Abstract
The aim of this study was to evaluate PCR and fluorescent in situ hybridization (FISH) techniques for detecting Arcobacter and Campylobacter strains in river water and wastewater samples. Both 16S and 23S rRNA sequence data were used to design specific primers and oligonucleotide probes for PCR and FISH analyses, respectively. In order to assess the suitability of the methods, the assays were performed on naturally and artificially contaminated samples and compared with the isolation of cells on selective media. The detection range of PCR and FISH assays varied between 1 cell/ml (after enrichment) to 103 cells/ml (without enrichment). According to our results, both rRNA-based techniques have the potential to be used as quick and sensitive methods for detection of campylobacters in environmental samples.
The family Campylobacteraceae includes the genera Arcobacter and Campylobacter, characterized as fastidious gram-negative, non-spore-forming, motile, microaerobic spiral-shaped organisms (29). At the moment, most species of this family are considered of great concern for public health, and thermotolerant campylobacters, particularly Campylobacter jejuni and Campylobacter coli, are the most common human enteric pathogens causing acute bacterial diarrhea worldwide (8). Foods of animal origin and drinking water are widely regarded as the main source of food-borne infection due to the presence of those organisms as part of the intestinal flora of many animals (22). Although the majority of cases are sporadic, some outbreaks involving up to 3,500 individuals have been related to drinking untreated or inadequately chlorinated water (18).
It has been suggested that the distribution of sewage sludge to land may be one of the routes by which thermophilic campylobacters reenter the human food chain (3). Previous studies have shown that sewage and sewage sludge, respectively, contain campylobacters in concentrations of 102 to 105 CFU/100 ml and 101 to 103 CFU/100 ml, respectively (12, 27).
Little is known about the epidemiology of Arcobacter species. The fact that they have been frequently isolated from ill animals, chicken carcasses, and humans with enteritis strongly suggests that Arcobacter species may be important human pathogens (23). It has been suggested that water may play an important role in the transmission of these organisms (24), and drinking water has been cited as a major risk factor in acquiring diarrheal illness associated with Arcobacter (21).
Arcobacter species have been found in sewage and activated sludge, with frequencies varying from 41 to 80% (26), suggesting high implications for animal and human health. However, more extensive studies must be done to assess the real risk for public health.
Isolation of campylobacters may require about 4 to 5 days due to slow growth and lack of a suitable selective medium (31). Besides, campylobacters as food-borne pathogens are often stressed by nonfavorable conditions such as nutrient starvation, pH in food, or temperature variation, and they would generally be transformed into nonculturable coccoid forms (15).
Arcobacter is frequently misidentified as atypical Campylobacter when relying on conventional plating methods and phenotypic tests due to their lack of sensitivity (14). This may lead to an important underestimation of the true incidence of Arcobacter species in environmental samples and human illness.
Over the last decade, molecular techniques such as PCR-based systems have been applied to develop improved detection methods for campylobacters in stool and food samples (19). The ability of PCR to amplify specific regions of DNA has been used to identify certain campylobacters. A prerequisite for designing primers in any diagnostic assay is the availability of genomic sequence information, and 16S and 23S rRNA gene sequence data are widely used as a basic tool for the development of PCR assays for identifying bacteria.
Due to its high sensitivity, specificity, and rapid results, PCR is presented as an alternative to conventional methods. However, environmental samples may contain inhibitory substances with a significant effect on the activity of the Taq polymerase enzyme (10). Direct PCR amplification of campylobacters from water samples has proved to be difficult due to the presence of only low numbers of these bacteria in environmental resources (9). Therefore, a short preenrichment step and subsequent purification of the isolated bacterial DNA are required prior to perform a PCR (28).
To improve the efficiency of detection methods, in recent years, rRNA probe hybridization without cultivation has been widely adopted for detection of specific bacterial groups in mixed populations (2). Fluorescent in situ hybridization (FISH) with rRNA oligonucleotide probes has been used for detection and identification of different microorganisms, including Campylobacter species (20).
The FISH assay is a rapid detection method without culture, less prone to inhibitory substances, which can be used in association with PCR techniques. In this work, we report the development of a PCR assay for direct detection of Arcobacter and thermotolerant Campylobacter species in water and activated-sludge samples. In addition, a rapid in situ hybridization protocol using partial 16S rRNA gene sequence as a probe was developed to detect campylobacters in naturally and artificially contaminated samples under restrictive conditions.
