Abstract
Enterocin AS-48 is a cyclic peptide produced by Enterococcus faecalis S-48 whose genetic determinants have been identified in the conjugative plasmid pMB2. A region of 7.8 kb, carrying the minimum information required for production of and immunity against AS-48, had been previously cloned and sequenced in pAM401 (pAM401-52). In this region, the as-48A structural gene and as-48B, as-48C, as-48C1, as-48D, and as-48D1 genes and open reading frame 6 (ORF6) and ORF7 had been identified. The sequence analysis carried out in this work in the BglII B fragment (6.6-kb) from pMB2 cloned downstream from the last ORF identified (ORF7) revealed the existence of two new ORFs, as-48G and as-48H, necessary for full AS-48 expression. Thus, JH2-2 transformants obtained with the pAM401-81 plasmid became producers and resistant at the wild-type level. Tn5 disruption experiments in the last genes, as-48EFGH, were not able to reproduce these expression levels, confirming that expression of these genes is necessary to get the phenotype conferred by the wild-type pMB2 plasmid. The as-48EFGH operon encodes a new ABC transporter that could be involved in producer self-protection. On the basis of the observed similarities, As-48G would be the ATP-binding domain, the deduced amino acid sequences of As-48E and As48-H could be assigned as transmembrane subunits, and As-48F, with an N-terminal transmembrane segment and a coiled-coil domain, strongly resembles the structure of some known ABC transporter accessory proteins whose localization in the cell is discussed. This cluster of genes is expressed by two polycistronic mRNAs, T2 and T3, in JH2-2(pAM401-81) in coordinate expression. Our results also suggest that expression of T3 could be regulated, because in JH2-2(pAM401EH) transformants, T3 was not detected, suggesting that these genes do not by themselves confer immunity, in accordance with the requirement for the as-48D1 gene for immunity against AS-48.
In recent years there has been renewed interest in the search for bacteriocins produced by lactic acid bacteria due to their high efficiency in controlling the growth of pathogenic or harmful microorganisms, which makes them attractive as food preservatives. Considerable progress has been made toward a better genetic characterization of these substances, but to date, the use of bacteriocins, other than nisin and pediocin, is still at an experimental stage. Nevertheless, they may have future application in enhancing the safety and extending the shelf life of many inherently perishable foods.
Enterocin AS-48 is a cyclic globular peptide of cationic nature, without modified residues and very stable in a wide range of pHs and temperatures. AS-48 is active against many gram-positive bacteria (encompassing undesirable and pathogenic microbes, such as Clostridium, Bacillus, Staphylococcus, and Listeria), as well as some gram-negative bacteria (11, 24). The latter group, especially enterobacteria, are much more resistant at physiological pH, probably due to the protective effect of the outer membrane, requiring acid or basic pH or combined treatments with heat and/or permeabilizing agents (1). We have previously shown that bacteriocin AS-48 interacts with the cytoplasmic membranes of sensitive bacteria to induce ion permeation, accompanied by the collapse of the membrane potential, probably by means of a molecular electroporation mechanism (12, 14). Because of these characteristics, AS-48 offers promising perspectives for biotechnological applications; thus, the possibility of transferring AS-48 production to microorganisms of industrial interest prompted us to explore the as-48 biosynthetic gene cluster.
In previous studies, it has been shown that the basal level of the AS-48 phenotype (production and immunity) requires the coordinated expression of six genes identified in the as-48 region (as-48ABCC1DD1) (23). However, the cooperation of a new operon, as-48EFGH, described in this work, seems to be necessary for full expression of the AS-48 phenotype. We have verified that the new operon identified encodes a new ABC transporter that could be involved in the self-protection mechanism of these organisms against exogenously administered AS-48 (resistance). These subjects, in addition to the characterization of functional domains in the different proteins identified and the analysis of the transcription of this cluster, are discussed.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The plasmid-free strain Enterococcus faecalis JH2-2 (17) was used in cloning experiments and heterologous production of AS-48. E. faecalis JH2-2(pAM401-81) was a construction obtained by cloning in frame the BglII B fragment of pMB2 into pAM401-52 (23). This construction (pAM401-81) was transferred by electroporation to E. faecalis JH2-2. Enterococci were grown overnight without aeration at 37°C in buffered brain heart infusion (BHI-B). Escherichia coli strain DH5α (Bethesda Research Laboratory) was used as an intermediate host for cloning. It was grown with shaking in Luria broth at 37°C. Plasmid pAM401 was used as an E. coli-E. faecalis shuttle vector (40). All plasmids used and constructed in this work are listed in Table 1. Antibiotics were added at the following concentrations: ampicillin, 50 μg/ml; chloramphenicol, 20 μg/ml; tetracycline, 10 μg/ml; and kanamycin, 30 μg/ml.
TABLE 1.
