Abstract
Waters impacted by fecal pollution can exact high risks to human health and can result in financial losses due to closures of water systems used for recreation and for harvesting seafood. Identifying the sources of fecal pollution in water is paramount in assessing the potential human health risks involved as well as in assessing necessary remedial action. Recently, various researchers have used the ribotyping method to identify sources of bacterial indicators (Escherichia coli and enterococci) in environmental waters. While these studies have identified genotypic differences between human- and animal-derived indicators that are capable of differentiating organisms isolated from humans and various animal hosts, most have focused on organisms collected from a confined geographic area and have not addressed the question of whether these ribotype profiles are watershed specific or if they can be applied universally to organisms from other geographic locations. In this study, E. coli isolates were obtained from humans, beef cattle, dairy cattle, swine, and poultry from locations in northern, central, and southern Florida and were subjected to ribotyping analysis. The intent was to determine (i) if ribotype profiles are capable of discriminating the source of E. coli at the host species level and (ii) if the resulting fingerprints are uniform over an extended geographic area or if they can be applied only to a specific watershed. Our research indicated that, using a single restriction enzyme (HindIII), the ribotyping procedure is not capable of differentiating E. coli isolates from the different animal species sampled in this study. Results indicate, however, that this procedure can still be used effectively to differentiate E. coli as being either human or animal derived when applied to organisms isolated from a large geographic region.
Fecal pollution affects the quality and safety of many water systems and can originate from a variety of human and nonhuman sources. Human fecal material is generally considered to be of greater risk to human health as it is more likely to contain human enteric pathogens (e.g., Shigella spp., Salmonella enterica serovar Typhi, hepatitis A virus, and Norwalk group viruses). However, other enteric pathogens are shared with animals (e.g., various serotypes of Salmonella and Escherichia coli). Many of these human pathogens are not readily detectable in the environment by conventional methods as they are often present in very low numbers; furthermore, many of them have a very low infectious dose, which renders even a low prevalence in polluted waters hazardous to human health. Therefore, the prediction of their presence and potential associated health risks is typically performed by the detection of established indicators of fecal pollution.
E. coli has long been used as an indicator of fecal pollution (2). It has good characteristics as an indicator, such as not normally being pathogenic to humans and being present at concentrations much higher than those of the pathogens it predicts. However, it is well established that E. coli is not limited to humans; it also exists in the intestines of many other warm-blooded animals (5). Consequently, when it is detected in water with conventional bacteriological tests, its source and the full extent of potential human health risks cannot be determined.
Testing methods capable of identifying E. coli or other indicator organisms as being derived from a specific host aid in the assessment of the potential health risks associated with their presence in a specific watershed. Multiple methods have been developed for this purpose, and this area of research has been collectively termed microbial source tracking.
One of these methods, ribotyping, has been used by several researchers to discriminate between closely related strains of bacteria as well as to track sources of fecal contamination (1, 3, 4, 6-11). While these studies have shown genotypic differences between human- and animal-derived indicators, most have focused on isolates collected from a confined geographic area and have not addressed the question of whether these profiles are watershed specific or if they can be applied universally to organisms from other geographic locations.
In this study, E. coli isolated from humans, beef cattle, dairy cattle, swine, and poultry were collected from locations in northern, central, and southern Florida and subjected to ribotyping analysis. The intent was to determine if ribotype profiles (i) were capable of discriminating the source of E. coli at the host species level and (ii) were specific for a particular animal source in a specific confined or broad geographical region.
MATERIALS AND METHODS
Collection of fecal samples from livestock and humans.