The purpose was to compare the detection methods available for campylobacters, to investigate the occurrence of these organisms in water and activated sludge, and to determine if sludge flocks could act as an environmental reservoir of campylobacters.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
A total of four Arcobacter strains, nine Campylobacter strains, and 15 additional strains belonging to other bacterial genera were used to examine primer and probe specificity (Table 1). Strains Arcobacter butzleri NCTC 12481 and C. jejuni NCTC 11168 were used for inoculating samples and sensitivity tests.
TABLE 1.
Arcobacter primers and probe specifity tests
| Bacterium | Straina | Result of:
|
|
|---|---|---|---|
| PCRb | FISHc | ||
| Arcobacter butzleri | NCTC 12481 | + | + |
| Arcobacter cryaerophilus | NCTC 11885 | + | + |
| Arcobacter nitrofigilis | NCTC 11885 | + | + |
| Arcobacter skirrowii | NCTC 12713 | + | + |
| Campylobacter coli | NCTC 11366 | − | − |
| Campylobacter fetus | NCTC 10842 | − | − |
| Campylobacter helveticus | NCTC 12430 | − | − |
| Campylobacter hyointestinalis | NCTC 11608 | − | − |
| Campylobacter jejuni | NCTC 11168 | − | − |
| Campylobacter jejuni | NCTC 11828 | − | − |
| Campylobacter jejuni | NCTC 12506 | − | − |
| Campylobacter lari | NCTC 11352 | − | − |
| Campylobacter upsaliensis | NCTC 11845 | − | − |
| Clostridium perfringens | ATCC 13124 | − | − |
| Enterobacter faecalis | DSM 20478 | − | − |
| Enterobacter faecium | DSM 20477 | − | − |
| Escherichia coli | NCTC 12900 | − | − |
| Helicobacter pylori | GEHO 1 | − | − |
| Helicobacter pylori | GEHO 2 | − | − |
| Helicobacter pylori | GEHO 3 | − | − |
| Helicobacter pylori | GEHO 4 | − | − |
| Helicobacter pylori | GEHO 5 | − | − |
| Helicobacter felis | ATCC 49179 | − | − |
| Helicobacter mustelae | ATCC 43772 | − | − |
| Listeria monocytogenes | ATCC 19113 | − | − |
| Pseudomonas aeruginosa | ATCC 10145 | − | − |
| Salmonella typhimurium | NCTC 12117 | − | − |
| Wollinella succinogenes | NCTC 11488 | − | − |
Abbreviations used for culture collection: ATCC, American Type Culture Collection; DSM, Deutsche Sammlung Von Mikroorganismen; GEHO, strains kindly provided by General Hospital, Valencia, Spain; NCTC, National Collection of Type Cultures.
Primers ARCO1 and ARCO2.
With the probe ARC94.
Arcobacter strains were grown on 5% sheep blood agar plates under aerobic conditions at 30°C for 24 to 72 h. Campylobacter strains were cultured on 5% sheep blood agar plates under microaerophilic conditions (5% O2, 10% CO2, 85% N2) at 37°C for 24 to 48 h. All the isolates were stored in glycerol broth (10% [vol/vol] glycerol in 1% [wt/vol] nutrient broth number 2 [NB] [catalog no. CM67; Oxoid]) with glass beads at −80°C until required.
Preparation of samples for preliminary assays.
Overnight cultures of A. butzleri NCTC 12481 and C. jejuni NCTC 11168 were serially diluted to give 10 to 108 CFU/ml and used to inoculate 10 ml of sterile water and 10 ml of Arcobacter- and Campylobacter-free activated-sludge samples (negative detection by PCR and by culture). Samples were shaken for 1 h at 160 rpm to enable bacteria to attach to sludge particles. The amount of cells of each dilution was calculated following plating on 5% sheep blood agar plates for 48 h. For Arcobacter enrichment, 1 ml of the inoculated samples was incubated in 5 ml of NB at 30°C under aerobic conditions. For Campylobacter detection, 1 ml was inoculated in Preston selective broth (catalog no. SR117E; Oxoid) at 37°C in a microaerophilic atmosphere during 24 h. For PCR and FISH detection, samples aliquots were taken after 1, 6, 17, and 24 h of enrichment.
PCR analysis.
