Abstract
The likelihood that products prepared from raw meat and milk may act as vehicles for antibiotic-resistant bacteria is currently of great concern in food safety issues. In this study, a collection of 94 tetracycline-resistant (Tcr) lactic acid bacteria recovered from nine different fermented dry sausage types were subjected to a polyphasic molecular study with the aim of characterizing the host organisms and the tet genes, conferring tetracycline resistance, that they carry. With the (GTG)5-PCR DNA fingerprinting technique, the Tcr lactic acid bacterial isolates were identified as Lactobacillus plantarum, L. sakei subsp. carnosus, L. sakei subsp. sakei, L. curvatus, and L. alimentarius and typed to the intraspecies level. For a selection of 24 Tcr lactic acid bacterial isolates displaying unique (GTG)5-PCR fingerprints, tet genes were determined by means of PCR, and only tet(M) was detected. Restriction enzyme analysis with AccI and ScaI revealed two different tet(M) allele types. This grouping was confirmed by partial sequencing of the tet(M) open reading frame, which indicated that the two allele types displayed high sequence similarities (>99.6%) with tet(M) genes previously reported in Staphylococcus aureus MRSA 101 and in Neisseria meningitidis, respectively. Southern hybridization with plasmid profiles revealed that the isolates contained tet(M)-carrying plasmids. In addition to the tet(M) gene, one isolate also contained an erm(B) gene on a different plasmid from the one encoding the tetracycline resistance. Furthermore, it was also shown by PCR that the tet(M) genes were not located on transposons of the Tn916/Tn1545 family. To our knowledge, this is the first detailed molecular study demonstrating that taxonomically and genotypically diverse Lactobacillus strains from different types of fermented meat products can be a host for plasmid-borne tet genes.
For several decades, studies on the selection and dissemination of antibiotic resistance have focused mainly on clinically relevant bacterial species. More recently, many investigators speculated that commensal bacteria may act as reservoirs of antibiotic resistance genes similar to those found in human pathogens (30; S. B. Levy and A. A. Salyers, Reservoirs of Antibiotic Resistance [ROAR] Network, http://www.healthsci.tufts.edu/apua/Roar/roarhome.htm, 2002). Such commensal reservoir bacteria can be present in the intestines of farmed animals exposed to antibiotics (2, 38) and may subsequently contaminate the raw meat produced from these animals even when hygiene regulations are respected. Several examples of antibiotic-resistant lactic acid bacteria isolated from raw meat exist (20, 26, 28, 37). Fermented foods prepared from raw meat or milk can therefore be considered potential vehicles for the spread of antibiotic-resistant lactic acid bacteria along the food chain to the consumer (35).
Genes conferring resistance to tetracycline, chloramphenicol, erythromycin, and vancomycin have been detected and characterized in Lactococcus lactis (27) and enterococci (15, 35) isolated from fermented meat and milk products. In contrast, no molecular data are available on the occurrence of antibiotic resistance genes in lactobacilli present in fermented food products. Members of the genus Lactobacillus also constitute an important part of the natural microflora associated with fermented products and are indigenous to the animal and human gastrointestinal tract. These properties make lactobacilli, along with enterococci, interesting indicator organisms with which to study the molecular ecology of antibiotic resistance determinants in food fermentation industries.
Previously, we isolated tetracycline-resistant (Tcr) strains of various Lactobacillus species from different types of modified atmosphere-packed fermented dry sausage (FDS) sold in Belgian retail shops (13). All Lactobacillus isolates collected in this way were resistant to at least 64 μg of tetracycline per ml as determined with the de Man, Rogosa, and Sharpe (MRS) agar dilution method (13). The aim of the present investigation was to perform a molecular characterization of the tet genes conferring the high-level phenotypic resistance to tetracycline in these isolates. For this purpose, an extended collection of Tcr Lactobacillus isolates was subjected to (GTG)5-PCR fingerprinting (14) to reveal their taxonomic identity and genotypic diversity. Based on these results, a selected subset of different strain types were characterized further to assess the identity, the heterogeneity, and the locations of their tet genes.