Plasmids used in this studya
| Plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| pGEM-3Zf(+) | Ampr; cloning vector | Promega |
| pSL1180 | Ampr; cloning vector | Amersham Biosciences |
| pAM401 | Cmr Tcr; bifunctional E. coli-E. faecalis cloning vector | 23 |
| pMB2 | 68 kb; AS-48+ AS-48r | 22 |
| pAM401-52 | Cmr; 7.8-kb SphI-BglII-BglII fragment from pMB2 in pAM401; (as-48ABCC1DD1 and ORF6-ORF7); AS-48+ AS-48r | 23 |
| pAM401-76 | Cmr; 1.9-kb SphI-BglII fragment and BglII B fragment (6.6 kb) from pMB2 in pAM401; AS-48− AS-48s | This work |
| pSL-B | BglII B fragment (6.6 kb) from pMB2 cloned in pSL1180 | This work |
| pSL-C | BglII C fragment (6.4 kb) from pMB2 cloned in pSL1180 | This work |
| pAM401-81 | Cmr; 25-kb D (SphI-BglII) and C and B (BglII) fragment from pMB2 cloned in frame in pAM401; AS-48+ AS-48r | This work |
| pAM401EH | Cmr; cloning of as-48EFGH by deletion of the SphI fragment (8.2 kb, including as-48A to D1 genes) in pAM401-81::Tn5D1-E mutant; AS-48− AS-48s | This work |
Cmr, chloramphenicol resistance; AS-48+, AS-48 production; AS-48−, AS-48 activity not detected; AS-48r, AS-48 resistance; AS-48s, AS-48 sensitive; Ampr, ampicillin resistance; Tcr, tetracycline resistance.
Antimicrobial-activity assays.
Production of AS-48 by E. faecalis transformants and mutants was assayed by spotting 2 μl from a liquid overnight culture onto BHI agar (1.5%), followed by incubation at 37°C for 16 h. The plate was then overlaid with 5 ml of BHI-B- soft agar (BHI, 1.5%; agar, 0.75%) containing a 2% inoculum of sensitive E. faecalis JH2-2 cells and incubated at 37°C for 12 to 18 h before the results were read. The degrees of sensitivity of the different mutants obtained were determined as described previously (23).
Plasmids and DNA manipulations.
DNA cloning and E. coli transformations were performed according to standard protocols (21). E. faecalis plasmid DNA was extracted according to the method of Anderson and McKay (4). E. faecalis cells were transformed by electroporation as described previously (10). Chloramphenicol-resistant transformants were screened for AS-48 production by replica plating and overlaid with a sensitive strain. DNA sequencing was performed with the ABI PRISM (Applied Biosystems) Dye Terminator cycle-sequencing ready-reaction kit (Perkin-Elmer). Synthetic oligonucleotides were provided by Amersham-Pharmacia Biotech.
Generation of transposon insertional mutant.
Transposition of Tn5 into pAM401-81 was accomplished by introducing the plasmid into E. coli RYC1000 (23). Selected pAM401-81::Tn5-mutagenized DNAs were transferred to E. faecalis JH2-2 by electroporation, and the transformants obtained were screened for AS-48 production and immunity.
Northern blot analysis.
Total RNA of E. faecalis was isolated as described by Tomita et al. (36) and separated electrophoretically in a 1% agarose-formaldehyde gel (10 μg/lane). RNA was transferred by capillary blotting onto nylon membranes (Hybond-N+; Amersham-Pharmacia Biotech) in 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) overnight and fixed by UV illumination for 3 min. The filters were incubated overnight at 50°C in a buffer with a high sodium dodecyl sulfate concentration (7% sodium dodecyl sulfate, 50 mM sodium phosphate buffer, 50% formamide, 5× SSC, 2% blocking reagent, 0.1% N-laurylsarcosine, pH 7) containing 100 ng of specific probes. The probes used in the Northern blot experiments were labeled in a thermocycling reaction using the deoxynucleotide triphosphate-labeling mix of a digoxigenin (DIG)-DNA labeling and detection kit obtained from Boehringer (Mannheim, Germany) with pAM401-81 as a template. The following primers were used: as-48C1-2 (5′-GGTAGGAGTAAAGTAATG-3′ ) and as-48D-3 (5′-AGGTTCATCTAATAATAC-3′) for as-48D and as-48F-8 (5′-CGGTGAAACAATGATTGA-3′) and as-48G-1 (5′-GGAACGACTTCAGGGTT-3′) for as-48FG. DIG-labeled RNA-DNA hybrids were detected with the DIG DNA detection kit (Boehringer Mannheim) following the manufacturer's instructions. A lane was cut off and stained with ethidium bromide, and the ribosomal RNAs were used as the molecular weight reference.
Prediction of transmembrane protein segments.
The prediction of transmembrane protein segments (TM) was carried out by six different methods: TopPredII (39) (default settings, as implemented in SMART, TMPred (16), TMAP (28), MEMSAT (19) (default settings), PHD (21), and SOSUI (15). PHD also provided homology analysis and the secondary-structure prediction of the sequences.
Identification of domains and functional assignment of sequences.
For the identification of domains and functional assignment of sequences, SMART (33), SCANPROSITE (5), and GENEQUIZ (18) were used. For homology searching, we employed FASTA, WU-BLAST2, and PSI-BLAST (3, 25). REPRO (13) and SAPS (6) were used to obtain internal protein sequence repeats and statistical analysis of the sequences. The tentative models for the sequences with homologues of known structure were obtained with Swiss-Model (26, 27). Multiple alignments were obtained with MULTALIN (8), and the Gribskov profile analysis was carried out through the W2H interface of the Genetics Computer Group (34).
Nucleotide sequence accession numbers.
The nucleotide sequences of as-48ABCC1DD1 and as-48EFGH from the pMB2 plasmid were submitted to EMBL and were given the accession numbers Y12234 and AJ438950, respectively.
RESULTS AND DISCUSSION
Identification of the complete gene cluster involved in the AS-48 phenotype.