An amount of 1 ml of each sample was used for DNA extraction, following the CTAB method (30). Arcobacter detection was performed using primers ARCO1, 5′-GTCGTGCCAAGAAAAGCCA-3′ (forward), and ARCO2, 5′-TTCGCTTGCGCTGACAT-3′(reverse) (5). PCR primers to amplify a 439-bp 16S rRNA fragment from thermotolerant campylobacters were designed as described previously (20). Forward primer CAM 220 (5′-GGTGTAGGATGAGACTATATA-3′) corresponded to nucleotides 206 to 226, and reverse primer CAM 659 (5′-TTCCATCTGCCTCTCCCY-3′) corresponded to nucleotides 638 to 622 (Campylobacter sp. 16S rRNA gene numbering scheme).
For Campylobacter PCR assay, a final reaction volume of 50 μl was made by addition of 5 μl of each sample, 200 ng of each primer, a 0.2 mM concentration of each deoxynucleoside, 1.5 mM MgCl2, and 2 U of Taq polymerase (New England Biolabs, Inc., Beverly, Mass.). The amplification consisted of an initial DNA denaturing step at 95°C for 5 min, followed by a 33-cycle reaction (94°C for 1 min, 58°C for 1 min, 72°C for 2 min). The cycling included a final extension step at 72°C for 2 min to ensure full extension of the product (20). PCRs for detection of Arcobacter were carried out as described previously (5).
All PCRs were performed with an automatic thermal cycler (PHC-3 thermal cycler; Techne Corporation, Cambridge, United Kingdom). PCR products were analyzed by electrophoresis at 100 V for 1 h through 1% (wt/vol) SeaKem LE agarose (FMC Bioproducts) gels. Amplimers were visualized by staining with ethidium bromide under UV light. A 100-bp DNA ladder was used as a molecular weight marker.
FISH analysis.
For FISH analysis, a volume of 1 ml of each sample was centrifuged (1,000 × g at 4°C for 10 min), resuspended in PBS buffer (130 mM sodium chloride, 10 mM sodium phosphate [pH 7.2]), and fixed with three volumes of 4% paraformaldehyde for 2 h at 4°C. Subsequently, fixed samples were centrifuged again, washed with PBS buffer, and finally resuspended in 1:1 PBS-ethanol (vol/vol) as previously described (4).
Campylobacter oligonucleotide probe CAM 1, complementary to a 16S rRNA region of thermotolerant Campylobacter species has been previously described and evaluated (7, 20). The ARC94 probe, complementary to a 16S rRNA region of genus Arcobacter (25), was tested for specificity previous to its use (Table 1). Probes were synthesized and labeled by MGW Biotech (Mannheim, Germany) with 5(6)-carboxyfluorescein-N-hydroxysuccinimide ester (FLUOS) and tetramethylrhodamine-5-isothiocyanate (TRITC). An aliquot of 20 μl of fixed sample was placed on a gelatin-coated slide, air dried, dehydrated (50, 80, or 100% ethanol), and hybridized as described previously (1). To provide a specific hybridization to the target organisms, a final concentration of formamide was established at 20% in the hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl [pH 7.6]) and the NaCl concentration was established at 80 mM in the washing buffer (20 mM Tris-HCl, 0.01% sodium dodecyl sulfate, 5 mM EDTA).
The EUB338 universal probe, complementary to a region of 16S rRNA of the domain Bacteria, was used as a positive control to detect all bacteria present in the sample (1).
Slides were mounted with FluoroGuard antifade reagent (Bio-Rad) and visualized by epifluorescence Olympus microscopy BX50 with filters U-MWB, U-MWIB, and U-MWIG. Phase-contrast and fluorescence micrographs were captured on Fuji P1600 color film.
Detection of Arcobacter and Campylobacter on uninoculated water samples.
In order to evaluate the effectiveness of the PCR and FISH protocols for detecting Arcobacter and Campylobacter species on naturally contaminated samples, a total of 10 fresh water samples were obtained from the Turia River (Valencia, Spain), during a period of 3 months (September to December, 2000), with a periodicity of one sample per week. Representative samples of residual waters were collected from a secondary wastewater treatment plant (832,000 population equivalents). Samples were obtained from the influent, effluent (water), and aeration tank (activated sludge), and a total of 10 samples (S1 to S10) from each location were collected and analyzed. All the samples were placed into sterile glass bottles, refrigerated, transported to the laboratory immediately, and processed without further delay.
(i) Detection without enrichment.
Three hundred milliliters of each river and wastewater sample was centrifuged (1,000 × g) and resuspended in 1 ml of PBS buffer. Portions (500 μl) of this were used for PCR and FISH detection before enrichment. Aliquots of 100 μl of each sample were plated on modified CCDA-Preston selective agar (catalog no. CM739; Oxoid) with cefoperazone (16 g/liter for Campylobacter) and ABM medium supplemented with 5% of 5-fluorouracil (for Arcobacter) and subsequently incubated at 30 and 37°C, respectively, for 48 to 72 h.
