Abstract
A real-time reverse transcription-PCR system has been used to monitor the expression of an aflatoxin biosynthetic gene of Aspergillus flavus in wheat. Therefore, total RNA was isolated from infected wheat samples, reverse transcribed and subjected to real-time PCR. In parallel all samples were analyzed by high-pressure liquid chromatography for aflatoxin B1 production. The primer-probe system of the real-time PCR was targeted against nor-1, a gene of the aflatoxin biosynthetic pathway. By application of this method the nor-1 transcription was quantified during the course of incubation. After 4 days of incubation nor-1 mRNA could be detected for the first time. The amount of nor-1 mRNA increased rapidly, and the maximum was achieved after 6 days. Then, starting very slowly, the mRNA was degraded until day 8, and this was followed by a very fast degradation, reaching nondetectable levels at days 9 and 10. First traces of aflatoxin B1could be detected between the 5th and 6th day of incubation. The aflatoxin concentration reached its maximum after 9 days of incubation and remained constant for the whole period of observation. To ensure that differences in the nor-1 mRNA concentration were due to different expression levels, the expression of the constitutively expressed β-tubulin gene (benA56) has also been monitored. The expression of benA56 remained constant during the whole incubation time. As a parameter for fungal growth, the number of nor-1 gene copies was determined during the course of incubation. The numbers of nor-1 gene copies increased at the beginning of the incubation and reached a plateau at day 5. They correlate well with the viable counts albeit at a higher level.
Aspergillus flavus and A. parasiticus are the most important aflatoxin-producing filamentous fungi. They can occur in several plant products, like spices, cereals, and oily seeds (8, 9, 13). Aflatoxins are secondary metabolites with a high carcinogenic potential, especially in liver tissue. In addition the aflatoxins possess an acute toxicity at higher concentrations. The high health risk caused by aflatoxins leads to strict concentration limits in different countries.
Biosynthesis and the genetic background of aflatoxin production are well elucidated. An overview about the genetic background of the aflatoxin biosynthetic pathway has been given by Woloshuk and Prieto (20) and Brown et al. (1). The genes of both important aflatoxinogenic species, A. flavus as well as A. parasiticus, are very homologous (23). The knowledge of sequences of aflatoxin biosynthetic genes was a prerequisite for the development of molecular diagnostic tools for the detection of aflatoxin biosynthetic fungi in plant derived foods. Two different PCR systems for rapid detection of aflatoxinogenic fungi have been described (5, 16). One is a multiplex system targeted against three genes of the aflatoxin biosynthetic pathway, in particular nor-1, ver-1, and omt-1 (5). In the other system three genes are also targeted (aflR, nor-1, and ver-1); however, they are targeted in single reactions. These PCR systems have been used for the detection of A. flavus in wheat (16) and figs (4). Both PCR systems, however, have the drawback that the results which can be achieved are not directly correlated to the presence of aflatoxin in foods or to aflatoxin production itself. It is a well-known fact that the environment and the cultural conditions may have a strong influence on the biosynthesis of mycotoxins (7, 17). Ellis et al. (3) analyzed the influence of water activity, pH, temperature, and atmosphere composition on aflatoxin production. According to these results all parameters had a significant impact on the growth of fungal cells and thereby on aflatoxin biosynthesis. A review about nutritional factors influencing the aflatoxin biosynthesis has been written by Luchese and Harrigan (10). Even accompanying microorganisms can have an influence on the production of aflatoxin (6). These external influences can reduce aflatoxin biosynthesis without drastic change of the growth rate of the fungal mycelium. Besides others, this is the main reason why the detection of aflatoxin gene-specific DNA fragments in a PCR-based diagnostic assay for aflatoxinogenic aspergilli is not really significant with respect to the mycotoxicological safety of the product. Mycotoxin biosynthetic genes may be active or inactive, depending on the environmental conditions. A much better approach would be the detection of mRNA which is specific for an aflatoxin biosynthetic gene. In case this mRNA is detected, it is ensured that the aflatoxin biosynthetic gene is actively transcribed and it can be assumed that aflatoxin will be produced under the conditions analyzed.
