Abstract
Objectives: The paper (1) introduces health sciences librarians to the main concepts and principles of the Semantic Web (SW) and (2) briefly reviews a number of projects on the handling of biomedical information that uses SW technology.
Methodology: The paper is structured into two main parts. “Semantic Web Technology” provides a high-level description, with examples, of the main standards and concepts: extensible markup language (XML), Resource Description Framework (RDF), RDF Schema (RDFS), ontologies, and their utility in information retrieval, concluding with mention of more advanced SW languages and their characteristics. “Semantic Web Applications and Research Projects in the Biomedical Field” is a brief review of the Unified Medical Language System (UMLS), Generalised Architecture for Languages, Encyclopedias and Nomenclatures in Medicine (GALEN), HealthCyberMap, LinkBase, and the thesaurus of the National Cancer Institute (NCI). The paper also mentions other benefits and by-products of the SW, citing projects related to them.
Discussion and Conclusions: Some of the problems facing the SW vision are presented, especially the ways in which the librarians' expertise in organizing knowledge and in structuring information may contribute to SW projects.
INTRODUCTION
The Web has transformed the concept of information. The Web continues to grow in size and quantity, and, above all, it continues to change in structural complexity and underlying architecture.
The rise of extensible markup language (XML) and metadata standards such as Dublin Core (DC), along with the initiatives of the World Wide Web Consortium (W3C) to create a “Semantic Web,” point toward a new world of Web-based information, a world in which information will be machine understandable and machine readable. In the vision of the Semantic Web (SW) outlined by Berners-Lee et al. in a milestone article [1], intelligent search programs (also called “software agents”) are able to draw sophisticated inferences from metadata attached to Web-based information. This vision (and its underlying technology) shows great promise, though its impact has yet to be fully realized. To this end, much research work is currently being carried out and significant funds are being allocated, both in Europe and the United States.
This paper has two purposes: First, the authors wish to provide a high-level, comprehensive introduction to the main concepts of SW technology, from the perspective of medical librarians, while pinpointing links to the concepts used in medical information science. Second, the authors provide a brief review of several applications and running projects, involving the handling of medical information, that use or plan to use SW technology. The authors emphasize the way the expertise of information scientists can contribute to these projects. It is the authors' strong belief that medical librarians will continue to play an essential role in shaping the future of health information and that their expertise is a great asset even in projects whose core belongs to artificial intelligence.
SEMANTIC WEB (SW) TECHNOLOGY
Most of today's Web content is suitable for human consumption. Typical uses of the Web today involve humans seeking and consuming information, searching and getting in touch with other humans. The software tools to support these activities are not particularly well developed; in fact, the main ones remain search engines. Moreover, the technology of these tools remains roughly the same, and Web content outgrows technological progress. In particular, information retrieval is not very well supported. The major obstacle is that, at present, the meaning of the Web content is not machine accessible [2], in the sense that computers cannot interpret words, sentences, and the relationships between them.
The main goal of the SW is to add logic to the current Web, in other words, express the meaning of data, the properties of objects, and the complex relationships existing between them by a series of formal rules, which would make information accessible to machines. Machine accessibility should be understood as representing information in such a way that it is possible to make queries based on the meaning (i.e., semantics) of the data, independent of the form in which the information is presented.
Languages such as XML and other standards derived from it achieve this aim, but only in a limited way. Consider the following three XML tags:
1. <article name=“Duodenal ulcer”>
<author>Smith</author>
</article>
2. <author name=“Smith”>
<article>Duodenal ulcer</article>
</author>
3. <bibRecord>
<author>Smith</author>
<article>Duodenal ulcer</article>
</bibRecord>
All the above tags express the same meaning: an article called “Duodenal ulcer” authored by Smith. However, for a machine, the three representations are different, because the order of encapsulation (nesting) of the tags is different. The above representation has very little semantics.
XML is, however, sufficient for many applications, where structures are relatively simple and the enforcement of a strict syntax or format can be assured. Numerous examples of such representations exist. The most familiar perhaps is the display of PubMed records in XML format, but the fact is that unless the exact structure of the XML is respected, namely the document type description (DTD) specified by the National Library of Medicine (NLM), the code is not validated.
The SW approach is to add another layer on top of the XML standard, adding more meaning to the encoded information. This extra layer is represented by standards such as Resource Description Framework (RDF)/RDF Schema (RDFS) and Web Ontology Language (OWL).
It is important to note that SW should be seen as “an extension of the current Web” [1], not a replacement. That is, sites that use more advanced “semantic” knowledge representation techniques continue to coexist with sites built using simple XML or hypertext markup language (HTML).