MATERIALS AND METHODS
Bacterial isolates.
In this study, a total of 94 Tcr lactic acid bacterial isolates recovered from 14 batches of nine different FDS types were included. A type is defined here as a specific product variety distributed under a specific commercial brand. Each type is designated with a number (FDS-01, FDS-02, FDS-06, FDS-07, FDS-08, FDS-09, FDS-11, FDS-12, and FDS-14). A batch is defined as a group of sausages belonging to the same type that were produced at one time and is indicated with a letter from A to E.
The isolation of 52 Tcr lactic acid bacterial isolates was reported before (13). This collection was extended with 42 new Tcr lactic acid bacterial isolates recovered similarly from other batches of fermented dry sausage end products. Isolates were recovered on the basis of colony morphology rather than relative abundance in order to obtain a set of isolates with the highest diversity. All these fermented dry sausages were prepared with a starter culture. All isolates were stored in a bead storage system (Microbank system; Pro-LAB Diagnostics, Wirral, United Kingdom) at −80°C and grown on MRS agar (Difco Co., Detroit, Mich.) at 30°C under microaerophilic conditions (3.75% CO2-5% O2-7.5% H2-83.75% N2).
(GTG)5-PCR fingerprinting.
All Tcr lactic acid bacterial isolates were identified at the species level and typed at the intraspecies level with high-resolution rep-PCR fingerprinting with the (GTG)5 primer as previously described (14).
Antibiotic susceptibility testing and MIC determination.
A modified version of the Kirby-Bauer disk diffusion method (4a), in which Mueller-Hinton medium was replaced with MRS agar, was used for antibiotic sensitivity testing. Oxoid (Basingstoke, United Kingdom) susceptibility test disks of ampicillin (25 μg), chloramphenicol (30 μg), clindamycin (10 μg), erythromycin (10 μg), penicillin G (10 U), rifampin (30 μg), and tetracycline (30 μg) were applied to inoculated MRS plates with the Oxoid disk dispenser. The diameters of the inhibition zones were measured with a digital caliper (digital 2; Mauser, Ludwigsburg, Germany) following 16 to 18 h of incubation of the antibiograms at 30°C. For each of the antibiotics tested, classification of the isolates into sensitive and resistant groups was based on resistance histograms (i.e., number of strains versus size of the inhibition zone). Cutoff values to differentiate among resistant and susceptible groups were defined on the basis of the bimodal distribution of the population in the resistance histograms.
The MIC of tetracycline was determined by applying an Etest strip (AB Biodisk) on an inoculated MRS plate according to the manufacturer's instructions. The Etest strip was read following 16 to 18 h of incubation at 30°C.
DNA preparation and manipulations.
Total genomic DNA from each isolate was extracted and purified as described previously (14). Isolation of plasmid DNA was based on the alkaline lysis method of Anderson and McKay (3). Restriction endonuclease digestions of the tet(M) gene, agarose gel electrophoresis, and Southern blotting were carried out by standard procedures (31). Labeling of DNA probes with horseradish peroxidase with the ECL direct nucleic acid labeling kit (RPN3000; Amersham Biosciences) was performed according to the manufacturer's instructions.
PCR detection of tet, erm, and int genes.
The PCR assay mix (total volume, 50 μl) contained 20 pmol of each primer (Table 1), 1× PCR buffer (Applied Biosystems, Warrington, United Kingdom), each deoxynucleoside triphosphate at a concentration of 200 μM, and 1 U of AmpliTaq DNA polymerase (N808-0160; Applied Biosystems, Warrington, United Kingdom). A 50-ng portion of purified total DNA was used as a template. In a first PCR assay, tet genes encoding ribosomal protection proteins (RPP) were detected with degenerate primers DI and DII (7). If positive for RPP genes, additional PCR assays were performed with primers specific for tet(M), tet(O), and tet(S) (Table 1). Next to the RPP tet genes, isolates were also tested for the presence of the tetracycline efflux genes tet(K) and tet(L) and for the transposon integrase gene (int gene) of the Tn916/Tn1545 family (Table 1). One strain that expressed erythromycin resistance was analyzed with erm(B)-specific primers as described previously (19).