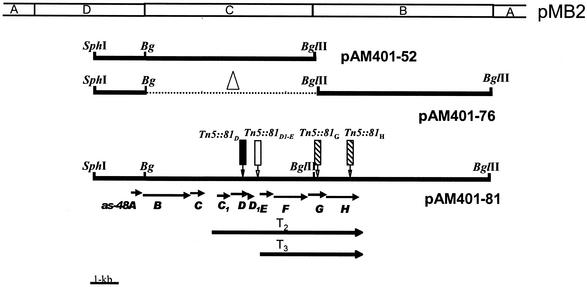
The essential genes involved in the enterocin AS-48 phenotype had been previously identified in the D (SphI-BglII) and C (BglII) fragments of the pMB2 plasmid, which were cloned in pAM401 and expressed in E. faecalis JH2-2(pAM401-52) (23). Analysis of these transformants revealed the presence of six genes involved in production of and immunity against AS-48 (as-48A, as-48B, as-48C, as-48C1, as-48D, and as-48D1), as well as two additional ORFs, ORF6 and -7, of unknown function (Fig. 1) (23). However, the levels of production and resistance of these transformants against high AS-48 concentrations were lower than those produced by the wild-type strain S-48 and the A-48-32 mutants (Table 2). These results suggest that additional genes are necessary for complete expression of the AS-48 phenotype. For this reason, the 6.6-kb BglII B fragment from pMB2 (Fig. 1) was cloned in frame into pAM401-52. For this purpose, we obtained an intermediary plasmid (pAM401-76) by ligation of the BglII B fragment from pSL-B with the D fragment from pAM401-52 (obtained by digestion with BglII-BamHI) (Table 1). Finally, ligation in frame of the C fragment from pSL-C into the pAM401-76 plasmid, previously digested with BglII, yielded plasmid pAM401-81 (25 kb). The proper orientation of the D, C, and B fragments was confirmed by restriction analysis. This plasmid was transferred by electroporation to E. faecalis JH2-2, and the phenotype of the transformants is summarized in Table 2. JH2-2(pAM401-81) transformants produced zones of inhibition similar to those of the wild-type strain A-48-32 (16-mm diameter versus the 9-mm diameter produced by pAM401-52 transformants) and were also resistant to 18 μg of pure AS-48 added exogenously (versus 3 μg for pAM401-52).
FIG. 1.
Physical map of the as-48 gene cluster cloned in E. faecalis JH2-2(pAM401-81). There is a succession of 10 genes, all running in the same direction, extended over 10.4 kb. The arrows indicate relative directions of transcription. The diagram displays a partial restriction map and the positions of Tn5 insertions obtained in this study (solid box, nonproducer strain; shaded box, hypoproducer strain; open box, wild-type phenotype). Bg, BglII. T2 and T3 represent the transcriptional units encompassing the as-48C1DD1EFGH and as-48EFGH genes, respectively.
TABLE 2.
Production and resistance against AS-48 in E. faecalis harboring plasmids used in this work
| Bacterial strain | Plasmid | AS-48 production (mm)a | Sensitivity to AS-48 at (μg/5 μl)b:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0.5 | 1 | 3 | 6 | 9 | 18 | |||
| E. faecalis JH2-2 | pMB2 (control) | 16 | R | R | R | R | R | R |
| pAM401 | S | S | S | S | S | S | ||
| pAM401-76 | S | S | S | S | S | S | ||
| pAM401-52 | 9 | R | R | R | S | S | S | |
| pAM401-81 | 16 | R | R | R | R | R | R | |
| pAM401EH | S | S | S | S | S | S | ||
| pAM401-81::Tn5D1-E | 16 | R | R | R | R | R | R | |
| pAM401-81::Tn5G | 11 | R | R | R | S | S | S | |
| pAM401-81::Tn5H | 11 | R | R | R | S | S | S | |
| E. faecalis A-48-32 | pMB2 (control) | 16 | R | R | R | R | R | R |
To test AS-48 sensitivity, 5-μl spots containing different amounts of purified AS-48 were applied on lawns seeded with standard innocula of E. faecalis JH2-2 harboring the different plasmids. The plates were examined for inhibition halos (diameters are given) after 18 h of incubation at 37°C.
R, resistant; S, sensitive. Production of AS-48 was determined against E. faecalis JH2-2(pAM401), using A-48-32 (wild-type phenotype) as a control.
The nucleotide sequence of the BglII B fragment cloned in pAM401-81 has been determined. According to the codon usage of Enterococcus, computer analysis revealed two new ORFs, as-48G and as-48H, within the B fragment newly sequenced on both strands, which were transcribed in the same direction as the rest of the as-48 cluster (Fig. 1). Potential ribosome binding sites (GACGG and AAGGAG) were identified preceding the two new ORFs.
ORF6 (as-48E) is predicted to encode a protein (169 residues) with a theoretical molecular mass of 19 kDa and a pI of 10.2 (23). In the intergenic region of 204 nucleotides (nt) after as-48D1, a putative promoter, TGTAAA (−35) and TATCTA (−10) (separated by 20 nt), is followed by 100 nt of untranslated sequence preceding ORF6, which encodes As-48E. In this region, moreover, there is a series of conserved sequences, TATGTA, TATTAG, and TATGAA, separated by 9 nt of unknown significance. ORF7 (as-48F) overlaps ORF6 and encodes a 407-residue protein with a molecular mass of 45.1 kDa and a pI of 5.4, the only acidic protein of this cluster of genes (23). ORF8 (as-48G) also overlaps ORF7. This ORF spans 683 bp and encodes a predicted protein of 227 amino acids (As-48G) with a pI of 7.8. The 3′ end of as-48G overlaps ORF9 (as-48H) by 3 bp. This fourth ORF (1,199 bp) might code for a protein of 399 amino acids (As-48H). Interestingly, as-48E, as-48F, as-48G, and as-48H are contiguous, since their initiation codons overlap and no terminator sequence was found among them. This type of structure, which has been found in other operons, could induce a translational coupling of these genes that could ensure an appropriate ratio of their products (38).