Composite fecal samples were collected from swine, poultry, dairy cattle, and beef cattle farms in three geographical regions of Florida over seasonal time intervals. Samples from dairy cattle farms were collected from retention ponds containing stall flush water located in Greenville (north), Hague (central), and Okeechobee (south). The dairy farms were at least 100 miles apart (maximum, 200 miles). Samples from beef cattle farms were collected from composite manure pits and flush water retention ponds in Lake City (north), Alachua (central), and Okeechobee (south). The beef farms were at least 50 miles apart (maximum, 200 miles). Samples from swine farms were collected from retention ponds located in Grand Ridge (north), Gainesville (central), and Dade City (south). Swine farms were at least 80 miles apart (maximum, 230 miles). Samples from chicken farms were collected from retention ponds located in Bushnell (north), Dade City (central), and Zolfo Springs (south). Poultry farms were at least 30 miles apart (maximum, 110 miles). Water samples were collected from at least three locations within the retention ponds, and at least three separate samples from composite manure pits were collected from each farm (where applicable). Human isolates were obtained directly from human volunteers, residential septic systems, and sewage lines that have no animal impact. Human-impacted sewage lines were identified as those located at points directly adjacent to buildings on the University of Florida campus. Samples were collected from outflow pipes which were not impacted by storm water runoff. After collection, all samples were stored at 4°C, transported to the laboratory in refrigerated (4°C) coolers, and processed within 24 h. A summary of the types of isolates and samples taken is shown in Table 1.
TABLE 1.
Numbers and sources of E. coli isolates used in this study
| Source | Total no. of isolates | No. of isolates (northern, central, southern) | Sample type(s) |
|---|---|---|---|
| Human | 84 | 21, 48, 15 | Septic tanks, fecal samples |
| Beef | 85 | 33, 22, 30 | Lagoon, compost pit |
| Dairy | 82 | 24, 35, 23 | Lagoon |
| Swine | 80 | 26, 26, 28 | Lagoon |
| Poultry | 70 | 21, 30, 19 | Lagoon |
Isolation of E. coli.
Fecal samples were streaked onto MacConkey agar plates (Difco) within 24 h of collection. Plates were incubated at 37°C for 24 h, and lactose-positive colonies were picked and subcultured into Luria broth (Difco) containing 4-methylumbelliferyl-β-d-glucuronide (MUG) substrate (Sigma, Inc.). MUG-positive isolates were presumed to be E. coli and were verified by using the IMViC series of tests (indole, methyl red, Voges-Proskauer, citrate). Isolates exhibiting ++−− IMViC profiles were confirmed as E. coli.
Ribotype profile database.
Over 3,000 human- and nonhuman-derived E. coli isolates had been collected previously and were used in the establishment of an original database for isolate classification by ribotype profile and discriminate analysis (8; unpublished source isolates). This database was tested for use in discriminating human versus animal isolates in the latter portion of this study.
DNA extraction.
E. coli isolates were grown overnight in Luria-Bertani broth, and DNA was extracted with a Masterpure DNA purification kit (Epicentre, Madison, Wis.) according to the manufacturer's instructions.
Determination of DNA concentration.
DNA concentration was determined with a TKO 100 fluorometer according to the manufacturer's instructions.
Restriction enzyme digestion.
Approximately 1 μg of DNA was digested with the HindIII restriction enzyme (Roche Molecular Biochemicals) according to the manufacturer's instructions. Digested DNA was separated on a 1.0% agarose gel at 30 V for 16 h in 1× Tris-borate-EDTA buffer, stained with ethidium bromide, and viewed under UV light.
Southern blot analysis.
After electrophoresis of restriction-digested DNA, agarose gels containing restricted DNA were depurinated in 0.2 M HCl for 10 min, denatured in 0.5 M NaOH-1.5 M NaCl for 35 min, and neutralized for 45 min in a buffer containing 0.5 M Tris-HCl (pH 7.2), 1.5 M NaCl, and 0.1 mM disodium EDTA. DNA was blotted from gels onto nylon membranes (Bio-Rad) with a vacuum blotting system (VacuGene XL) and fixed with shortwave UV light for 5 min.
Probe preparation.
E. coli 16S and 23S rRNA (Sigma, Inc.) was reverse transcribed into cDNA with avian reverse transcriptase and labeled with digoxigenin (DIG)-dUTP according to the manufacturer's instructions (Roche Molecular Diagnostics, Mannheim, Germany).
Hybridization and detection.
Membranes were prehybridized at 65°C for 30 min in 20 mM Na2HPO4-7% sodium dodecyl sulfate (SDS) (pH 7.2) and then hybridized in the same solution containing the DIG-labeled probe at 65°C for 16 h. After hybridization, membranes were washed twice for 60 min, each time with 20 mM Na2HPO4-5% SDS (pH 7.2) at 65°C, followed by two washes for 30 min with 20 mM Na2HPO4-1% SDS (pH 7.2) at 65°C. Membranes were then reacted with an alkaline phosphatase-conjugated anti-DIG antibody and visualized by using nitroblue tetrazolium and 5-bromo-4-chloro-3-indolylphosphate for colorimetric detection according to the manufacturer's instructions (Roche Molecular Diagnostics).