In this report we describe the application of a real-time reverse transcription (RT)-PCR system for a quantitative monitoring of the expression of the aflatoxin biosynthetic gene nor-1 and its correlation to aflatoxin production in a wheat matrix by A. flavus.
MATERIALS AND METHODS
Strains and growth conditions.
Throughout the analysis, the aflatoxinogenic strain A. flavus BFE96 derived from the culture collection of the Federal Research Center for Nutrition has been used. This strain is able to produce aflatoxin B1 (AFB1). The strain was generally grown in malt extract medium under agitating conditions or on malt extract plates (Merck, Darmstadt, Germany) supplemented with glucose (5 g/liter) at 30°C.
Inoculation and incubation of wheat samples.
Wheat samples (25 g) were inoculated with 103 freshly prepared spores/g and incubated at 30°C. The wheat samples were incubated in open petri dishes, which in turn were placed in a large vessel. A wetted filter paper was added to increase the moisture content of the whole system. During the course of incubation the wheat samples were thoroughly mixed every day, to ensure an uniform distribution of the fungal cells throughout the complete sample. At certain time intervals, samples (2.0 g) were withdrawn and divided into four parts of 0.5 g each. One part was used for real-time PCR to determine the copy number of the nor-1 gene, and another part was used for the isolation of total RNA and subsequent real-time RT PCR to determine the nor-1 mRNA copy number. The third part was used for determining the number of CFU, and the last part was used for monitoring for the aflatoxin concentration by high-pressure liquid chromatography (HPLC).
Determination of CFU.
The number of CFU was determined by standard 10-fold dilutions and plating on malt extract medium.
Determination of aflatoxin production by TLC.
For qualitative determination of aflatoxin production A. flavus BFE96 was grown at 30°C on malt extract agar plates for 10 days. An equal amount of fungal mycelium for each sample was transferred into a microreaction tube, and 500 μl of chloroform was added. The fungal mycelia were extracted for 20 min at room temperature on a rotary shaker, the growth of the mycelia was retarded, and the chloroformic extract was evaporated to dryness in a Speed Vac concentrator. The residues were dissolved in 10 μl of chloroform and spotted onto a thin-layer chromatography (TLC) plate (Silica Gel 60; Merck).
As the mobile phase, toluol-ethylacetate-acetic acid (50:30:4, vol/vol/vol) was used. Pure aflatoxin (Sigma, St. Louis, Mo.) was used as a standard. The spots were visualized under UV light (366 nm).
Determination of aflatoxin production by HPLC.
The AFB1 concentration was determined by HPLC. All samples were threefold extracted with chloroform, and this was followed by evaporation at 36°C under nitrogen gas; then the samples were finally dissolved in methanol. The samples were filtered through a Teflon filter (pore size, 0.2 μm; Chromafil; Macherey+Nagel, Düren, Germany) before they were used in HPLC analysis. Forty-microliter aliquots of these filtered extracts were injected for the quantitative determination of the AFB1 concentration.
The HPLC system consisted of a model L-7100 HPLC-pump (Merck/Hitachi, Darmstadt, Germany), a model L-7200 autosampler combined with a Peltier sample cooler (Merck/Hitachi) and a model HP 1050 diode-array detector (Hewlett-Packard, Böblingen, Germany). The chromatograms were digitally processed by the ChemStation software system (Hewlett-Packard).
For analysis of AFB1, a reversed-phase C18 column (LiChroCART 250-4 RP-18 [5.0 μm]; Merck) protected by a guard column (LiChroCART 4-4 RP-18 [5.0 μm]; Merck) was used with an isocratic mobile phase of acetonitrile-methanol-H2O (25:25:50, vol/vol/vol) at a flow rate of 1.0 ml/min. The presence of AFB1 was monitored by the diode array detector at a wavelength of 365 nm.
Isolation of fungal DNA from pure cultures.