As-48EFGH is a new multicomponent ABC transporter.
A homology search for the As-48EFGH proteins revealed similarities to proteins from different ABC transporters. Moreover, we found a high degree of homology between the DNA sequence from as-48EFGH and bacFGHI from bacteriocin 21, a bacteriocin encoded by the pheromone-responsive plasmid pPD1 with 100% homology to AS-48 (37).
As-48EFGH constitutes a new ABC transporter, with three subunits and an accessory factor. This ABC system could have its counterpart in the proteins YknXYZ and Yvr(M)NOP, both ABC transporters from Bacillus, and to a lesser extent in LolCDE from E. coli, which are part of the ATP-dependent transport system involved in the release of targeted lipoproteins from the inner to the outer membrane. As-48-G, YknY, YvrO, and LolD are the ATP-binding subunits. The membrane subunits are As-48H and As-48E, YknZ, YvrN and YvrM, and LolC and LolE. Finally, As-48F, YknX, and YvrP are the accessory proteins for the ABC transporters, which apparently have no equivalents in the lipoprotein-releasing system of E. coli.
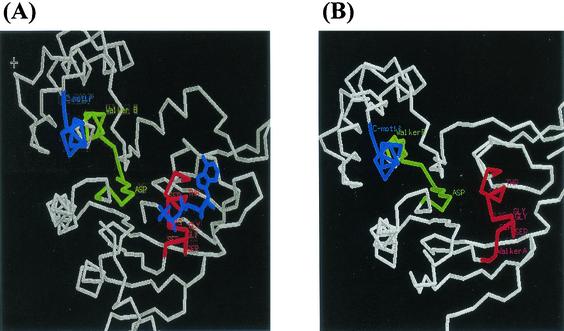
The deduced translation product of ORF8 (As-48G) is a partially hydrophilic protein lacking any extensive hydrophobic stretches capable of forming transmembrane segments. Results obtained with SCANPROSITE confirm the existence of two ATP-GTP-binding motifs: the sequences GPSGSGKSTLINL (residues 38 to 45) and RALINHPKFIIADEPT (less well conserved at positions 142 to 156), corresponding to the A and B motifs found in the ABC transporters (9). The Walker B site is immediately preceded by a highly conserved sequence, the C motif, LSGGQ(Q)QR, in which the second Gln is a conservative change in relation to the Arg or Lys of the signature sequence LSGGQ(RK)QR. It has been suggested that this region is involved in the transduction of the energy of ATP hydrolysis to the conformational changes in the integral membrane domains required for translocation of the substrate. The highest degree of homology found with WU-BLAST and FASTA was with transporters from Bacillus, YvrO (P = 10−54) and YknY (P = 10−52). The similarity to the lipoprotein-releasing system protein LolD from several organisms (Vibrio cholerae, Haemophilus influenzae, and E. coli) also displayed very high significance scores. In the case of As-48G, a putative structure can be ventured. In fact, the multiple alignment of As-48G with a representative set of homologous ABC transporters and the homologous sequence in PDB (1B0U, the ATP-binding subunit of the histidine permease HisP from Salmonella enterica serovar Typhimurium) shows four conserved regions that may have functional relevance: (i) the segment 31 to 47, which is an extension of the Walker A motif; (ii) the segment 80 to 90, around the sequence RNxxxGFIFQ and structurally between Walker A and Walker B; (iii) the segment 126 to 174, around the ABC signature and the contiguous Walker B motif; and (iv) the segment 194 to 205, around MVTHNPEV, close to the C-terminal end. Based on this homology with the structure 1B0U, we used Swiss-Model to obtain a tentative structure of As-48G (Fig. 2). The figure shows the A, B, and C motifs in the HisP structure and the equivalent regions in the proposed structure for As-48G. The labeled amino acids are those that satisfy the consensus sequences of the motifs.
FIG. 2.
Hypothetical structure proposed for As-48G protein (B), based on its homology with the structure of the ATP-binding subunit of the histidine permease HisP from S. enterica serovar Typhimurium (A), using Swiss-Model.
Although the hydrophobic domain of ABC transporters has six TM, transporters that have only four TM, encoded by individual genes, have also been described. This would be the case for our ABC system, in which the products of the as-48E and as-48H genes, with only four transmembrane α-helices each, display high homology with membrane proteins associated with transport processes, as shown below.
For As-48H, all the methods used predicted the existence of four transmembrane sequences, the first close to the N-terminal end and three grouped at the C-terminal end. The analysis of internal repeats by REPRO indicates some coincidences that could be due to the lower compositional diversity of the TM. In this sense, there are remarkable similarities between TM1 and TM3 and between TM2 and TM4, in both cases with the same orientation in the membrane. A different orientation occurs in the alignment of TM1 and TM4, although it seems more significant.
