strains or mixtures of strains was studied in reaction mixture IV (with Glu). The results obtained for the association of L. casei 2576 with L. lactis subsp. cremoris NCDO763 (data not shown) were similar to those obtained for the association of L. paracasei subsp. paracasei INF15D with the same Lactococcus strain (Fig. 3). In the presence of Glu, GDH-positive strains of L. paracasei subsp. paracasei INF15D and L. casei 2756 (data not shown), degraded amino acids mainly to keto acids and hydroxy acids, with the exception of Phe, where formation of phenylacetate was relatively dominating. However, GDH-positive strains produced significantly (P ≤ 0.05) more carboxylic acids from Met and Leu when compared with the reaction mixture containing α-KG (Fig. 1 and 3). L. lactis subsp. cremoris NCDO763 only slightly degraded the amino acids and produced mainly the corresponding carboxylic acids. Combining each of GDH-positive lactobacilli with L. lactis subsp. cremoris NCDO763 resulted in high production of carboxylic acids, especially from Leu and Met. In contrast, the degradation by the mixture of the GDH-negative L. paracasei 1244 strain with L. lactis subsp. cremoris was less than that observed with L. lactis subsp. cremoris NCDO763 alone, probably, because the quantity of Lactococcus cells in the cell mixture was half of the quantity used in the assay with L. lactis subsp. cremoris NCDO763 alone.
DISCUSSION
Most of the major aroma compounds of Cheddar and Gouda cheeses are derived from the catabolism of aromatic amino acids, branched-chain amino acids and Met (3, 33). Although NSLAB often have been associated with flavor development in Cheddar cheese, recent studies on amino acid catabolism did not reveal any particular ability of NSLAB to convert amino acids to aroma compounds when compared with starter lactococci (1, 13, 30). The amino acid conversion, in both lactobacilli and lactococci, is essentially initiated by aminotransferases, and the level of aminotransferase activities measured in lactobacillus strains have been shown to be lower than that reported for L. lactis subsp. cremoris NCDO763 (1, 30). The present study also showed that at pH 5.5 and in the presence of α-KG, Lactobacillus strains degraded less amino acids and produced less aroma compounds from Leu, Phe and Met than L. lactis subsp. cremoris NCDO763. The lactobacilli used, produced high levels of keto acids and hydroxy acids, which are not aroma compounds, while L. lactis subsp. cremoris NCDO786 mainly produced carboxylic acids, which are potent aroma compounds. Former studies report that under cheese like conditions, at high concentrations of salt (4 to 5% NaCl in the water phase) and low pH (5.0 to 5.5), the activities of intracellular enzymes, including aminotransferases, are considerably reduced (7, 11, 28). In our study, salt addition (1.5% NaCl) caused some decrease in the degradation of Phe and Leu; however, the main effect was on the conversion of keto acids to carboxylic acids in incubations with the Lactococcus strain. Overall, in the reaction mixtures containing α-KG, the combination of lactobacilli with lactococci did not highly affect the formation of aroma compounds, when compared with L. lactis subsp. cremoris NCDO763 alone.
However, our study revealed an interesting feature of some lactobacillus strains. Two lactobacillus strains that exhibited GDH activity were capable of degrading amino acids in a reaction mixture containing Glu. In contrast, L. paracasei strain without GDH activity did not degrade amino acids in the same reaction mixture. These results combined with the results obtained in another study on strains of L. plantarum and L. lactis demonstrate a high correlation between GDH activity in lactic acid bacteria and their ability to catabolize amino acids in the presence of Glu (24). In our study, the GDH-positive lactobacilli produced more carboxylic acids in the medium containing Glu than in the medium containing α-KG. A similar observation was previously reported for a genetically modified L. lactis strain, overproducing a GDH (20). So far, the mechanism of such behavior is not yet understood, but it may be related to a change in the intracellular oxido-reduction potential caused by the oxidative deamination of Glu to α-KG.
Moreover, our study demonstrated the beneficial effect of combining GDH-positive Lactobacillus with L. lactis subsp. cremoris NCDO763 in the formation of carboxylic acids in a reaction mixture containing Glu. The carboxylic acids, such as isovaleric acid are very potent aroma compounds that contribute considerably to cheese flavor (27). We propose, therefore, the following model for the cooperation between GDH-positive lactobacilli and L. lactis subsp. cremoris NCDO763 in formation of carboxylic acids from amino acids. The GDH-positive lactobacilli initiated amino acid conversion by transaminating amino acids to α-keto acids and also partially reduced α-keto acids to hydroxy acids via a reversible reaction. Subsequently L. lactis subsp. cremoris NCDO763 completed the conversion by transforming α-keto acids and hydroxy acids to carboxylic acids. It is rather unlikely that α-KG produced by lactobacilli was transported out of the cells and then utilized by L. lactis subsp. cremoris NCDO763. Actually, α-KG initially produced inside the cells of GDH-positive lactobacilli was probably rapidly used in transamination reactions that regenerate Glu. Furthermore, even if α-KG was exported by lactobacilli to the medium and then taken up by Lactococcus strain, its concentration in lactococcal cell would become very low and probably not sufficient for amino acid transamination.