The isolation of DNA from pure fungal strains was performed according to a modified method originally described by Yelton et al. (22). For this purpose the strains were grown for 72 to 96 h under shaking conditions (180 rpm) in malt extract broth. The mycelium was harvested by filtration, transferred to a mortar, frozen in liquid nitrogen, and ground. The powder was resuspended in lysis buffer (50 mM EDTA, 0.2% sodium dodecyl sulfate [pH 8.5]) and immediately heated to 68°C for 15 min. After centrifugation for 15 min at 15,000 × g, a 7-ml volume of the supernatant was transferred into a new centrifuge tube and 1 ml of sodium acetate (4 M) was added. This solution was placed on ice for 1 h and centrifuged for 15 min at 15,000 × g. After centrifugation the supernatant was transferred into a fresh tube. The solution was phenol extracted and the isolated DNA was precipitated by the addition of 2.5 volumes of ethanol.
Isolation of DNA from infected wheat samples.
A 0.5-g amount of the infected wheat sample was used for the isolation of DNA for real-time PCR. The sample was frozen in liquid nitrogen and ground to powder in a mortar. The DNA from these samples were isolated with the DNeasy kit essentially as described by the manufacturer (Qiagen, Hilden, Germany).
Isolation of total RNA from pure cultures and from wheat samples.
An amount of 0.5 g of the infected wheat sample was frozen in liquid nitrogen and ground to powder in a mortar. An amount of 200 mg of the powder was used for isolation of total RNA. For that purpose the E.Z.N.A. Fungal RNA kit (Peqlab, Erlangen, Germany) has been used according the recommendations of the manufacturer. An 80-μl volume of the RNA preparation was treated with 2 μl of DNase I (2.5 Kunitz units/μl; Qiagen) for degradadtion of traces of genomic DNA. The solution was incubated for 60 min at 37°C and subsequently for 10 min at 65°C to inactivate the DNase. An aliquot of the RNA was separated on an agarose gel, to check the integrity of the RNA. The RNA gel was prepared as described by Sambrook et al. (14).
cDNA synthesis.
For cDNA synthesis 8 μl of the DNase I-treated total RNA was used along with the Omniscript reverse transcription kit (Qiagen). The reaction mixture was composed essentially as described by the manufacturer and incubated at 37°C for 1 h. The cDNA was either directly used for real-time PCR or stored at −20°C.
Real-time PCR.
The real-time PCRs were performed in a GeneAmp 5700 sequence detection system (PE Applied Biosystems, Foster City, Calif.). The TaqMan system with two primers and an internal fluorescence labeled probe was used instead of the SYBR green system which has the disadvantage to be less specific and to be prone to the formation of primer dimers which can interfere with the results. The primers and the internal probe used in the reaction were recommended by the Primer Express software (version 1.0; PE Applied Biosystems). They were targeted against sequences of the nor-1 gene of the aflatoxin biosynthetic pathway. The primer-probe set had the following nucleotide sequence: nortaq-1, 5′-GTCCAAGCAACAGGCCAAGT-3′; nortaq-2, 5′-TCGTGCATGTTGGTGATGGT-3′; norprobe 5′-TGTCTTGATCGGCGCCCG-3′ (labeled with the fluorescence marker 6-carboxyfluorescein at the 5′end and with 6-carboxy-tetrametyl-rhodamine at the 3′ end) enclosing an amplicon of 66 bp from nucleotide 782 to 847 according the published sequence of the nor-1 gene (19). For the PCR the TaqMan reagent kit (PE Applied Biosystems) was used according to the recommendations of the manufacturer. For each reaction 1 μl of the DNA sample solution was mixed with 50 μl of the PCR stock solution containing 5 μl of 10 × TaqMan buffer A, 7 μl of 25 mM MgCl2, 1 μl of each deoxynucleoside triphosphate mixture (10 mM dATP, dCTP, and dGTP and 20 mM dUTP), 0.5 μl of the primers and probe (each 0.5 μM), 0.5 μl of uracil-N-glycosylase (1 U/μl), 0.19 μl of AmpliTaq Gold (5 U/μl), and 29.8 μl of sterile deionized water. The PCR was performed in MicroAmp reaction tubes. After an incubation of 2 min at 50°C to allow for uracil-N-glycosylase cleavage, AmpliTaq Gold polymerase was activated by an incubation step for 10 min at 95°C. All 35 PCR cycles were performed according the following temperature regimen: 95°C for 20 s, 55°C for 20 s, and 72°C for 30 s.