With a reliability of 0.99, GENEQUIZ assigns a functional homology with the hypothetical ATP-binding transport protein YbjZ from E. coli; analysis with PHD shows homology with the YcfU, FtsX, and YcfW proteins, related to families of GTP-binding proteins. Alignment of As-48H and several of their most homologous ABC transporters shows that the best-conserved regions are TM1 and TM2, the loop between TM2 and TM3, and the end of TM4 (Fig. 3). The conserved loop between TM2 and TM3 may constitute a recognition site for the ATP-binding subunit, in our case, As-48G. Although to a lesser extent, there are also similarities in the soluble regions of these sequences. Some of these homologues (like YvrN or YknZ from Bacillus) have a length of ∼600 amino acids and also include an ATP-binding domain. Interestingly, the TM assignments for these proteins coincide with the consensus prediction for As-48H, which clearly indicates that all these proteins form part of a particular family. Considering that all of the proteins homologous to As-48H appear in Interpro to belong to the family DUF214, our observation would offer a putative characterization for this group of sequences. The homology with ABC transporters with an N-terminal ATP-binding domain demonstrates that the membrane domains of these proteins, which have an equivalent in As-48H, can function together with a soluble ATP-binding transporter. The distance between the first and the second TM is in all cases ∼230 residues, and the rest of the architecture seems equivalent.
FIG. 3.
Multiple sequence alignment of As-48H with several related proteins using MULTALIN (9). The sequences used are Q9K885 (from Bacillus halodurans), YknZ and YvrN (from Bacillus subtilis [BACSU]), Q9K6Q4 (an ABC transporter from B. halodurans), Q9PHQ9 (from Campylobacter jejuni), Q9K0N7 (from N. meningitidis), and LolC (from E. coli [ECO]).
For As-48E, all the methods predicted four TM with both N- and C-terminal ends in the cytoplasmic side of the membrane. The secondary-structure prediction provided by PHD assigns an external helix between the first and second TM. Results obtained with BLAST show homology of As-48E with the intracellular septation protein IspZ from H. influenzae, an efflux protein from Campylobacter (interestingly, belonging to the DUF7 family, which contains some proteins that mimic the effect of GroE overexpression), and other proteins, like LctE from Lactococcus lactis, a putative ABC transporter also involved in lacticin 481 immunity (32). Viewing the TM as a helical wheel, there is a notable presence of five residues of Ser or Thr on the same face of the helix in TM3, which could be tentatively interpreted as an indication of the internal wall of a putative pore (data not shown).
The product of as-48F is the sole acidic protein found in the as-48 gene cluster, with a full hydrophilic profile, except for an N-terminal hydrophobic segment. In fact, all the methods applied agree on the prediction of one TM close to the N terminus. According to SMART, GENEQUIZ, and PHD, there is also a coiled-coil region in the molecule (amino acids 106 to 150 and 190 to 217). A functional assignment as an ABC transporter-like protein is provided by GENEQUIZ (reliability, 0.7) and by PHD, PSI-BLAST, and WU-BLAST, which identify the homology with the ABC transporters YknX (P = e−12) and YvrP (P = e−12) and with the protein AcrA/AcrE (P = 8e−7) of the HlyD family. SMART also finds a similarity to the nucleotide exchange factor GrpE (amino acids 113 to 268; E = 0.12), which stimulates the ATPase activity of the DnaK chaperone. All of these proteins are anchored by an N-terminal transmembrane region and present a coiled-coil region. The alignment of As-48F with the most homologous proteins showed some markedly conserved regions, such as the segment 77 to 90, and especially from residue 360 to the C-terminal end. These sequences clearly should be considered as suitable targets for further functional studies by mutagenesis. We have also compared our sequence to a representative sample of the so-called accessory proteins. Our results have shown the existence of several conserved regions in As-48F, including CbnD, an accessory protein from Carnobacterium, which is the closest sequence in this alignment. All these accessory proteins present, like As-48F, an N-terminal transmembrane segment and a coiled-coil region. When an alignment of the soluble portion of As-48F (residues 61 to 460) with other homologous sequences was used for the construction of a Gribskov profile, it was found that proteins of the family AcrA/AcrE are similar, with significant Z-score values. This is the case for AcrA/AcrE from Neisseria (Z = 27.1), AcrE from Aquifex (Z = 17.4), and AcrA from E. coli (Z = 16.2), but also for the hemolysin secretion protein HlyD from Synechocystis (Z = 14.8), the PrtE protein from Erwinia (Z = 13.1), HlyD from E. coli (Z = 10.5) and Salmonella (Z = 9.5), the ABC exporter accessory protein SapE from Lactobacillus sakei (Z = 9.1), and LcnD from L. lactis (Z = 8.04). A remarkable similarity was also found for the heat shock protein GroES from several origins (Z = 6.4), which could be related to the similarity obtained with GrpE. When several secondary-structure-predictive methods were used with As-48F, all of them were found to agree on the existence of a mainly helical region in the region 110 to 200, which is followed by a largely beta domain (approximately from residue 220 to the end of the protein) (data not shown). In a similar way, the protein GrpE mentioned above also presents the organization α + β.