RESOURCE DESCRIPTION FRAMEWORK (RDF)
RDF is a description language, accepted as a W3C standard <http://www.w3c.org/RDF/>, which aims to capture the semantics of data represented on the Web. It is important to note that while RDF uses XML as an underlying representation, it does not conceptually depend on XML syntax. More formally stated, RDF/RDFS are at a different level of abstraction than XML.
The RDF standard is based on several fundamental concepts. The first such concept is “resources.” Resources are the basic objects or “things” that are to be described in the domain (e.g., articles, books, authors, electronic resources, etc.). All resources are identified through a unique, global identifier called the universal resource identifier (URI). In most applications, the URI is the uniform resource locater (URL) of a Web page, a part of a Web page (e.g., anchor URL), or a link to a document available on a Web server. However, the URI is a more general concept than a Web link—the only condition is that it uniquely identifies a resource. In a library setting, the MARC catalog record for a book, for example, could be considered a URI.
Another essential concept is “classes,” which are basically collections of objects and things with common properties. It is very important to distinguish between a class and an instance of a class or an object. For example, “article” is the class, while “Duodenal ulcer” is an object or instance in the “article” class. Classes can be further divided into subclasses; for example, the “article” class is a subclass of “Printed Resource,” which in turn is a subclass of the more general class “Resource” (which may contain both printed and electronic resources). This is similar to the concept of hierarchical classification in information science.
A third important concept is “properties.” Properties describe relations between other classes or resources. For example, the “article” class may have the properties “has_author,” “appears_in_journal,” “has_publication_date,” and so on. To some degree, this relationship can be seen as the facets of facetted classification, though the analogy should not be taken too literally.
Having defined both classes and properties, “statements” can be derived. Statements in RDF have the basic form: subject-predicate-object, where the subject is an RDF resource, the predicate is a RDF property, and the object is another RDF property or a literal (a name, a number, code, etc.). To use the above example, statements such as: “Duodenal ulcer has_author Smith,” or “Duodenal ulcer has_publication_date 01.01.2001” can be made up. In proper RDFS terminology (see “RDF schema” below), the subject is called the domain of the property, while the object is called the range.
RDF SCHEMA
RDF Schema is a standard (associated with RDF) for describing the structure of the given information, where structure is specific to a particular domain. This standard is fundamentally different from standards such as DC, which are used to specify only content. In DC, the fields that must be filled are prespecified— namely, dc:Creator, dc:Date, etc.—the user cannot change these fields but simply assigns values to them, the same way a user does not change the fields of a catalog record.
In contrast, RDFS is a standard specifying the structure that an RDF document must have, namely, the classes, properties, and relations between them that are allowed to appear in an RDF specification. Then RDF is used to specify the content (i.e., the actual information). Returning to the above example, in RDFS, users can specify structures such as: an instance from “article” class has_author having an instance from “author” class or an instance of “article” class has_publication_date having an instance of “date.” In RDF, this structure can then be instantiated as “Duodenal ulcer has_author Smith” or “Duodenal ulcer has_publication_date 01.01.2001.”
ONTOLOGIES
The notion of ontology has its roots in philosophy. Aristotle defined it as “the science of being qua being,” where the word “qua” means “with regard to the aspect of” [3]. A number of definitions for this term have been derived in relation to its use in computer science. According to Sowa [4], ontology investigates “the categories of things that exist or may exist” in a particular domain and produces a catalog that details the types of things and the relations between those types that are relevant for that domain. This catalog of types is an ontology.
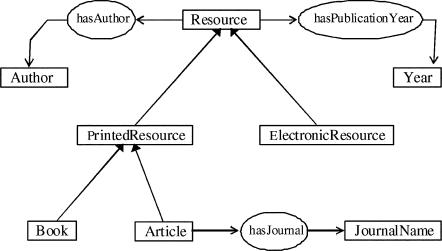
In other words, an ontology is the attempt to formulate an exhaustive and rigorous conceptual schema (i.e., a map of concepts and their relationships) in a given domain. The most commonly cited definition has been given by Gruber: “an explicit specification of conceptualization” [5]. Sabou has provided a clearer one, with the same terms: “a shared conceptualization of a domain” [6]. The emphasis here is on the term “shared,” because all parties using the information have to agree to use the same representation. For example, when different people classify articles under some system of categories and subcategories, the categories are known and agreed on by everyone. Figure 1 provides an example of a possible ontology for the library domain.
Figure 1.
Sample ontology for library information sources
In Figure 1, two types of elements appear: classes, represented as rectangles, and properties of classes, represented as ovals. Note that there are two types of arrows as well, corresponding to the two main types of relations allowed by RDF/RDFS. The thicker, closed arrows are “subclass of” (also called “is_a”) relationships. For example, any instance of the class “book” is a “Printed Resource” and therefore is also a “Resource.” This corresponds to a hierarchical structure between classes.