To generate the standard curve a larger PCR fragment of the nor-1 gene, generated with the primers nor1 and nor2 (5) was used. The concentration of this standard PCR product was determined in a fluorometer (DyNa Quant 200; Pharmacia, Uppsala, Sweden), and the number of copies was calculated. These stock solutions were diluted serially by a factor of 10, and an aliquot of the dilutions was used as a copy number standard during each setup of the real-time PCR. The concentration of unknown samples was calculated by the GeneAmp 5700 system according the generated standard curve. All reactions were carried out at least twice.
The same reaction conditions were used with the β-tubulin gene (benA56) as the target sequence. In this case the primers and probe bentaq1, 5′-CTTGTTGACCAGGTTGTCGAT-3′; bentaq2, 5′-GTCGCAGCCCTCAGCCT-3′; and benprobe, 5′-CGATGTTGTCCGTCGCGAGGCT-3′ were used for the real-time PCR. For the production of the PCR fragment used as a concentration standard a conventional PCR was carried out with the following primers: ben1, 5′-GGCCAGTCCGGTGCTGG-3′, and ben2, 5′-GTTGTCGATACAGAAGG-3′. The sequences of all used primers and probes were deduced from a part of the sequence of the A. flavus β-tubulin gene (benA56) published in GenBank under the accession number AF036801.
RESULTS
Determination of growth kinetics by viable cell counts (CFU) and nor-1 gene copy number.
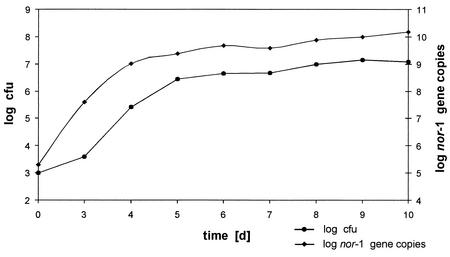
To follow the growth kinetics of A. flavus BFE96 in wheat, at certain time intervals samples were analyzed for the number of viable cells and for the copy number of the nor-1 gene in the wheat samples (Fig. 1). An increase in CFU could first be detected after 3 days of incubation. The CFU values reached a maximum after 5 days of and stayed constant during the complete observation period. A similar behavior could be observed with the nor-1 copy number. It increased until day 5. After that time the number of the nor-1 gene remained nearly constant or increased only very slightly. The copy numbers of the nor-1 gene however, were generally higher than the CFU numbers by two orders of magnitude.
FIG. 1.
Growth kinetics of A. flavus BFE96 in wheat, determined by plate count technique (•) or quantitative determination of the copy number of the nor-1 gene by real-time PCR (♦). The values are given in log counts per gram of wheat.
Aflatoxin production in wheat.
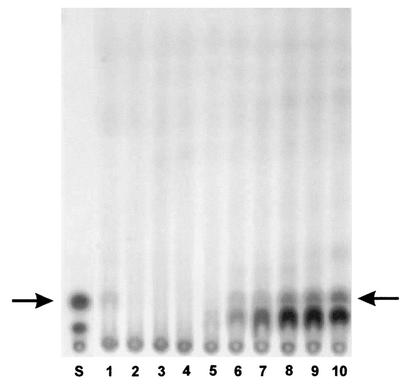
To determine the kinetics of AFB1 production by A. flavus BFE96 in wheat, samples were withdrawn serially from the incubated wheat and subjected to TLC analysis. First tests proved A. flavus BFE96 to be a strong producer of AFB1. Biosynthesis of AFB1 could first be detected after 6 days of incubation (Fig. 2). With the same kinetics a second unknown metabolite occurred. The results clearly demonstrate that A. flavus BFE96 is able to produce aflatoxin on wheat under the incubation conditions used.