Statistical analysis of ribotype profiles as predictors of source.
Ribotype banding profiles were read by hand, and DNA fragments were translated into binary code, where the presence or absence of bands at a specific length was recorded as a 1 or 0, respectively. Binary codes were examined by using statistical discrimination methodology as implemented in SAS (SAS Institute, Inc., Cary, N.C.). Discriminate-analysis results were summarized, and the performance of the discriminating functions was defined by the average rate of correct classification and the percentages of correctly classified and misclassified isolates in a classification table created by using 10-fold cross-validation. An initial analysis examined the ability of ribotype profiles to predict the source of an isolate as to the primary farm animal (beef cattle, dairy cattle, poultry, and swine). A second analysis examined the ability of ribotype profiles to predict the source of an isolate from a comparison of the ribotypes of the livestock and human isolates collected in this study to our preexisting ribotype database. This second analysis demonstrated the degree to which human source isolates differ from farm animal and nonhuman source isolates in ribotype profiles and helps to identify a subset of ribotype profiles that might be useful as indicators of human source E. coli.
RESULTS
Over 1,800 E. coli were isolated from dairy cattle, beef cattle, swine, and poultry from north, central, and south Florida farm ponds during the spring, summer, fall, and winter seasons, and 317 of these were analyzed by ribotyping. In addition, 84 human isolates were ribotyped and subjected to discriminate analysis. The ribotype profiles were not successful in discriminating E. coli isolated from the four animal types as shown in Table 2. For this analysis, 34 ribotype profile bands were entered into a quadratic discriminate model with prior probabilities of group membership assumed to be proportional to group frequency (beef, 26.8%; dairy, 25.9%; poultry, 25.2%; swine, 22.1%). The beef and dairy isolates were collectively classified as dairy, and nearly one-half of the poultry and swine isolates are also classified as dairy. The overall misclassification rate was 65.3%. Individual misclassification rates were 94, 20, 67.5, and 81.4% for beef, dairy, poultry, and swine, respectively.
TABLE 2.
Species-level classification of ribotype profiles generated from E. coli isolated from livestock
| Source (no. of isolates) | % (no.) of isolates classified as:
|
|||
|---|---|---|---|---|
| Beef | Dairy | Poultry | Swine | |
| Beef (85) | 6 (5) | 72 (61) | 21 (18) | 1 (1) |
| Dairy (82) | 5 (4) | 80 (66) | 10 (8) | 5 (4) |
| Poultry (80) | 6 (5) | 56 (45) | 33 (26) | 5 (4) |
| Swine (70) | 4 (3) | 47 (33) | 30 (21) | 19 (13) |
Attempts were made using stepwise discriminate analysis techniques to find alternate discrimination models using fewer ribotype bands to achieve the same level of success. Most subset models produced higher misclassification rates, with more isolates being classified as dairy. Animal misclassification as a function of location (north, central, and south Florida ponds and farms) was also looked at. The pattern of misclassification was consistent for all locations (data not shown).
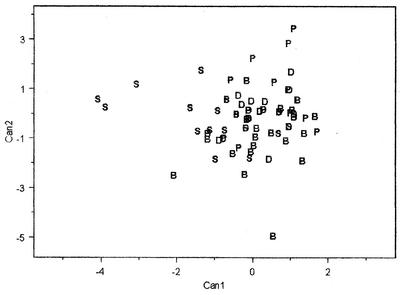
Canonical discriminate analysis was used to illustrate the difficulty of classification among the farm pond isolates. For this analysis, a set of linear functions that best separated the classes in the directions of most variability was determined. The coefficients from each linear function were used with the indicator of presence or absence of a ribotype band to develop a score for the isolate. If the animal groups were well separated, scatter plots of the canonical scores would show well-separated scatters for the classes. Figure 1 shows the scatter plot of the ribotype data from the livestock isolates. The four animal classes are not visually separated, illustrating the difficulty in separating the livestock groups. The problem is primarily due to the fact that the more common ribotype profiles are often found multiple times in each type of animal. Most isolates were classified as dairy; however, most isolates have ribotype profiles that look alike. Therefore, the dairy label is used for convenience and not as an indication of anything unique regarding the dairy livestock class.