The second type of relationship is the “Domain-Property-Range” relationship, followed through the thinner, open arrows (e.g., resources have an author or have, as publication year, some instance of year). Note that properties defined for larger classes also apply to all their subclasses. For example, because the authors have specified in Figure 1 that all resources can have an author, this means that “Printed Resources” and “Electronic Resources” also have one and, hence, books and articles. However, the property “hasJournal” only applies to the subclass of journal articles and does not apply (at least in this ontology) to books or electronic resources.
This example is over-simplified, of course. Real-life ontologies (and in particular those in the biomedical domain) may have hundreds of classes and properties.
AN EXAMPLE USING EXTENSIBLE MARKUP LANGUAGE TAGS
The concepts from the above three sections can be exemplified by providing the tags corresponding to a few of the classes and properties represented in Figure 1. The purpose of this article is not to give the full syntax of RDF or RDFS tags; however, it is important to make the above example more concrete.
First, recall that the RDFS is used to present structure, while the RDF is used to input actual information for such a structure. Consider the ontology structure from Figure 1. Definition in RDFS would include tags such as the following. The topmost tag defines the XML domain of the tags used.
<rdf:RDF xmlns:rdf=“http://www.w3c.org/…/rdf”
xmlns:rdfs=“http://www.w3c.org/…/rdfs”>
<rdfs:Class rdf:about=“Article”>
<rdfs:subClassOf rdf:resource=“PrintedResource”>
</rdfs:Class>
<rdf:Property rdf:about=“hasAuthor”>
<rdfs:domain rdf:resource=“Resource”>
<rdfs:range rdf:resource=“Author”>
</rdf:Property>
…. </rdf:RDF>
Once the ontology structure has been defined in RDFS tags, it can be stored in a file, called “libStructure.rdfs,” on a local server. The classes and properties defined in this file can be reused by considering them as belonging to a self-defined domain, called “lib.” The heading of the RDF file will contain a reference to this information.
<rdf:RDF xmlns:rdf=“http://www.w3c.org/…/rdf”
xmlns:lib=http://www.umfcluj.ro/…/libStructure.rdfs”>
<rdf:Description rdf:ID = “Duodenal ulcer”>
<rdf:type>
<rdfs:Class rdf:resource=“Article”>
</rdf:type>
<bib:hasJournal rdf:resource=“Gastroenterology”>
<bib:hasAuthor rdf:resource=“Smith”>
<bib:hasPublicationYear rdf:resource=“2005”>
</rdf:Description>
… </rdf:RDF>
UTILITY IN INFORMATION RETRIEVAL
The first (and probably most important) benefit of such encoding is that it enables more complex information retrieval, using a specialized query language called Resource Query Language (RQL). Could this be achieved through simple XML? The answer is no, at least not in a straightforward way. For example, the following two tags:
1) <BibliographicResource>
<Title>“Duodenal ulcer”</Title>
<Author>Smith</Author>
<PublicationYear>2001</PublicationYear>
</BibliographicResource>
2) <JournalArticle>
<Title>“Treatment of duodenal ulcer”</Title>
<Author>Smith</Author>
<PublicationYear>2002</PublicationYear>
</JournalArticle>
The first tag (i.e., “BibliographicResource”) may refer to an unpublished clinical report. For simplicity, suppose users already know that the author and date are at the second level of nesting in XML. A query can now be made on the above structure to retrieve all bibliographic resources that have the author Smith and appeared after 2000. This would return only the first record, the only one explicitly marked as a Bibliographic Resource, and not the second one.
For human understanding, it is obvious that journal articles are also bibliographic sources of information. However, a machine does not know this, unless its underlying representation (or ontology) contains this information, hence the benefits of more semantically meaningful representations. Furthermore, the benefits of using more semantic representations increase as the size and complexity of the information to be represented increases. The ontologies used in the biomedical domain, with hundreds of classes and properties, often entail such complexity.
The central idea of all these structures, rules, and standards is to reinforce the formal logic and tighten the “meaning” to a point where it can no longer escape correct computer interpretation.
BEYOND RDF: MORE ADVANCED SW LANGUAGES
RDF and the corresponding RDFS are the simplest of Web languages that can be called “semantic.” RDF has already been accepted by the W3C as a standard and is most likely to be encountered in a practical setting.