To assign a function to As-48F, it might be necessary to determine its localization in the cell. Interestingly, most of the methods used for topology prediction locate the soluble part of As-48F in the cytoplasm. However, the results obtained with WU-BLAST clearly suggest homology of the soluble segment of As-48F with the proteins YvrP and YknX from Bacillus and, to a lesser extent, with AcrA/AcrE from Neisseria meningitidis, whose outer location has been proposed due to their similarity with the HlyD protein from E. coli. However, the homology of As-48F with the nucleotide exchange factor GrpE can be explained only by assuming a cytoplasmic localization for the protein (in accordance with the results of Tmpred, MEMSAT, and TMAP). In this location, As-48F could stimulate the ATPase activity of As-48G, accelerating the release of ADP and allowing As-48G to recycle more efficiently. The similarity obtained with GroES also supports this putative function.
Tn5 insertion mutants.
In order to discover the function of the new ABC transporter As-48EFGH described above, we obtained mutants with altered AS-48 expression after Tn5 insertion into some ORFs from JH2-2(pAM401-81). The precise locations of the Tn5 insertions were determined by EcoRI and BamHI restriction and nucleotide sequence analyses.
The phenotypes of mutants with Tn5 insertions into the new ORFs identified in this work (as-48G and as-48H) indicated that they were still capable of producing enterocin, although in lesser amounts (11 mm of halo), suggesting that they are not directly involved in AS-48 biosynthesis. However, the resistance of these mutants was clearly reduced (as much as 16.6%) (Table 2).
Interestingly, Tn5 insertion in the intergenic region as-48D1-E (mapped 147 bp upstream of as-48E; pAM401-81::Tn5D1-E mutant) (Fig. 1) showed a wild-type phenotype without any polar effect on the expression of the last gene (Table 2). This result could be interpreted as the last four genes constituting an independent transcription unit (see the discussion below) and, of course, requiring their own promoter. In order to confirm this fact, we constructed pAM401EH by the deletion of an 8.2-kb SphI fragment (including the as-48A to -D1 genes) in a pAM401-81::Tn5D1-E mutant, using the internal SphI site from Tn5 (position 2080). However, JH2-2(pAM401EH) transformants did not show resistance to AS-48 (Table 2), suggesting that these genes do not by themselves confer immunity.
Transcriptional analysis in the as-48EFGH region.
The expression of the as-48EFGH gene cluster was studied by Northern analysis using total RNA extracted by the hot-phenol method from transformants of JH2-2 harboring pAM401-81 or pAM401EH or from some mutants obtained in this study. PCR-amplified specific fragments from the as-48 region were used as probes.
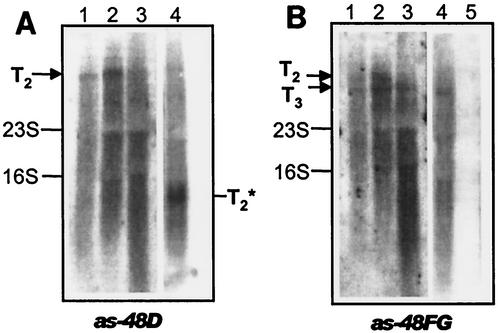
Transcription analysis carried out previously in JH2-2(pAM401-52) demonstrated the existence of a T2 transcript for as-48C1DD1 genes of 2.7 kb (23). However, analysis of this transcript in JH2-2(pAM401-81) using probes from the as-48C1D genes showed that T2 also transcribes the last four ORFs described in this work (as-48C1DD1EFGH) (Fig. 4A, lanes 1, 2, and 3), with a size of 6.4 kb. Expression of T2 at different times (1.5 to 5 h) revealed that this mRNA is only present in the very early phase of exponential growth (1.5 to 3 h) and disappears after 5 h of growth. As expected, when the experiment was carried out using RNA from a JH2-2(pAM401-81::Tn5D-E) mutant, a smaller band corresponding to a truncated T2 was detected (Fig. 4A, lane 4). The major new finding was obtained when probes from the as-48FG genes were used. In this case, two transcripts were detected: the T2 described above (for the as-48C1DD1EFGH genes) and a new mRNA (T3) with a size of 5.4 kb, corresponding to the transcription of the last four genes (as-48EFGH) (Fig. 4B, lanes 1, 2, and 3), confirming the existence of its own promoter. The T3 signal was not due to the processing of T2, because it was present in the insertion mutant pAM401-81::Tn5D1-E (Fig. 4B, lane 4). In addition, T3 is abundant throughout the logarithmic growth phase of the cell (from 1.5 to 5 h). These results corroborate the importance of these gene products in the cell, whose expression is safeguarded by the two mRNAs T2 and T3.
FIG. 4.
Transcriptional analysis of the as-4EFGH gene cluster carried out in JH2-2(pAM401-81) and several mutants. The DIG-PCR fragments of the different genes used as probes are indicated at the bottom of each panel. Approximately 10 μg of total RNA was loaded in each lane. The growth conditions are described in Materials and Methods. Total RNAs were extracted at different times of growth. (A) Lanes 1, 2, and 3, total RNA from E. faecalis JH2-2(pAM401-81) at 1.5, 3, and 5 h of exponential growth; lane 4, E. faecalis JH2-2(pAM401-81::Tn5D1-E) at 3 h of growth hybridized with a probe derived from the as-48D gene. T2∗ is a truncated T2 transcript. (B) Lanes 1, 2, and 3, total RNA from E. faecalis JH2-2(pAM401-81) at 1.5, 3, and 5 h of exponential growth; lane 4, total RNA from JH2-2(pAM401-81::Tn5D1-E; lane 5, RNA from JH2-2(pAM401EH) at 5 h of growth, hybridized with a probe derived from the as-48FG genes.