(ii) Detection with enrichment.
From each river water sample, a 300-ml aliquot was filtered through 0.45-μm-pore-size membrane filters (Whatman, Maidstone, England). For residual water samples only 100 ml was filtered, due to the presence of suspended solids, which made the filtration difficult. The membranes were aseptically rolled and transferred to 100 ml of Preston selective broth for Campylobacter detection and to 100 ml of NB supplemented with 5% 5-fluorouracil for Arcobacter identification. Both enrichment broths were incubated in aerobic and microaerobic conditions at 37 and 30°C, respectively, for 48 h. For activated sludge, 25-ml samples were inoculated directly in enrichment broths without previous filtration.
After 24 h of incubation, 1-ml aliquots of each enrichment broth were used for PCR and FISH analysis. In order to improve efficiency of FISH detection, sludge samples were treated with tetrasodium pyrophosphate (17) to disperse the sludge particles.
For cultural detection, portions of 0.1 ml of each enrichment broth were platted on modified CCDA-Preston selective agar (catalog no. CM739; Oxoid) with cefoperazone (16 g/liter) and ABM medium supplemented with 5% 5-fluorouracil and subsequently incubated at 30 and 37°C, respectively, for 48 to 72 h.
RESULTS
PCR preliminary assay.
Alignment of GenBank published sequences of Arcobacter with other related organisms shows that the pair of primers used in this study is suitable for PCR detection of Arcobacter species. As shown in Table 1, PCRs using primers ARCO1 and ARCO2 were positive for the four Arcobacter strains and negative for the remaining bacterial genera tested.
The detection limits of PCR assays for Arcobacter and Campylobacter in inoculated water and sludge samples without enrichment or after 24 h of incubation are shown on Table 2.
TABLE 2.
Detection limits of PCR in inoculated samples
| Sample | Detection limit (CFU/ml)
|
|
|---|---|---|
| Arcobacter | Campylobacter | |
| River water | ||
| Without enrichment | 103 | 102 |
| After 24-h enrichment | 1 | 10 |
| Sludge | ||
| Without enrichment | 103 | 103 |
| After 24-h enrichment | 102 | 102 |
Incubation periods longer than 24 h did not improve the detection level for both genera, in any type of sample. Based on these results, a 24-h enrichment step was always performed when environmental samples were analyzed.
FISH preliminary assays.
Under stringent conditions, the ARC94 probe was able to detect all Arcobacter species tested, while other bacteria yielded negative results (Table 1).
Despite the fact that sludge samples showed a moderate nonspecific fluorescence background, Arcobacter and Campylobacter cells could be easily detected. The signals obtained with both probes were strong compared against background yellow signals due to nonspecific probe attachment to sludge flocks.
The sensitivity levels of FISH assays for Arcobacter and Campylobacter in inoculated water and sludge samples with or without enrichment (24 h of incubation) are shown in Table 3. Based on these results, a 24-h enrichment step was always performed when environmental samples were analyzed.
TABLE 3.
Detection limits of FISH in inoculated samples
| Sample | Detection limit (CFU/ml)
|
|
|---|---|---|
| Arcobacter | Campylobacter | |
| River water | ||
| Without enrichment | 102 | 103 |
| After 24-h enrichment | 1 | 102 |
| Sludge | ||
| Without enrichment | 103 | 104 |
| After 24-h enrichment | 102 | 103 |
Campylobacter detection in noninoculated samples.
In river water samples, a total of 6 out of 10 samples yielded the expected Campylobacter PCR 439-bp band after enrichment in Preston selective broth (Table 4).
TABLE 4.
Detection of Campylobacter in noninoculated river water samples
| Sample | Detection of Campylobacter by:
|
Culture result | |
|---|---|---|---|
| PCR | FISH | ||
| W1 | + | + | C. coli |
| W2 | − | − | |
| W3 | − | − | |
| W4 | − | − | |
| W5 | + | − | |
| W6 | − | − | |
| W7 | + | + | |
| W8 | + | − | |
| W9 | + | + | C. coli |
| W10 | + | − | C. coli |
Analysis of wastewater samples yielded PCR-positive results for Campylobacter in 12 out of 30 samples (Table 5). In three cases (S6, S7, and S8 samplings), contamination was detected in samples from influent, activated sludge, and effluent sites. In S1 sampling, Campylobacter was only detected in influent sample. Finally, in the case of S9, PCR was positive in activated sludge and effluent samples.