It should be mentioned that RDF has limitations regarding the type of structures that can be represented by it. More powerful representation languages for the SW include: Defense Advanced Research Projects Agency (DARPA) Agent Mark-Up Language (DAML), developed with DARPA's support in the United States, and Ontology Inference Layer (OIL), developed in Europe, at the Artificial Intelligence Department of the Free University (VU) of Amsterdam. Considerably more powerful than RDFS, these two languages were later united and a “lightweight” version was proposed as a possible standard to the W3C, called Web Ontology Language (OWL). Programs also exist to “write” in ontology languages, such as Protégé, developed by Stanford Medical Informatics, and a national resource for biomedical ontologies and knowledgebases supported by NLM. Examples of the type of representations that cannot be specified in RDF/RDFS but are possible in OWL are:
Cardinality restrictions: For example, users might want to say that a printed resource may have more than one author but only one publication year.
Disjointedness of classes: For example, users may wish to enforce that a resource is classified either as printed or electronic.
Other special characteristics of properties, such as “uniqueness,” a book can only have one International Standard Book Number (ISBN); “inverse properties,” for example, “is published by” applied to a book is the inverse of the property “publisher of” applied to a publishing house; or “local scope,” restricting the domain of the property to certain classes.”
Describing and exemplifying OWL or DAML+OIL is beyond the scope of this introductory article. The interested reader should consult Antoniou and Van Harmelen's Semantic Web Primer [2].
SW APPLICATIONS AND RESEARCH PROJECTS IN THE BIOMEDICAL FIELD
The very nature of the SW concept makes it very difficult, at least at this time, to point to a Website that illustrates what it is exactly and how it works [7]. The practical applications of the SW are far from being consolidated, they are still at level of research work and some concern exists that it may turn out to be far more valuable in concept than in practice. However, a number of existing applications and research projects in the biomedical and health sciences are based on SW technology or incorporate it to an important extent. This section aims to briefly review a number of them to give readers an appreciation of what the important issues are. Given the crucial role in information retrieval of biomedical vocabularies, terminology, and taxonomies [8], most of these projects are related to their formalization, namely defining the classes of contained entities and, especially, establishing the relations among them to develop them into ontologies.
The Unified Medical Language System
Developed by NLM, the Unified Medical Language System (UMLS) incorporates the highest number of resources in the biomedical field: it includes the Metathesaurus, Semantic Network, Specialist Lexicon, and lexical programs. It is the closest to the concept of ontology, especially due to the Semantic Network with its 135 semantic types <http://www.nlm.nih.gov/research/umls/META3_current_semantic_types.html>, which are high-level categories covering more than 1 million concepts from the more than 100 vocabularies and terminologies. It has 2 major hierarchies, one for “Entity” (including “Physical Object” and “Conceptual Entity”) and one for “Event” (including “Activity” and “Phenomenon or Process”). Beside the directly hierarchical is_a relationship, 53 kinds of defined relationships are used to represent more than 6,700 hierarchical and associative relations among the semantic types. UMLS is continually developing, and research work is carried out regarding its coverage, content, organization, and compatibility with other ontologies [9–12].
As an ontology, the UMLS plays an important role in developing other products and projects, among which it is important to mention The Semantic Knowledge Representation (SKR) Project, which aims “to develop programs to provide usable semantic representation of biomedical free text by building on resources currently available at the library” (such as UMLS knowledge sources) [13].† The Medical Text Indexer [14], developed as part of this project, is a program for producing indexing recommendations for Medical Subject Headings (MeSH), which can be used (and have been used at NLM since 2002) both for semiautomatic and fully automatic indexing purposes.
Generalised Architecture for Languages, Encyclopaedias and Nomenclatures in Medicine
Generalised Architecture for Languages, Encyclopaedias and Nomenclatures in Medicine (GALEN): <http://www.opengalen.org> is a project of the European Union aimed at providing terminology resources for clinical systems. It is not based on a previously existing thesaurus or taxonomy, therefore formalization was possible from the beginning, and the terms are coordinated when they are included in the system. As stated on the Website, “GALEN is trying to make it easier to build useful and usable clinical applications, to support clinicians in their day-to-day work.” The “clinical terminology,” the GALEN Common Reference Model, aims to represent “only sensible medical concepts” and categories that can be classified automatically [15]. It also uses existing coding and classification systems as well as natural language processing techniques [16]. The supported languages are English, French, Italian, Dutch, German, Finnish, and Swedish.
The HealthCyberMap
The HealthCyberMap <http://healthcybermap.semanticWeb.org> “aims to map selected parts of health information resources in cyberspace in novel semantic ways to improve their retrieval and navigation” [17, 18]. It uses the DC metadata, including UMLS terms in the “subject” element.