Transcription analysis carried out in the JH2-2(pAM401EH) transformants showed that the genes were not transcribed under any of the tested growth conditions (Fig. 4B, lane 5), and no increase in resistance against AS-48 could be demonstrated (Table 2), confirming that this plasmid did not confer self-protection. These results are in accordance with the requirement for the as-48D1 gene for immunity against AS-48, previously demonstrated (23), and suggest that expression of T3 could be regulated, although systems of translation signals involving histidine kinases and cytosolic proteins have not been identified in the as-48 cluster. However, there are some molecular features in the promoter region of as-48E (a series of conserved sequences separated by 9 nt) that suggest the existence of expression by means of transcriptional control, as occurs with other bacteriocins that have been described (29). In this way, T3 expression could be dependent on regulatory proteins belonging to an additional transcription unit(s), but AS-48 is apparently not a signaling molecule for T3 transcription, because in mutants lacking the structural gene, transcription of T2 and T3 could be detected (results not shown).
Conclusions.
In the as-48 gene cluster, two ABC systems coexist: a previously identified as-48C1D transporter (23), devoted to the cleavage and export of newly synthesized bacteriocin and providing low levels of immunity, which in any case could be replaced by the presence of the second ABC transporter described in this work, as-48EFGH, which would be mainly related to higher resistance against exogenously administered AS-48. For this reason, As-48EFGH is proposed to represent a multicomponent ABC system involved in self-protection against AS-48, in addition to the as-48D1 immunity gene, operating as a second immunity mechanism. The close proximity of the last four ORFs, three subunits (as-48EGH) and an accessory factor (as-48F), which ensures the production of these proteins at the equimolecular ratio, seems to confirm this hypothesis. The existence of two ABC systems has also been described in the cluster genes of nisin, subtilin, lacticin 481, epidermin, mutacin II, and mersacidin (2, 7, 20, 29, 31, 35) and could represent the conservation of a general resistance mechanism with secretion and protection functions during evolution, probably interacting cooperatively with the immunity protein. This type of transporter has been described in bacteriocins that produce pores in the cytoplasmic membrane, and its function seems to be to keep the bacteriocin concentration in the cytoplasmic membrane below the critical level necessary for pore formation (14, 30), either by active transport of the mature molecules to the insides of the cells or by active extrusion of bacteriocin molecules. Finally, the activity proposed in this work for As-48F, accelerating the release of ADP from As-48G, would make the self-protection of this ABC transporter against AS-48 more efficient.
Acknowledgments
We thank A. Pedro Collado for his help in the transcription analysis.
This work was supported by the Spanish Dirección General de Investigación Científica y Técnica (Project BIO98-0908-CO2-01) and Plan Andaluz de Investigación (CVI 160).
REFERENCES
- 1.Abriouel, H., E. Valdivia, A. Gálvez, and M. Maqueda. 1998. Response of Salmonella choleraesuis LT2 spheroplasts and permeabilized cells to the bacteriocin AS-48. Appl. Environ. Microbiol. 64:4623-4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altena, K., A. Guder, C. Cramer, and G. Bierbaum. 2000. Biosynthesis of the lantibiotic mersacidin: organization of a type B lantibiotic gene cluster. Appl. Environ. Microbiol. 66:2565-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schaffer, J., Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson, D. G., and L. L. McKay. 1983. A simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Appel, R. D., A. Bairoch, and D. F. Hochstrasser. 1994. A new generation of information retrieval tools for biologists: the example of the Expasy WWW server. Trends Biochem. Sci. 19:258-260. [DOI] [PubMed] [Google Scholar]
- 6.Brendel, V., P. Bucher, L. Nourbakhsh, B. E. Blaisdell, and S. Karlin. 1992. Methods and algorithms for statistical analysis of protein sequences. Proc. Natl. Acad. Sci. USA 89:2002-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, P., F. Qi, J. Novak, and P. Caufield. 1999. The specific genes for lantiobiotic mutacin II biosynthesis in Streptococcus mutans T8 are clustered and can be transferred en bloc. Appl. Environ. Microbiol. 65:1356-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Corpet, F. 1998. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fath, M. J., and R. Kolter. 1993. ABC transporters: bacterial exporters. Microbiol. Rev. 57:995-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiedler, S., and R. Wirth. 1991. Transformation of Enterococcus faecalis and Enterococcus faecium by electroporation, p. 301. In G. M. Dunny, P. P. Clearly, and L. L. McKay (ed.), Genetics and molecular biology of streptococci, lactococci and enterococci. American Society for Microbiology, Washington, D.C.