FIG. 1.
Spatial plot of the first two canonical dimensions for ribotype profiles of E. coli isolated from livestock. B, beef; P, poultry; D, dairy; S, swine.
Ribotype profiles from the human and nonhuman isolates were originally used to construct a discrimination function as discussed by Parveen et al. (8). This function was used in this project to determine the fraction of farm pond isolate ribotype profiles that would be considered as more likely to come from a human source than a nonhuman source. As shown in Table 3, 78.6% (n = 249) of farm pond isolates were classified as likely to come from a nonhuman source and the remaining 21.4% (n = 68) were classified as human source. In addition, the human isolates collected as part of this study were correctly classified 84.5% (n = 71) of the time and misclassified 15.5% (n = 13) of the time. This further supports the idea that it may be possible to differentiate human- from animal-derived E. coli over a broad geographic region via the single-enzyme (HindIII) ribotyping procedure. The additional results from this study suggest that it will be much more difficult to discriminate between E. coli isolated from multiple nonhuman sources by using this methodology.
TABLE 3.
Classification of ribotype profiles generated from E. coli isolated from humans and livestock as human or nonhumana
| Source (no. of isolates) | % (no.) of isolates classified as:
|
|
|---|---|---|
| Nonhuman | Human | |
| Beef (85) | 84.7 (72) | 15.3 (13) |
| Dairy (82) | 80.5 (66) | 19.5 (16) |
| Poultry (80) | 71.3 (57) | 28.5 (23) |
| Swine (70) | 77.1 (54) | 22.9 (16) |
| Animal (317) | 78.6 (249) | 21.4 (68) |
| Human (84) | 15.5 (13) | 84.5 (71) |
Results of comparing ribotype profiles from livestock- and human-derived E. coli isolates to a preexisting ribotype database.
DISCUSSION
The intent of this study was to evaluate the ability of a single-enzyme ribotyping protocol to differentiate between E. coli isolates from various livestock over a broad geographic range. Although this protocol has been used within a confined watershed with success (4, 8), no information that evaluates the usefulness of this procedure over a larger geographic range is available in the literature. The results of this study indicate that this procedure may not be useful for this purpose. However, although a two-enzyme protocol was not evaluated in our present study, recent research indicates that such a protocol may be useful for this purpose and should be the focus of additional investigation (4, 9). The alternative procedure is more costly and labor intensive, however, which is an inherent drawback of the ribotyping method. In the present study, E. coli isolates were collected from southern, central, and northern Florida from beef, dairy, poultry, and swine farms. Ribotype profiles were generated from each type of animal in each geographic location until no profile variation was observed. These profiles were then cross-referenced within and among animal sources, and assessments as to whether they provided discriminatory information were made. Overlap of ribotype profiles within and among animal groups was significant. Reasons for the significant overlap in ribotype profiles, which subsequently resulted in an inability to differentiate sources of E. coli using this procedure, are not known. However, one significant difference between this study and a previous study by Carson et al. (1) is the diversity of the sample collection and, in particular, the type of samples collected. Whereas Carson et al. collected fecal samples predominantly from central Missouri from a relatively small number of individuals, we collected E. coli from a larger geographic region. Furthermore, the samples collected as part of that study were composite fecal samples from lagoons or compost pits. In our study, care was taken to ensure that samples were collected from farms housing only one type of animal. It is possible, however, that fecal material from other animals (e.g., birds) could also be present in the samples, which could have potentially caused an overlap in results obtained by ribotyping analysis. We believe, however, that the majority of organisms would have originated from the indicated animal simply due to the relative abundance of feces entering the lagoons or compost pits. This type of sample collection procedure was chosen because it is likely that these samples would contain isolates having the most potential environmental impact. In addition, it is likely that this type of sample would contain isolates that have been subjected to various external stressors, which would result in collection of organisms more likely to survive and more representative of those one would expect to find in the environment. Our results show significant overlap of ribotype profiles, perhaps indicating an ability of a subset of E. coli found within a variety of animal hosts to thrive in the environment. Therefore, a possible conclusion of the present study is that a combination of geographic and environmental variation may play a significant role in affecting the ability of ribotyping to identify sources of E. coli in the environment.