LinkBase
LinkBase <http://www.landcglobal.com/pages/linkbase.php> claims to be “the world's largest formal medical ontology, i.e., a conceptual computer-understandable representation of medicine.” It is currently developed and maintained by the modeling team of Language and Computing as a commercial product, after extensive research funded by the European Community. LinkBase contains over 1 million language-independent medical and general purpose concepts that can be expressed both by standard terminologies and natural language expressions. The medical concepts are language independent and are linked to about 3 million terms in various languages (English, French, Spanish, etc.). The concepts are linked together into a semantic network using approximately 450 different link types for expressing formal relationships [19]. First DataBank, Healthgate, and WebMD are listed among their clients.
National Cancer Institute Thesaurus and Ontology
This ontology is “a public domain description logic-based terminology produced by the [National Cancer Institute (NCI)] and distributed as a component of the NCI Center for Bioinformatics caCORE distribution” [20]. It is based on the UMLS Metathesaurus, with a public version available at http://ncimeta.nci.nih.gov. Research is underway to convert the NCI Thesaurus to OWL-Lite. In September 2005, the ontology (downloadable version at http://www.mindswap.org/2003/CancerOntology) included more than 500,000 triples.
Another very important benefit that the SW initiative claims is proof of how an answer was derived, the fact that the querying application could potentially do some reasoning about how “believable” a fact is. At the very least, derived facts could be attributed to a source, and, over time, applications could be developed that rate sources as to their integrity, reliability, and so on. This is especially important for the medical field, where information must be ensured the highest possible level of trust [21].
MedCIRCLE
According to its Website, MedCIRCLE <http://www.medcircle.org> is
a collaboration of European health subject gateways or rating services—builds on, expands and continues work on rating health information on the Internet piloted within the MedCERTAIN project. Both projects, MedCIRCLE and MedCERTAIN, are complementary Semantic Web projects with the overall objective to develop and promote technologies able to guide consumers to trustworthy health information on the Internet, to establish a global Web of trust for networked health information, and to empower consumers to “filter” or positively select high quality health information on the Web.
It is based on the Health Information Disclosure, Description and Evaluation Language (HIDDEL) vocabulary, which allows the description of health information in terms of quality and trust [22, 23]. The Catalog and Index of French-language Medical Resources (CISMeF) is a partner in this project [24].
Another project that is already beyond the status of prototype has been reported by Joubert et al. [25] and refers to modeling and implementing Web portals providing access to certified and high-quality information in the health domain. It is based on UMLS and the existing ARIANE project. Semantic relationships are used to automatically translate queries that can be “run” in PubMed, Health on the Net (HON) databases, CISMeF, or other highly authorized sources.
Other lines of research, which have the advantage of dealing with unstructured data, aim to construct ontologies and taxonomies automatically or enrich existing ones through automatic techniques such as statistical clustering [16, 26]. The authors point out, however, that generally text mining, natural language processing, and other algorithms for automatic information retrieval (for example, those used by Google) are not traditionally considered SW techniques (because they are not strictly based on formal logic). However, much recent work has focused on extracting semantic representations from free text and existing, partially structured, resources (e.g., the Semantic Knowledge Representation Project Website [13]).
Also, semantic indexing is already reported for medical educational resources [27] or digital audiovisual resources [28], while other SW features are put to work for building lexical resources in languages other than English [29, 30].
THE SW WITH A PINCH OF SALT
The SW is not the first attempt to control knowledge [31]. Universalist ambitions (e.g., universal bibliographic control, developed by the library and information science community in the 1970s) existed before [32]. Although the protocols and standards have a solid foundation in formal, symbolic artificial intelligence, so far the human mind has proved too elusive to model in all its complexity. Generalizations, ambiguity, and fuzzy relationships are problems inherent in any classification scheme, and the world will be always described in terms of these properties. Nelson [10] expressed this by the quotation, “the beauty of poetry is the despair of science.” On the other hand, Berners-Lee sees this “beauty of poetry” as “human mumblings” [21].
Traditional SW approaches generally aim to achieve 100% accuracy, by using formal logic to capture the meaning of information beyond a point where it cannot escape correct computer interpretation. However, providing information already fully structured according to an agreed ontology can be a very labor-intensive task. In fact, most existing information comes in unstructured (e.g., free text, HTML) or semi-structured formats (e.g., MeSH). Some projects, such as NLM's Semantic Knowledge Representation Project, aim to bridge this gap by constructing programs that build semantic representations automatically based on biomedical free text, making use of existing resources.
Another problem encountered by SW proponents is the fact that ontologies used to represent information in open environments such as the Web do not generally coincide (e.g., the field “date” in one ontology may be represented by “year,” “month,” “day” in another). This problem is addressed though a line of work called ontology merging.