FIG. 2.
TLC analysis of the aflatoxin production of A. flavus BFE96 in wheat. Lane S, aflatoxin standard, indicated by an arrow; lanes 1 to 10, analyzed wheat samples after 1 to 10 days of incubation, respectively.
Correlation of the transcription of the nor-1 gene to aflatoxin production.
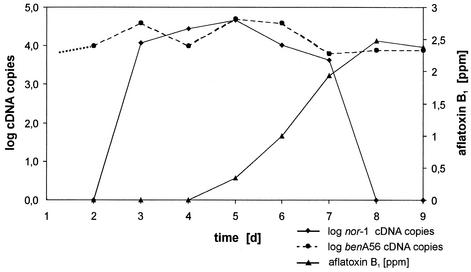
The RNA isolated from each time point was reverse transcribed and used for real-time PCR. From aliquots of the same samples the AFB1 concentration was determined by HPLC. The results are shown in Fig. 3. First nor-1 mRNA appeared at day 4. A high level of the mRNA concentration was achieved quite fast. From day 4 to 6 only a slight increase in the nor-1 mRNA level was observed. After that time the mRNA concentration started to decrease slowly until day 8 and than rapidly down to non detectable levels at day 9.
FIG. 3.
Production of nor-1 (♦) and benA56 (•) mRNA determined by quantitative real-time RT-PCR and production of AFB1 (▴) by A. flavus BFE96 determined by HPLC in wheat. The dotted part of the benA56 curve is extrapolated and indicates the lag phase where no real-time PCR signal could be obtained.
First amounts of AFB1 have been measured after 6 days of incubation. The concentration of AFB1 increased constantly until day 9, then it remained constant during the observation period. The results clearly demonstrate that there was a transient induction of the nor-1 gene before aflatoxin could be determined. After the degradation of the nor-1 mRNA no further increase in the concentration of aflatoxin could be observed.
Comparison of expression of the β-tubulin gene (benA56) to that of the nor-1 gene.
The expression of the benA56 gene was analyzed in parallel to the nor-1 gene. The benA56 gene is a housekeeping gene whose expression is less dependent on the growth phase than the expression of a gene involved in secondary metabolism. A more-or-less uniform expression of the benA56 gene is an indication for the functionality of the approach and the reliability of the RNA isolation procedure. Copy numbers of the cDNAs of the benA56 gene obtained during the time course of the experiment are shown in Fig. 3 in relation to the expression of the nor-1 gene. Before day 3, during the lag phase (Fig. 1), no expression signal could be obtained from both genes. The expression of the benA56 gene is roughly constant during the whole subsequent growth phases. In contrast, nor-1 was clearly induced during the last part of the log phase. It reaches its maximum at the beginning of the stationary phase and later on it decreased to non detectable levels. These comparison indicates that the obtained expression profile of the nor-1 gene is specific for that gene and not due to artifacts.
DISCUSSION
PCR systems for the detection of mycotoxinogenic fungi in food samples have been described for several species (2, 5, 12, 16). The results obtained with these systems however are not directly correlated to mycotoxin production. A step further towards the control of mycotoxin production in a food product is the described combined monitoring system for quantitative determination of the growth kinetics of A. flavus and for the expression of the aflatoxin biosynthetic gene nor-1. The growth data obtained by real-time PCR correlate well with the growth data obtained by the conventional plate count technique, although the former numbers are higher. In a previous publication (11) the possible reasons for this difference have been discussed in detail. Knowing the relationships between the CFU and real-time PCR data, the latter can be used to assess the mycological status of a food product in a much shorter time than with the conventional method.