TABLE 5.
Detection of Campylobacter in noninoculated wastewater samples
| Sampling | Origin | Detection of Campylobacter by:
|
Culture result | |
|---|---|---|---|---|
| PCR | FISH | |||
| S1 | Influent | + | + | C. coli |
| S1 | Sludge | − | − | |
| S1 | Effluent | − | − | |
| S2 | Influent | − | − | |
| S2 | Sludge | − | − | |
| S2 | Effluent | − | − | |
| S3 | Influent | − | − | |
| S3 | Sludge | − | − | |
| S3 | Effluent | − | − | |
| S4 | Influent | − | − | |
| S4 | Sludge | − | − | |
| S4 | Effluent | − | − | |
| S5 | Influent | − | − | |
| S5 | Sludge | − | − | |
| S5 | Effluent | − | − | |
| S6 | Influent | + | − | |
| S6 | Sludge | + | + | |
| S6 | Effluent | + | + | C. coli |
| S7 | Influent | + | + | C. coli |
| S7 | Sludge | + | + | |
| S7 | Effluent | + | + | C. coli |
| S8 | Influent | + | + | C. coli |
| S8 | Sludge | + | − | |
| S8 | Effluent | + | − | C. coli |
| S9 | Influent | − | − | |
| S9 | Sludge | + | − | |
| S9 | Effluent | + | − | C. coli |
| S10 | Influent | − | − | |
| S10 | Sludge | − | − | |
| S10 | Effluent | − | − | |
When FISH analysis was performed in river water samples (Table 4), only three samples yielded positive results for Campylobacter CAM probe hybridization following 24 h of enrichment. Analysis of wastewater samples allowed the detection of Campylobacter cells in seven samples (Table 5). In one case (S7 sampling), contamination was detected in samples from influent, activated sludge, and effluent sites. In S6 sampling, hybridization assay was positive in activated sludge and effluent samples. Finally, in S1 and S8 samplings, Campylobacter was only detected in influent sample.
Campylobacter strains were isolated in three river water and seven wastewater samples. Biochemical tests identified all those isolates as C. coli.
Cultural detection of Campylobacter from sludge samples was difficult because of the massive growth of competitive biota in both Preston and ABM selective media used for isolation. So, most of the samples were considered negative as characteristic colonies could not be observed.
Arcobacter detection in noninoculated samples.
In river water samples, nine samples were positive for Arcobacter DNA fragment, from both Preston and ABM enrichment broths (Table 6).
TABLE 6.
Detection of Arcobacter in noninoculated river water samples
| Sample | Detection of Arcobacter by:
|
Culture result | |
|---|---|---|---|
| PCR | FISH | ||
| W1 | + | + | Arcobacter sp. |
| W2 | + | + | Arcobacter sp. |
| W3 | − | + | |
| W4 | + | + | |
| W5 | + | + | |
| W6 | + | + | |
| W7 | + | + | |
| W8 | + | + | |
| W9 | + | + | Arcobacter sp. |
| W10 | + | + | Arcobacter sp. |
Arcobacter was detected in 28 wastewater samples (Table 7). PCR was negative from effluent in both S2 and S5 samples.
TABLE 7.