However, despite the fact that some of the initial ambitions from the SW vision [1] have yet to materialize and many problems still exist, SW techniques have proved very successful in applications where the structure of the information to be represented and retrieved is highly complex, such as the biomedical domain.
WHAT HAS THE LIBRARIAN TO DO WITH IT?
Controlled vocabularies, taxonomies, classification systems, information retrieval, quality-controlled gateways, metadata, and knowledge management constitute an integral part of the profession. It could even be argued that many of the underlying problems currently faced in SW research have been studied for years by librarians, long before the emergence of the Web itself.
For example, faceted classification (originally proposed by Ranganathan in the 1930s) uses concepts such as classes and facets to allow multiple, flexible ways to label and retrieve information sources. This approach resembles the multiple classes and properties used in RDF/RDFS (though the latter protocols have the clear advantage of being grounded in formal logic). There is much revived interest in the faceted approach, and its benefits are being explored in the context of the Web. The recently reported “concise medical taxonomy” of the American Medical Association [33] explored the capacity of faceted indexing to create more concise, Web-friendly displays. However, as already known in the library community, faceted classification never really took off in real life. One of the main reasons it did not take off was that it appealed much less to human [sic!] logic than a hierarchical approach. Most of the taxonomies used today are hierarchical and, therefore, conceptually simpler. Arguably, hierarchies provide less expressivity and flexibility but have the major advantage that standards are much easier to understand and apply uniformly by all parties.
Librarians and library activities can contribute in many ways to the SW on a practical level [7, 34, 35]:
get well acquainted with metadata standards such as DC and the various ways in which they may be expressed (HTML, XML, RDF)
learn more about the concept of ontologies and about information representations using XML or RDF
participate in projects that aim to use SW technology; librarian expertise will be highly valued
The authors argue that both the medical library and SW communities have a lot to learn from each other. On the one hand, the SW approach provides novel ways to represent and retrieve information. These approaches are grounded in formal artificial intelligence, which assures that the representations used are logically consistent and their meaning is also accessible to machines.
Nevertheless, the importance of medical librarians in this process is not only unlikely to decrease, but their role will become even more crucial. Developing the various ontologies and medical taxonomies cannot lead to any useful real-life applications without major input from librarians. As Soergel [36] points out in relation to WordNet, the largest and best known existing ontology, “a wonderful system whose construction would have profited from applying experience in thesaurus construction … and standard methods for classification.” He also shows that: “large and useful systems are being built with more effort than necessary,” because often the existing experience from classification science fails to be used. The authors argue that the two directions of work, so far largely parallel, are probably converging. The number of research projects under way confirms the authors' supposition.
This paper attempts to cover only the most fundamental concepts and abstracts away many technical details of the standards. There are challenges that must be addressed before using such techniques in medical library settings. A great deal of further research is needed to explore them.
Footnotes
* Based on an invited tutorial lecture delivered at the Ninth European Association for Health Information and Libraries Conference in Santander, Spain, September 2004. The original course is available in two parts at http://www.chu-rouen.fr/documed/eahilsantander.html and http://www.chu-rouen.fr/documed/semanticWeb.html, respectively.
† See also the report from the National Library of Medicine at http://lhncbc.nlm.nih.gov/lhc/docs/reports/2003/tr2003004.pdf.
Contributor Information
Ioana Robu, Email: irobu@umfcluj.ro.
Valentin Robu, Email: V.Robu@cwi.nl.
Benoit Thirion, Email: Benoit.Thirion@chu-rouen.fr.