The system was also applicable for quantification of nor-1 mRNA. According to our results nor-1 mRNA could be detected about 48 h before the first AFB1 could be determined. The nor-1 gene was induced at day 4 under the incubation conditions used. The amount of mRNA reached its maximum very rapidly and remained at this maximum for 4 days. After this time a rapid decrease of the nor-1 mRNA could be observed. The produced AFB1 could be detected just 6 days after the inoculation, 2 days later than the induction of the nor-1 gene. Its concentration reached a maximum after 10 days and stayed constant (the aflatoxin concentration at day 15 were roughly the same as at day 10 [data not shown]). This is an indication that no more aflatoxin is synthesized after degradation of the nor-1 mRNA.
A clear difference between the expression pattern of the benA56 and the nor-1 gene could be determined. Whereas the benA56 gene was constitutively expressed during the whole time of observation, the nor-1 gene was clearly induced for 5 days. These different expression patterns reflect the difference between the expression of a “housekeeping” gene of the primary metabolism and a gene of a secondary metabolic pathway.
In a similar approach, Xu et al. (21) used a fusion construct of the promoter region from the nor-1 gene to the gus gene as a reporter gene, to monitor the expression of the nor-1 gene in peanut pods. For their analysis they used an A. parasiticus strain transformed with a plasmid carrying the nor-1/uidA fusion gene. The produced GUS activity was monitored and correlated to AFB1 production. The authors found a time shift of 12 h between the first occurrence of GUS activity and AFB1 appearance. The fact that in our case the time shift between induction of the nor-1 gene and aflatoxin production lasts 48 h may be explained by the different experimental approaches or growth conditions.
It was demonstrated that the described real-time PCR system is able to completely characterize the mycological status of wheat as a model food matrix. The infection of a wheat sample by an aflatoxinogenic Aspergillus could be quantitatively determined very quickly by monitoring the copy number of the nor-1 gene. A comparable system for the detection and quantification of the tri5 gene of trichothecene-producing fusaria has been described by Schnerr et al. (15).
In addition the system described is useful for monitoring the expression of the aflatoxinogenic gene nor-1. Sweeney et al. (18) introduced a similar system directed against the regulatory aflR gene and the ord1 gene. They also found a correlation between expression of the aflatoxin biosynthesis genes and aflatoxin production. However, they did not quantify their results and they demonstrated the functionality of the system in media but not in a food matrix.
The system described here is a valuable tool for hazard analysis critical control point (HACCP) purposes, as it has the capacity to very exactly monitor the process of AFB1 biosynthesis and to elucidate the critical control points at which AFB1 production is possible.
Acknowledgments
This work was supported by the EU Project ERB IC15-CT98-0901.
REFERENCES
- 1.Brown, M. P., C. S. Brown-Jenco, and G. A. Payne. 1999. Genetic and molecular analysis of aflatoxin biosynthesis. Fungal Genet. Biol. 26:81-98. [DOI] [PubMed] [Google Scholar]
- 2.Doohan, F. M., D. W. Parry, P. Jenkinson, and P. Nicholoson. 1998. The use of species-specific PCR-based assays to analyse Fusarium ear blight of wheat. Plant Pathol. 47:197-205. [Google Scholar]
- 3.Ellis, W. O., P. J. Smith, B. K. Simpson, S. Khanizadeh, and J. H. Oldham. 1993. Control of growth and aflatoxin production of Aspergillus flavus under modified atmosphere packaging conditions. Food Microbiol. 10:9-21. [DOI] [PubMed] [Google Scholar]
- 4.Färber, P., R. Geisen, and W. H. Holzapfel. 1997. Detection of aflatoxinogenic fungi in figs by a PCR reaction. Int. J. Food Microbiol. 36:215-220. [DOI] [PubMed] [Google Scholar]