It is obvious that many interactions occur in cheese between microorganisms in flavor formation, however, our knowledge in this area is limited. This is mostly due to the complexity of the microbial ecosystem found in cheese. However, recently the formation of the malty aroma compound 3-methylbutanal has been achieved in milk culture by combining two lactococcal strains: one producing free amino acids and the other degrading leucine (2). Our study, performed under pH and salt conditions simulating a cheese ripening environment, demonstrates that the cooperation between starter culture and certain lactobacilli, within a metabolic pathway, is possible and can lead to formation of cheese aroma compounds. However, now it is necessary to carry out the cheese trials since the experimental conditions in vitro differ considerably from conditions found in cheese. Nevertheless, the results of this study may partially explain the flavoring properties of certain lactobacilli such as INF15D. This strain used as adjunct produced a good-quality Norvegia cheese (21). Experiments in sterile-cheese models using GDH-positive and GDH-negative lactobacilli are now in progress to determine the respective role of GDH activity and the cooperation with L. lactis in the formation of volatile aroma compounds from amino acids.
Acknowledgments
The financial grants from the French-Norwegian collaboration research program Aurora and the Research Council of Norway are highly acknowledged.
REFERENCES
- 1.Amarita, F., T. Requena, G. Taborda, L. Amigo, and C. Pelaez. 2001. Lactobacillus casei and Lactobacillus plantarum initiate catabolism of methionine by transamination. J. Appl. Microbiol. 90:971-978. [DOI] [PubMed] [Google Scholar]
- 2.Ayad, E. H. E., A. Verheul, W. J. M. Engels, J. T. M. Wouters, and G. Smit. 2001. Enhanced flavour formation by combination of selected lactococci from industrial and artisanal origin with focus on completion of a metabolic pathway. J. Appl. Microbiol. 90:59-67. [DOI] [PubMed] [Google Scholar]
- 3.Banks, J. M., M. Yvon, J. C. Gripon, M. A. de la Fuente, E. Y. Brechany, A. G. Williams, and D. D. Muir. 2001. Enhancement of amino acid catabolism in Cheddar cheese using α-ketoglutarate: amino acid degradation in relation to volatile compounds and aroma character. Int. Dairy. J. 11:235-243. [Google Scholar]
- 4.Beutler, H. O. 1985. Metabolites: lipids, amino acids and related compounds, p. 369-376. In H. U. Bergmeyer (ed.), Methods of enzymatic analysis, 3 ed., vol. VIII. Verlag Chemie, Deerfield Beach, Fla.
- 5.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilising the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Crow, V., B. Curry, and M. Hayes. 2001. The ecology of non-starter lactic acid bacteria (NSLAB) and their use as adjuncts in New Zealand Cheddar. Int. Dairy. J. 11:275-283. [Google Scholar]
- 7.Dias, B., and B. Weimer. 1998. Conversion of methionine to thiols by lactococci, lactobacilli, and brevibacteria. Appl. Environ. Microbiol. 64:3320-3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fenelon, M. A., P. O'Connor, and T. P. Guinee. 2000. The effect of fat content on the microbiology and proteolysis in Cheddar cheese during ripening. J. Dairy Sci. 83:2173-2183. [DOI] [PubMed] [Google Scholar]
- 9.Fieser, L., and M. Fieser. 1956. Organic chemistry. Chapman and Hall, London, United Kingdom.