REFERENCES
- Berners-Lee T, Hendler J, and Lassila O. The Semantic Web. a new form of Web content that is meaningful to computers will unleash a revolution of new possibilities. Scientific American [serial online]. 2001 May 17;284(5). [rev. 15 Jul 2004; cited 25 Apr 2005]. <http://www.sciam.com/article.cfm?articleID=00048144-10D2-1C70-84A9809EC588EF21>. [Google Scholar]
- Antoniou G, Van Harmelen F. A Semantic Web primer. Cambridge, MA: MIT Press, Apr 2004. (Preliminary version available at: <http://www.ics.forth.gr/isl/swprimer/>. [rev. 3 Jul 2004; cited 25 Apr 2005].). [Google Scholar]
- Ontology. [Web document]. WordIQ Dictionary and Encyclopedia. [rev. 13 Jul 2004; cited 25 Apr 2005]. <http://www.wordiq.com/definition/Ontology>. [Google Scholar]
- Sowa JF. Knowledge representation: logical, philosophical, and computational foundations. Pontiac Grove, CA: Brooks Cole Publishing Co, 2000. Quoted in: Jacob EK. Ontologies and the Semantic Web. Bull Am Soc Inform Sci Technol. 2003 Apr/May; 29(4):19–22. [Google Scholar]
- Gruber TR. A translation approach to portable ontology specifications. Technical report KSL 92-71. Sep 1992; revised Apr 1993. [Web document]. Knowledge Systems Laboratory. [cited 27 Nov 2005]. <http://tomgruber.org/writing/ontolingua-kaj-1993.pdf>. [Google Scholar]
- Web-based knowledge representation [course slides]. Free University of Amsterdam, 2002. [Google Scholar]
- Palmer SB. Does it work? what Semantic Web applications are there? In: Palmer SB, ed. The Semantic Web: an introduction. [Web document]. 2001–09. [rev. 8 Feb 2005; cited 25 Apr 2005]. <http://infomesh.net/2001/swintro/>. [Google Scholar]
- Lorence DP, Spink A. Semantics and the medical Web: a review of the barriers and breakthroughs in effective healthcare query. Health Info Libr J. 2004 Jun; 21(2):109–16. [DOI] [PubMed] [Google Scholar]
- Bodenreider O, Burgun A. Biomedical ontologies. [Web document]. In: Chen H, Fuller S, Hersh WR, Friedman C, eds. Medical informatics: advances in knowledge management and data mining in biomedicine. Springer-Verlag, 2005 (in press). (<http://mor.nlm.nih.gov/pubs/pdf/2005-chapter_medont-ob.pdf>. [rev. 18 Apr; cited 25 Apr 2005].). [Google Scholar]
- Nelson JS. MeSH, UMLS, and the Semantic Web. [Web document]. 1/13/02. [rev. 13 Aug 2004; cited 25 Apr 2005]. <http://www.nlm.nih.gov/mesh/presentations/taiwan2001/semanticweb/index.htm>. [Google Scholar]
- Kashyap V. The UMLS Semantic Network and the Semantic Web. [Web document]. In: AMIA 2003 Symposium Proceedings. 2003 Nov:351–5. [rev. 18 Aug 2004; cited 25 Apr 2005]. <http://lhncbc.nlm.nih.gov/lhc/docs/published/2003/pub2003061.pdf>. [PMC free article] [PubMed] [Google Scholar]
- The future of the UMLS Semantic Network: a workshop organized on April 7–8, 2005 at the US National Library of Medicine (NLM) in Bethesda, Maryland. [Web document]. [cited 27 Nov 2005]. <http://mor.nlm.nih.gov/snw/>. [Google Scholar]
- National Library of Medicine. Semantic knowledge representation project. [Web document]. The Library. [rev. 1 Sep 2005; cited 14 Sep 2005].<http://skr.nlm.nih.gov>. [Google Scholar]
- Aronson AR, Mork JG, Gay CW, Humphrey SM, and Rogers WJ. The NLM indexing initiative's medical text indexer. [Web document]. In: Fieschi M, Cuiera E, Y-CJ Li, eds. MEDINFO 2004: proceedings of the 11th World Congress on Medical Informatics. Amsterdam, The Netherlands: IOS Press, 2004. [rev. 1 Sep 2005; cited 14 Sep 2005]. <http://cmbi.bjmu.edu.cn/news/report/2004/medinfo2004/pdffiles/papers/5122Aronson.pdf>. [Google Scholar]
- Rector AL, Rogers JE, and Pole P. The GALEN high level ontology. [Web document]. In: Fourteenth International Congress of the European Federation for Medical Informatics, MIE-96; Copenhagen, Denmark; 1996. [rev. 24 Mar 2005; cited 25 Apr 2005]. <http://www.opengalen.org/technology/technology.html>. [Google Scholar]
- Biczyk do Amaral M, Roberts A, and Rector AL. NLP techniques associated with the OpenGalen ontology for semi-automatic textual extraction of medical knowledge: abstracting and mapping equivalent linguistic and logical constructs. AMIA Symposium 2000. [Web document]. [5 pp.]. [rev. 24 Aug 2004; cited 25 Apr 2005]. <http://www.amia.org/pubs/symposia/D200078.PDF>. [PMC free article] [PubMed] [Google Scholar]