- 11.Gálvez, A., M. Maqueda, M. Martínez-Bueno, and E. Valdivia. 1989. Bactericidal and bacteriolytic action of peptide antibiotic AS-48 against Gram-positive and Gram negative bacteria and other organisms. Res. Microbiol. 140:57-68. [DOI] [PubMed] [Google Scholar]
- 12.Gálvez, A., M. Maqueda, M. Martínez-Bueno, and E. Valdivia. 1991. Permeation of bacterial cells, permeation of cytoplasmic and artificial membrane vesicles, and channel formation on lipid bilayers by peptide antibiotic AS-48. J. Bacteriol. 173:886-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.George, R. A., and J. Heringa. 2000. The REPRO server: finding protein internal sequence repeats through the web. Trends Biochem. Sci. 25:515-517. [DOI] [PubMed] [Google Scholar]
- 14.Gónzalez, C., G. M. Langdon, M. Bruix, A. Gálvez, E. Valdivia, M. Maqueda, and M. Rico. 2000. Bacteriocin AS-48, a cyclic polypeptide structurally and functionally close to mammalian NK-lysin. Proc. Natl. Acad. Sci. USA 97:11221-11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, K., and W. Stoffel. 1993. TMbase—a database of membrane spanning protein segments. Biol. Chem. Hoppe-Seyler 347:166. [Google Scholar]
- 17.Ike, Y., R. C. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquistion of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iliopoulos, I., S. Tsoka, M. A. Andrade, P. Janssen, B. Audit, A. Tramontano, A. Valencia, C. Leroy, C. Sander, and C. A. Ouzounis. 29. December 2000, posting date. Genome sequences and great expectations. Genome Biol. 2:interactions0001.1-0001.3. [Online.] http://genomebiology.com/2000/2/1/interactions/0001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1994. A model recognition approach to the prediction of all-helical membrane protein structure and topology. Biochemistry 33:3038-3049. [DOI] [PubMed] [Google Scholar]
- 20.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 22.Martínez-Bueno, M., M. Maqueda, A. Gálvez, B. Samyn, J. van Beeumen, J. Coyette, and E. Valdivia. 1994. Determination of the gene sequence and molecular structure of the enterococcal peptide antibiotic AS-48. J. Bacteriol. 176:6334-6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martínez-Bueno, M., E. Valdivia, A. Gálvez, J. Coyette, and M. Maqueda. 1998. Analysis of the gene cluster involved in production and immunity of the peptide antibiotic AS-48 in Enterococcus faecalis. Mol. Microbiol. 27:347-358. [DOI] [PubMed] [Google Scholar]
- 24.Mendoza, F., M. Maqueda, A. Gálvez, M. Martínez-Bueno, and E. Valdivia. 1999. Antilisterial activity of peptide AS-48 and study of the changes induced in the cell envelope properties of an AS-48-adapted strain of Listeria monocytogenes. Appl. Environ. Microbiol. 65:618-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peitsch, M. C. 1996. ProMod and Swiss-Model: Internet-based tools for automated comparative protein modelling. Biochem. Soc. Trans. 24:274-279. [DOI] [PubMed] [Google Scholar]
- 27.Peitsch, M. C., and N. Guex. 1997. Swiss-Model and the Swiss-PDBViewer: an environment for comparative protein modeling. Electrophoresis 18:2714-2723. [DOI] [PubMed] [Google Scholar]
- 28.Persson, B., and P. Argos. 1994. Prediction of transmembrane segments in proteins utilising multiple sequence alignments. J. Mol. Biol. 237:182-192. [DOI] [PubMed] [Google Scholar]
- 29.Peschel, A., J. Augustins, T. Kupke, S. Stevanovic, and S. F. Gotz. 1993. Regulation of epidermine biosynthetic genes by EpiQ. Mol. Microbiol. 9:31-39. [DOI] [PubMed] [Google Scholar]
- 30.Peschel, A., and F. Gotz. 1996. Analysis of the Staphylococcus epidermidis genes epiF, -E, and -G involved in epidermin immunity. J. Bacteriol. 178:531-536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rincé, A., A. Dufuor, P. Uguen, J. P. Le Pennec, and D. Haras. 1997. Characterization of the lacticin 481 operon: the Lactococcus lactis genes lctF, lctE, and lctG encode a putative ABC transporter involved in bacteriocin immunity. Appl. Environ. Microbiol. 63:4252-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rost, B., P. Fariselli, and R. Casadio. 1996. Topology prediction for helical transmembrane proteins at 86% accuracy. Protein Sci. 5:1704-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schultz, J., R. R. Copley, T. Doerks, C. P. Ponting, and P. Bork. 2000. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28:231-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senger, M., T. Flores, K. Glatting, P. Ernst, A. Hotz-Wagenblatt, and S. Suhai. 1998. W2H: www interface to the GCG sequence analysis package. Bioinformatics 14:452-457. [DOI] [PubMed] [Google Scholar]
- 35.Siezen, R. J., O. P. Kuipers, and W. M. de Vos. 1996. Comparison of the lantibiotic gene clusters and encoded proteins. Antonie Leeuwenhoek 69:171-184. [DOI] [PubMed] [Google Scholar]
- 36.Tomita, H., S. Fujimoto, and Y. Ike. 1996. Cloning and genetic organization of the bacteriocin 31 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pY117. J. Bacteriol. 178:3583-3593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tomita, H., S. Fujimoto, K. Tanimoto, and Y. Ike. 1997. Cloning and genetic and sequence analysis of the bacteriocin 21 determinant encoded on the Enterococcus faecalis pheromone-responsive conjugative plasmid pPD1. J. Bacteriol. 179:7843-7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van de Guchte, M., J. Kok, and G. Venema. 1992. Gene expression in Lactococcus lactis. FEMS Microbiol. Rev. 88:73-92. [DOI] [PubMed] [Google Scholar]
- 39.von Heijne, G. 1996. Prediction of transmembrane protein topology, p. 1001-1109. In M. J. E. Sternberg (ed.), Protein structure prediction. A practical approach. IRL Press, Oxford, United Kingdom.
- 40.Wirth, R., F. Y. An, and D. B. Clewell. 1986. Highly efficient protoplast transformation system for Streptococcus faecalis and Escherichia coli-S. faecalis shuttle vector. J. Bacteriol. 165:831-836. [DOI] [PMC free article] [PubMed] [Google Scholar]