- 5.Geisen, R. 1996. A multiplex PCR reaction for the detection of aflatoxin and sterigmatocystin producing fungi. Syst. Appl. Microbiol. 19:388-392. [Google Scholar]
- 6.Gourama, H., and L. B. Bullermann. 1995. Antimycotic and antiaflatoxinogenic effect of lactic acid bacteria: a review. J. Food Protect. 57:1275-1280. [DOI] [PubMed] [Google Scholar]
- 7.Häggblom, P. E., and J. Ghosh. 1985. Postharvest production of ochratoxin A by Aspergillus ochraceus and Penicillium viridicatum in barley with different protein levels. Appl. Environ. Microbiol. 49:787-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jelinek, C. F., A. E. Pohland, and G. E. Wood. 1989. Worldwide occurrence of mycotoxins in foods and feeds—an update. J. Assoc. Anal. Chem. 72:223-230. [PubMed] [Google Scholar]
- 9.Llewellyn, G. C., R. L. Mooney, T. F. Cheatle, and B. Flannigan. 1992. Mycotoxin contamination of spices—an update. Int. Biodeterior. Biodegrad. 29:111-121. [Google Scholar]
- 10.Luchese, R. H., and W. F. Harrigan. 1993. Biosynthesis of aflatoxin. The role of nutritional factors. J. Appl. Bacteriol. 74:5-14. [DOI] [PubMed] [Google Scholar]
- 11.Mayer. Z., A. Bagnara, P. Färber, and R. Geisen. 2002. Quantification of the copy number of nor-1, a gene of the aflatoxin biosynthetic pathway by real time PCR and its correlation to the cfu of Aspergillus flavus in foods. Int. J. Food Microbiol., in press. [DOI] [PubMed]
- 12.Niessen, M. L., and R. F. Vogel. 1998. Group specific PCR detection of potential trichothecene-producing Fusarium species in pure cultures and cereal samples. Syst. Appl. Microbiol. 21:618-631. [DOI] [PubMed] [Google Scholar]
- 13.Pittet, A. 1998. Natural occurrence of mycotoxins in foods and feeds—an updated review. Rev. Vet. Med. 149:479-492. [Google Scholar]
- 14.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 15.Schnerr, H., L. Niessen, and R. F. Vogel. 2001. Real time detection of the tri5 gene in Fusarium species by LightCyclerTM-PCR using SYBR® Green I for continuous fluorescence monitoring. Int. J. Food Microbiol. 71:53-61. [DOI] [PubMed] [Google Scholar]
- 16.Shapira, R., N. Paster, O. Eyal, M. Menasherov, A. Mett, and R. Salomon. 1996. Detection of aflatoxinogenic molds in grains by PCR. Appl. Environ. Microbiol. 62:3270-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stander, M. A., P. S. Steyn, A. Lübben, A. Miljkovic, P. G. Mantle, and G. J. Marais. 2000. Influence of halogen salts on the production of the ochratoxins by Aspergillus ochraceus Wilh. J. Agric. Food Chem. 48:1865-1871. [DOI] [PubMed] [Google Scholar]
- 18.M. J. Sweeney, P. Pamies, and A. D. W. DobsonT. 2000. The use of reverse transcription-polymerase chain reaction (RT-PCR) for monitoring aflatoxin production in Aspergillus parasiticus 439. Int J. Food Microbiol. 56:97-103. [DOI] [PubMed] [Google Scholar]
- 19.Trail, F., P. K. Chang, J. W. Cary, and J. Linz. 1994. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxins by Aspergillus parasiticus. Appl. Environ. Microbiol. 60:4078-4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woloshuk, C. P., and R. Prieto. 1998. Genetic organization and function of the aflatoxin B1 biosynthetic genes. FEMS Microbiol. Lett. 160:169-176. [DOI] [PubMed] [Google Scholar]
- 21.Xu, H., S. Annis, J. Linz, and F. Trail. 2000. Infection and colonization of peanut pods by Aspergillus parasiticus and the expression of the aflatoxin biosynthetic gene nor-1 in infection hyphae. Physiol. Mol. Plant Pathol. 56:185-196. [Google Scholar]
- 22.Yelton, M. M., J. E. Hamer, and W. E. Timberlake. 1984. Transformation of Aspergillus nidulans by using a trpC plasmid. Proc. Natl. Acad. Sci. USA 81:1470-1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, J., P. K. Chang, J. W. Cary, M. Wright, D. Bhatnagar, T. Cleveland, G. A. Payne, and J. Linz. 1995. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]