- Boulos MN.. A first look at HealthCyberMap medical semantic subject search engine. Technol Health Care. 2004;12(1):33–41. [PubMed] [Google Scholar]
- Boulos MN, Roudsari AV, and Carson ER. Towards a semantic medical Web: HealthCyberMap's tool for building an RDF metadata base of health information resources based on the Qualified Dublin Core Metadata Set. Med Sci Monit. 2002 Jul; 8(7):MT124–36. [PubMed] [Google Scholar]
- Van Buggenhout C, Ceusters W. A novel view on information content of concepts in a large ontology and a view on the structure and the quality of the ontology. Int J Med Inform. 2005 Mar; 74(2–4):125–32. [DOI] [PubMed] [Google Scholar]
- Golbeck J, Fragoso G, Hartel F, Hendler J, Oberthaler J, and Parsia B. The National Cancer Institute's thesaurus and ontology. J Web Semantics [serial online]. 2003 Dec; 1(1)[32 pp.]. [rev. 6 Feb 2005; cited 25 Apr 2005]. <http://www.Websemanticsjournal.org/ps/pub/2004-6>. [Google Scholar]
- Berners-Lee T, Hendler J. Scientific publishing on the Semantic Web. Nature [serial online]. 2001 Apr;26;410(6832): 1023–4. [rev. 8 Jul 2004; cited 25 Apr 2005]. <http://www.nature.com/nature/debates/e-access/Articles/bernerslee.htm>. [DOI] [PubMed] [Google Scholar]
- Mayer MA, Darmoni SJ, Fiene M, Kohler C, Roth-Berghofer TR, Eysenbach G.. MedCIRCLE: collaboration for Internet rating, certification, labelling and evaluation of health information on the World-Wide-Web. Stud Health Technol Inform. 2003;95:667–72. [PubMed] [Google Scholar]
- Eysenbach G. An ontology of quality initiatives and a model for decentralized, collaborative quality management on the (semantic) World-Wide-Web. J Med Internet Res. 2001 Oct–Dec; 3(4):E34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion B, Loosli G, Douyere M, Darmoni SJ.. Metadata element set in a quality-controlled subject gateway: a step to a health Semantic Web. Stud Health Technol Inform. 2003;95:707–12. [PubMed] [Google Scholar]
- Joubert M, Dufour JC, Aymard S, Falco L, and Fieschi M. Designing and implementing health data and information providers. Int J Med Inform. 2005 Mar; 74(2–4):133–40. [DOI] [PubMed] [Google Scholar]
- Soualmia LF, Darmoni SJ. Combining different standards and different approaches for health information retrieval in a quality-controlled gateway. Int J Med Inform. 2005 Mar; 74(2–4):141–50. [DOI] [PubMed] [Google Scholar]
- Pouliquen B, Le Duff F, Delamarre D, Cuggia M, Mougin F, and Le Beux P. Managing educational resource in medicine: system design and integration. Int J Med Inform. 2005 Mar; 74(2–4):201–7. [DOI] [PubMed] [Google Scholar]
- Cuggia M, Mougin F, and Le Beux P. Indexing method of digital audiovisual medical resources with Semantic Web integration. Int J Med Inform. 2005 Mar; 74(2–4):169–77. [DOI] [PubMed] [Google Scholar]
- Zweigenbaum P, Baud R, Burgun A, Namer F, Jarrousse E, Grabar N, Ruch P, Le Duff F, Forget JF, Douyere M, and Darmoni S. UMLF: a unified medical lexicon for French. Int J Med Inform. 2005 Mar; 74(2–4):119–24. [DOI] [PubMed] [Google Scholar]
- Weske-Heck G, Zaiss A, Zabel M, Schulz S, Giere W, Schopen M, and Klar R. The German specialist lexicon. Proc AMIA Symp 2002:884–8. [PMC free article] [PubMed] [Google Scholar]
- Brooks TA. The Semantic Web, universalist ambition and some lessons from librarianship. Inform Res [serial online]. 2002;7(4). [rev. 24 Aug 2004; cited 2 May 2005]. <http://InformationR.net/ir/7-4/paper136.html>. [Google Scholar]
- Anderson D. Universal bibliographic control: a long-term policy, a plan for action. Pullach bei München, Germany: Verlag Dokumentation, 1974. [Google Scholar]
- McGregor B. Constructing a concise medical taxonomy. J Med Libr Assoc. 2005 Jan; 93(1):121–3. [PMC free article] [PubMed] [Google Scholar]
- Greenberg J, Sutton S, and Campbell DG. Metadata: a fundamental component of the Semantic Web. Bull Am Soc Inform Sci Techno. 2003 Apr/May; 29(4):16–8. [Google Scholar]
- Campbell DG, Fast KV.. Academic libraries and the Semantic Web: what the future may hold for research-supporting library catalogues. J Acad Libr. 2004;30(5):382–90. [Google Scholar]
- Soergel D. The rise of ontologies or the reinvention of classification. J Am Soc Inf Sci. 1999 Oct; 50(12):1119–20. [Google Scholar]