Detection of Arcobacter in noninoculated wastewater samples
| Sampling | Origin | Detection of Arcobacter by:
|
Culture | |
|---|---|---|---|---|
| PCR | FISH | |||
| S1 | Influent | + | + | Arcobacter sp. |
| S1 | Sludge | + | + | |
| S1 | Effluent | + | + | |
| S2 | Influent | + | + | |
| S2 | Sludge | + | + | |
| S2 | Effluent | − | + | |
| S3 | Influent | + | + | Arcobacter sp. |
| S3 | Sludge | + | + | |
| S3 | Effluent | + | + | Arcobacter sp. |
| S4 | Influent | + | + | |
| S4 | Sludge | + | + | |
| S4 | Effluent | + | + | |
| S5 | Influent | + | + | |
| S5 | Sludge | + | + | |
| S5 | Effluent | − | + | |
| S6 | Influent | + | + | |
| S6 | Sludge | + | + | Arcobacter sp. |
| S6 | Effluent | + | + | |
| S7 | Influent | + | + | Arcobacter sp. |
| S7 | Sludge | + | + | |
| S7 | Effluent | + | + | Arcobacter sp. |
| S8 | Influent | + | + | Arcobacter sp. |
| S8 | Sludge | + | + | |
| S8 | Effluent | + | + | Arcobacter sp. |
| S9 | Influent | + | + | Arcobacter sp. |
| S9 | Sludge | + | + | Arcobacter sp. |
| S9 | Effluent | + | + | Arcobacter sp. |
| S10 | Influent | + | + | Arcobacter sp. |
| S10 | Sludge | + | + | |
| S10 | Effluent | + | + | |
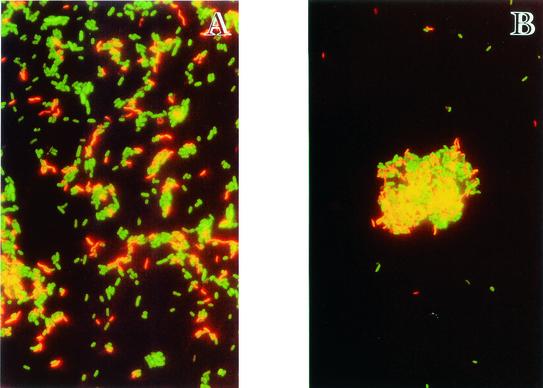
In river water, all the 10 samples were FISH positive for Arcobacter. Analysis of wastewater samples allowed the detection of Arcobacter cells in all the 30 analyzed samples, both from Preston and ABM enrichment broths. Figure 1 shows typical images of Arcobacter obtained from hybridization with fluorescence-labeled probes.
FIG. 1.
Detection of Arcobacter by whole-cell hybridization with fluorescence-labeled oligonucleotides by simultaneous application of probes ARC94-TRITC and EUB338-FLUOS in a river water sample (A) and in an activated-sludge sample (B).
Arcobacter colonies were isolated in 4 river water and in 12 wastewater samples. In all the cases, only identification to the genus level was achieved.
DISCUSSION
The comparison of results obtained using the molecular methods described in this study suggests that both PCR and FISH are suitable tools for the identification of campylobacters in water samples.
A complication when applying PCR in water microbiology is that DNA of nonculturable or dead cells may be present, thus yielding a false- positive reaction (2). In addition, inhibitory substances can inactivate the Taq polymerase (16). Nevertheless, PCR is a little more sensitive than FISH in samples containing a high number of campylobacters, and only a small portion of sample is required to obtain a successful amplification. Viability and PCR inhibitor problems were solved by a short enrichment step prior to the DNA isolation. The combination of PCR with a short enrichment step increases the level of viable cells, while the nonculturable or dead cells are diluted (10).
Eight samples that were positive for Campylobacter by PCR were negative by cultural analysis after 48 h of incubation. This may indicate that either Campylobacter was stressed, remaining viable without the capacity to grow on medium culture, or only bacterial DNA was present in the samples. When using FISH, three out of these eight samples were shown to contain Campylobacter cells. Similarly, 21 samples that were positive for Arcobacter by PCR were negative by conventional plating methods. FISH yielded positive results in all of the cases. These results show both, the great prevalence of Arcobacter in wastewater and surface water, and the inadequacy of available cultural methods for its detection. So, in the case of Arcobacter, PCR and FISH are especially useful in detecting the bacteria in environmental samples.
A previous study showed that primary sedimentation can remove more than 78% of the incoming campylobacters. Nevertheless, campylobacters are able to pass sewage treatment processes (27). The percentage of coccoid, nontypeable, and nonculturable campylobacters is increasing during clarification processes (11). Although no campylobacters are normally isolated from digested sludge it should not be called Campylobacter-free, as nonculturability cannot be equated to nonviability (13), and there is still uncertainty about the ability of campylobacters to survive in sludge in the viable but nonculturable form (6).
The FISH method has the advantage of not being inactivated by sample inhibitors even when a large amount of sample is processed (20). Besides, a protocol to obtain the DNA from bacteria is not necessary, and positive results may be directly observed in the sample. This method has also been reported to allow for the detection of viable but nonculturable forms which could not sometimes be detected by PCR due to the decrease of DNA content (2).
Samples of seeded activated sludge that were FISH positive for Arcobacter and Campylobacter strains prior to enrichment contained at least 104 cells/ml. When the seeded activated-sludge samples were incubated 24 h in Preston broth, the sensitivity level increased to 103 and 102 cells/ml for Campylobacter and Arcobacter, respectively. The signals obtained with CAM 654 were strong compared against background yellow signals due to nonspecific probe attachment to sludge flocks. The enrichment appeared to increase the level of viable cells. Additionally, high levels of naturally occurring activated-sludge microflora in flocks did not interfere with the FISH assay. In conclusion, the FISH method has potential as a quick and sensitive method for detection of Campylobacter cells in sludge samples and is relatively insensitive to false-positive results due to the presence of nonviable cells.