One significant result of this study was that ribotype profiles from E. coli isolated from animals still differed significantly from those obtained from human isolates. Therefore, it appears that this method may have far-reaching capacity for discriminating between E. coli isolates collected from animals and those collected from humans. Overall, the correct classification of animal-derived E. coli isolates as being either human or animal derived was greater than 78%, while the human-derived isolates collected as part of this study were correctly classified greater than 85% of the time. Although there is not an established standard of accuracy that has been defined for any bacterial source tracking method, any method with a correct rate of classification of over 50% has been considered as a worthwhile tool for predicting the potential sources of fecal pollution in environmental waters. Therefore, the results of this study indicate that the ribotyping procedure continues to have merit as a viable molecular tool to be used for this purpose.
Acknowledgments
We gratefully acknowledge the financial support of the U.S. Department of Agriculture (NMFF grant NA97FD0065).
Footnotes
Florida Agricultural Experiment Station Journal Series no. R-09019.
REFERENCES
- 1.Carson, A., B. L. Shear, M. R. Ellersieck, and A. Asfaw. 2001. Identification of fecal Escherichia coli from humans and animals by ribotyping. Appl. Environ. Microbiol. 67:1503-1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geldreich, E. E. 1966. Sanitary significance of fecal coliform in the environment. Water Pollution Control Research Series Publication WP-20-3. Federal Water Pollution Control Administration, U.S. Department of the Interior, Cincinnati, Ohio.
- 3.Hartel, P. G., W. I. Segars, N. J. Stern, J. Steiner, and A. Buchan. 1999. Ribotyping to determine host origin of Escherichia coli isolates in different water samples, p.377-382. In D. S. Olsen and J. P. Potyondy (ed.), Wildland hydrology. Technical Publication Series TPS-99-3. American Water Resources Association, Herndon, Va.
- 4.Hartel, P. G., J. D. Summer, J. L. Hill, J. V. Collins, J. A. Entry, and W. I. Segars. 2002. Geographic variability of Escherichia coli ribotypes from animals in Idaho and Georgia. J. Environ. Qual. 31:1273-1278. [DOI] [PubMed] [Google Scholar]
- 5.Orskov, F., and I. Orskov.1981. Enterobacteriaceae, p. 340-352. In A. I. Broude (ed.), Medical microbiology and infectious diseases. W. B. Saunders Corp., Philadelphia, Pa.
- 6.Parveen, S., N. C. Hodge, R. E. Stall, S. R. Farrah, and M. L. Tamplin. 2001. Phenotypic and genotypic characterization of human and nonhuman Escherichia coli. Water Res. 35:379-386. [DOI] [PubMed] [Google Scholar]
- 7.Parveen, S., R. L. Murphree, L. Edminston, C. W. Kaspar, K. M. Portier, and M. L. Tamplin. 1997. Association of multiple-antibiotic-resistance profiles with point and nonpoint sources of Escherichia coli in Apalachicola Bay. Appl. Environ. Microbiol. 63:2607-2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Parveen, S., K. M. Portier, K. Robinson, L. Edminston, and M. L. Tamplin. 1999. Discriminant analysis of ribotype profiles of Escherichia coli for differentiating human and nonhuman sources of fecal pollution. Appl. Environ. Microbiol. 65:3142-3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samadpour, M., and N. Chechowitz.1995. Little Soos Creek microbial source tracking: a survey. University of Washington Department of Environmental Health, Seattle.
- 10.Svec, P., I. Sedlacek, R. Pantucek, L. A. Devriese, and J. V. Doskar. 2001. Evaluation of ribotyping for characterization and identification of Enterococcus haemoperoxidus and Enterococcus moraviensis strains. FEMS Microbiol. Lett. 203:23-27. [DOI] [PubMed] [Google Scholar]
- 11.Tamplin, M. L., J. K. Jackson, C. Buchreiser, R. L. Murphree, K. M. Portier, V. Ganger, L. G. Miller, and C. W. Kaspar. 1996. Pulsed-field gel electrophoresis and ribotype profiles of clinical and environmental Vibrio vulnificus isolates. Appl. Environ. Microbiol. 62:3572-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]