- 10.Fox, P. F., P. L. H. McSweeney, and C. M. Lynch. 1998. Significance of non-starter lactic acid bacteria in Cheddar cheese. Aust. J. Dairy Technol. 53:83-89. [Google Scholar]
- 11.Gao, S., D. H. Oh, J. R. Broadbent, M. E. Johnson, B. C. Weimer, and J. L. Steele. 1997. Aromatic amino acid catabolism by lactococci. Lait 77:371-381. [Google Scholar]
- 12.Jimeno, J., M. J. Lazaro, and H. Sollberger. 1995. Antagonistic interactions between propionic acid bacteria and non-starter lactic acid bacteria. Lait 75:401-413. [Google Scholar]
- 13.Kieronczyk, A., S. Skeie, K. Olsen, and T. Langsrud. 2001. Metabolism of amino acids by resting cells of non-starter lactobacilli in relation to flavour development in cheese. Int. Dairy. J. 11:217-224. [Google Scholar]
- 14.Lynch, C. M., D. D. Muir, J. M. Banks, and P. McSweeney. 1999. Influence of adjunct cultures of Lactobacillus paracasei ssp paracasei or Lactobacillus plantarum on cheddar cheese ripening. J. Dairy. Sci. 82:1618-1628. [Google Scholar]
- 15.Martley, F. G., and V. L. Crow. 1993. Interactions between non-starter microorganisms during cheese manufacture and ripening. Int. Dairy J. 3:461-483. [Google Scholar]
- 16.Morishita, T., and M. Yajima. 1995. Incomplete operation of biosynthetic and bioenergetic functions of the citric acid cycle in multiple auxotrophic lactobacilli. Biosci. Biotechnol. Biochem. 59:251-255. [Google Scholar]
- 17.Muir, D. D., J. M. Banks, and E. A. Hunter. 1996. Sensory properties of cheddar cheese: effect of starter type and adjunct. Int. Dairy J. 6:407-423. [Google Scholar]
- 18.Perry, K. D., and W. A. McGillivray. 1964. The manufacture of normal and starter-free cheddar cheese under controlled bacteriological conditions. J. Dairy Res. 31:155-165. [Google Scholar]
- 19.Rapposch, S., F. Eliskases-Lechner, and W. Ginzinger. 1999. Growth of facultatively heterofermentative lactobacilli on starter cell suspensions. Appl. Environ. Microbiol. 65:5597-5599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rijnen, L., P. Courtin, J. C. Gripon, and M. Yvon. 2000. Expression of a heterologous glutamate dehydrogenase gene in Lactococcus lactis highly improves the conversion of amino acids to aroma compounds. Appl. Environ. Microbiol. 66:1354-1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skeie, S., C. Lindberg, and J. Narvhus. 2001. Development of amino acids and organic acids in Norvegia, influence of milk treatment and adjunct Lactobacillus. Int. Dairy J. 11:399-411. [Google Scholar]
- 22.Swearingen, P. A., D. J. O'Sullivan, and J. J. Warthesen. 2001. Isolation, characterization, and influence of native, non-starter lactic acid bacteria on Cheddar cheese quality. J. Dairy Sci. 84:50-59. [DOI] [PubMed] [Google Scholar]
- 23.Tammam, J. D., A. G. Williams, J. Noble, and D. Lloyd. 2000. Amino acid fermentation in non-starter Lactobacillus spp. isolated from Cheddar cheese. Lett. Appl. Microbiol. 30:370-374. [DOI] [PubMed] [Google Scholar]
- 24.Tanous, C., A. Kieronczyk, S. Helinck, E. Chambellon, and M. Yvon. 2002. Glutamate dehydrogenase activity: a major criterion for the selection of flavour-producing lactic acid bacteria strains. Antonie Leeuwenhoek 82:271-278. [PubMed] [Google Scholar]
- 25.Thomas, T. D. 1987. Cannibalism among bacteria found in cheese. N. Z. J. Dairy Sci. Technol. 22:215-219. [Google Scholar]
- 26.Tungjaroenchai, W., M. A. Drake, and C. H. White. 2001. Influence of adjunct cultures on ripening of reduced fat Edam cheeses. J. Dairy Sci. 84:2117-2124. [DOI] [PubMed] [Google Scholar]
- 27.Urbach, G. 1997. The flavour of milk and dairy products. 2. Cheese: contribution of volatile compounds. Int. J. Dairy. Technol. 50:79-89. [Google Scholar]
- 28.Weimer, B., B. Dias, M. Ummadi, J. Broadbent, C. Brennand, J. Jaegi, M. Johnson, F. Milani, J. Steele, and D. V. Sisson. 1997. Influence of NaCl and pH on intracellular enzymes that influence Cheddar cheese ripening. Lait 77:383-398. [Google Scholar]
- 29.Wijesundera, C., M. Roberts, and G. K. Y. Limsowtin. 1997. Flavour development in aseptic cheese curd slurries prepared with single-strain starter bacteria in the presence and absence of adjuncts. Lait 77:121-131. [Google Scholar]
- 30.Williams, A. G., J. Noble, and J. M. Banks. 2001. Catabolism of amino acids by lactic acid bacteria isolated from Cheddar cheese. Int. Dairy. J. 11:203-215. [Google Scholar]
- 31.Yvon, M., S. Berthelot, and J. C. Gripon. 1998. Adding α-ketoglutarate to semi-hard cheese curd highly enhances the conversion of amino acids to aroma compounds. Int. Dairy J. 8:889-898. [Google Scholar]
- 32.Yvon, M., E. Chambellon, A. Bolotin, and F. Roudot Algaron. 2000. Characterization and role of the branched-chain aminotransferase (BcaT) isolated from Lactococcus lactis subsp. cremoris NCDO 763. Appl. Environ. Microbiol. 66:571-577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yvon, M., and L. Rijnen. 2001. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 11:185-201. [Google Scholar]
- 34.Yvon, M., S. Thirouin, L. Rijnen, D. Fromentier, and J. C. Gripon. 1997. An aminotransferase from Lactococcus lactis initiates conversion of amino acids to cheese flavor compounds. Appl. Environ. Microbiol. 63:414-419. [DOI] [PMC free article] [PubMed] [Google Scholar]