Both PCR and FISH techniques described here are rapid, sensitive, and specific methods to detect and identify food-borne pathogens. Moreover, a combination of both methods could be an excellent tool to detect thermotolerant campylobacters in water samples.
Acknowledgments
We are grateful to H. Meier (GSF Institute, Neuherberg, Munich, Germany) for assistance in designing primers and probes. We thank E. Echevarría from EMARSA company, which allowed us access to and sampling at the Pinedo wastewater treatment plant.
This work was supported by grant ALI96-1080 from the Comisión Interministerial de Ciencia y Tecnología (CICYT) of Spain. Y.M. is the recipient of a grant from the Ministerio de Educación y Ciencia of Spain.
REFERENCES
- 1.Amann, R. I., L. Krumholz, and D. A. Stahl. 1990. Fluorescent oligonucleotide probing of whole cells for determinative phylogenetic and environmental studies of microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arimi, S. M., C. R. Fricker, and R. W. A. Park. 1988. Occurrence of thermophilic campylobacters in sewage and their removal by treatment processes. Epidemiol. Infect. 101:279-286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aznar, R., W. Ludwig, R. I. Amann, and K. H. Schleifer. 1994. Sequence determination of rRNA genes of pathogenic Vibrio species and whole-cell identification of Vibrio vulnificus with rRNA-targeted oligonucleotides probes. Int. J. Syst. Bacteriol. 44:330-337. [DOI] [PubMed] [Google Scholar]
- 5.Bastyns, K., D. Cartuyvels, S. Chapelle, P. Vandamme, H. Goossens, and R. Dwachter. 1995. A variable 23S rDNA region in a useful discriminating target for genus-specific and species-specific PCR amplification in Arcobacter species. Syst. Appl. Microbiol. 18:353-356. [Google Scholar]
- 6.Beumer, R. R., J. De Vries, and F. M. Rombouts. 1992. Campylobacter jejuni non-culturable cells. Int. J. Food Microbiol. 15:153-163. [DOI] [PubMed] [Google Scholar]
- 7.Buswell, C. M., Y. M. Herlihy, L. M. Lawrence, J. T. M. McGuiggan, P. D. Marsh, C. W. Keevil, and S. A. Leach. 1998. Extended survival and persistence of Campylobacter sp. in water and aquatic biofilms and their detection by immunofluorescent-antibody and rRNA staining. Appl. Environ. Microbiol. 64:733-741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frost, J. A. 2001. Current epidemiological issues in human campylobacteriosis. J. Appl. Microbiol. 90:85S-95S. [DOI] [PubMed]
- 9.Giesendorf, B. A. J., A. van Belkum, A. Koeken, H. Stegeman, M. H. C. Henkens, J. van der Plas, H. Goossens, H. G. M. Niesters, and W. G. V. Quint. 1993. Development of species-specific DNA probes for Campylobacter jejuni, C. coli, and C. lari by polymerase chain reaction fingerprinting. J. Clin. Microbiol. 31:1541-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez, J., J. L. Alonso, A. Fayos, I. Amoros, and R. J. Owen. 1995. Development of a PCR assay combined with a short enrichment culture for detection of Campylobacter jejuni in estuarine surface waters. FEMS Microbiol. Lett. 127:201-206. [DOI] [PubMed] [Google Scholar]
- 11.Höller, C. 1988. Long-term study of occurrence, distribution and reduction of Campylobacter spp. in the sewage system and wastewater treatment plant of a big town. Wat. Sci. Technol. 20:529-531. [Google Scholar]
- 12.Höller, C., and V. Schomakers-Revaka. 1994. A note: comparison of different homogenization procedures for detecting Campylobacter spp. in sewage sludges. J. Appl. Bacteriol. 77:591-596. [DOI] [PubMed] [Google Scholar]
- 13.Hsueh, P., L. Teng, P. Yang, S. Wang, S. Chang, S. Ho, W. Hsieh, and K. Luh. 1997. Bacteremia caused by Arcobacter cryaerophilus 1B. J. Clin. Microbiol. 35:489-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jacob, J., I. Feuerpfeil, and E. Schulze. 1996. PCR mediated DNA fingerprinting of atypical Campylobacter strains isolated from surface and drinking water. Zentbl. Bakteriol. 285:106-112. [DOI] [PubMed] [Google Scholar]
- 15.Jones, D. M., E. F. Sutcliffe, and A. Curry. 1991. Recovery of viable but non-culturable Campylobacter jejuni. J. Gen. Microbiol. 137:2477-2482. [DOI] [PubMed] [Google Scholar]
- 16.Jones, K., M. Betaieb, and D. R. Teldford. 1990. Seasonal variation of thermophilic campylobacters in sewage sludge. J. Appl. Bacteriol. 69:185-189. [DOI] [PubMed] [Google Scholar]
- 17.Kepner, R. L., and J. R. Pratt. 1994. Use of fluorochromes for direct enumeration of total bacteria in environmental samples: past and present. Microbiol. Rev. 58:603-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Koenraad, P. M. F. J., F. M. Rombouts, and S. H. W. Notermans. 1997. Epidemiological aspects of thermophilic Campylobacter in water-related environments: a review. Water Environ. Res. 69:52-63. [Google Scholar]
- 19.Lai-King, N. J., C. Isigidi, B. Kingombe, W. Yan, D. E. Taylor, K. Hiratsuka, N. Malik, and M. Garcia. 1997. Specific detection of Campylobacter jejuni by DNA hybridization and PCR. Appl. Environ. Microbiol. 63:4558-4563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moreno, Y., M. Hernández, M. A. Ferrús, J. L. Alonso, S. Botella, R. Montes, and J. Hernández. 2001. Direct detection of thermotolerant campylobacters in chicken products by PCR and in situ hybridization. Res. Microbiol. 152:577-582. [DOI] [PubMed] [Google Scholar]
- 21.Musmanno, R. A., M. Russi, H. Lior, and N. Figura. 1997. In vitro virulence factors of Arcobacter butzleri strains isolated from superficial water samples. Microbiologica 20:63-68. [PubMed] [Google Scholar]
- 22.Pearson, A. D., M. H. Greenwood, R. K. A. Feltham, T. D. Healing, J. Donaldson, D. M. Jones, and R. R. Colwell. 1996. Microbial ecology of Campylobacter jejuni in a United Kingdom chicken supply chain: intermittent common source, vertical transmission, and amplification by flock propagation. Appl. Environ. Microbiol. 62:4614-4620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Phillips, C. A. 2001. Arcobacters as emerging human foodborne pathogens. Food Control 12:1-6. [Google Scholar]
- 24.Rice, E. W., M. R. Rodgers, I. V. Wesley, C. H. Johnson, and S. A. Tanner. 1999. Isolation of Arcobacter butzleri from ground water. Lett. Appl. Microbiol. 28:31-35. [DOI] [PubMed] [Google Scholar]
- 25.Snaird, J., R. Amann, I. Huber, W. Ludwig, and K. H. Schleiffer. 1997. Phylogenetic analysis and in situ identification of bacteria in activated sludge. Appl. Environ. Microbiol. 63:2884-2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stampi, S., G. Deluca, O. Varoli, and F. Zanetti. 1999. Occurrence removal and seasonal variation of thermophilic campylobacters and Arcobacter in sewage sludge. Zentbl. Hyg. Umweltmed. 202:19-27. [PubMed] [Google Scholar]
- 27.Stelzer, W., J. Jacob, and E. Schulze. 1991. Environmental aspects of Campylobacter infections. Zentbl. Mikrobiol. 146:3-15. [PubMed] [Google Scholar]
- 28.Van Camp, G., H. Fierens, P. Vandamme, H. Goossens, A. Huyghebaert, and R. De Wachter. 1993. Identification of enteropathogenic Campylobacter species by oligonucleotide probes and polymerase chain reaction based on 16S rRNA genes. Syst. Appl. Microbiol. 16:30-36. [Google Scholar]
- 29.Vandamme, P., and J. De Ley. 1991. Proposal for a new family Campylobacteraceae. Int. J. Syst. Bacteriol. 41:451-455.
- 30.Wilson, K. 1987. Preparation of genomic DNA from bacteria p. 2.4.1. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. A. Smith, J. G. Seidman, and K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, N.Y. [DOI] [PubMed]
- 31.Winters, D. K., A. E. O'Leary, and M. F. Slavik. 1998. Polymerase chain reaction for rapid detection of Campylobacter jejuni in artificially contaminated foods. Lett. Appl. Microbiol 27:163-167. [DOI] [PubMed] [Google Scholar]