TABLE 1.
Primers for PCR detection of tet, erm, and int genes
| Primer pair | Gene(s) targeted | Sequencea | Annealing temp (°C) | Amplicon size (bp) | Positive control strain and reference | Reference for primers |
|---|---|---|---|---|---|---|
| DI | RPP | 5′-GAYACNCCNGGNCAYRTNGAYTT-3′ | 45 | 1,083 | pJI3 (22) | 7 |
| DII | 5′-GCCCARWANGGRTTNGGNGGNACYTC-3′ | |||||
| DI | tet(M) | 5′-GAYACNCCNGGNCAYRTNGAYTT-3′ | 55 | 1,513 | pJI3 (22) | 7 |
| TetM-R | 5′-CACCGAGCAGGGATTTCTCCAC-3′ | |||||
| TetS-FWT 1 | tet(S) | 5′-ATCAAGATATTAAGGAC-3′ | 55 | 573 | pVP2 (27) | 5 |
| TetS-RVT 2 | 5′-TTCTCTATGTGGTAATC-3′ | |||||
| TetO-FW 1 | tet(O) | 5′-AATGAAGATTCCGACAATTT-3′ | 55 | 781 | pAT121 (J. M. Collard, | 32 |
| TetO-RV 1 | 5′-CTCATGCGTTGTAGTATTCCA-3′ | personal communication) | ||||
| TetK-FW 1 | tet(K) | 5′-TTATGGTGGTTGTAGCTAGAAA-3′ | 55 | 348 | pAT102 (P. Courvalin, | J. M. Collard, personal communication |
| TetK-RV 1 | 5′-AAAGGGTTAGAAACTCTTGAAA-3′ | personal communication) | ||||
| TetL-FW 3 | tet(L) | 5′-GTMGTTGCGCGCTATATTCC-3′ | 55 | 696 | pAT103 (P. Courvalin, | J. M. Collard, |
| TetL-RV 3 | 5′-GTGAAMGRWAGCCCACCTAA-3′ | personal communication) | personal communication | |||
| Int-FW | int | 5′-GCGTGATTGTATCTCACT-3′ | 50 | 1,028 | Tn1545 (P. Courvalin, personal communication) | 9 |
| Int-RV | 5′-GACGCTCCTGTTGCTTCT-3′ | |||||
| ErmB-FW | erm(B) | 5′-CATTTAACGACGAAACTGGC-3′ | 55 | 405 | Tn1545 (P. Courvalin, personal communication) | 19 |
| ErmB-RV | 5′-GGAACATCTGTGGTATGGCG-3′ |
N = A, C, G, and T; R = A and G; W = A and T; Y = C and T.
All PCR amplifications were performed in a GeneAmp 9600 PCR system (Perkin-Elmer) with the following temperature program: initial denaturation at 94°C for 5 min; 30 cycles of 94°C for 1 min, annealing temperature (see Table 1) for 1 min, and 72°C for 2 min; and a final extension step at 72°C for 10 min. PCR products (5 μl) were separated by electrophoresis on a 1% agarose gel and visualized by ethidium bromide staining.
Sequencing of PCR products.
For tet(M)-positive isolates, the purified PCR products obtained with the DI and TetM-R primers were directly sequenced with the primers DI, DII, and TetM-R (Table 1). Sequencing was performed with a BigDye Terminator version 2 Ready Reaction cycle sequencing kit (Applied Biosystems, Warrington, United Kingdom) on an ABI Prism 310 genetic analyzer (Applied Biosystems, Warrington, United Kingdom). On-line similarity searches were performed with the BLAST (Basic Local Alignment Search Tool) family of programs in GenBank.
Nucleotide sequence accession numbers.
The sequences of the tet(M) genes described in this paper have been assigned GenBank accession numbers AY149574 to AY149597.
RESULTS
Identification of isolates.
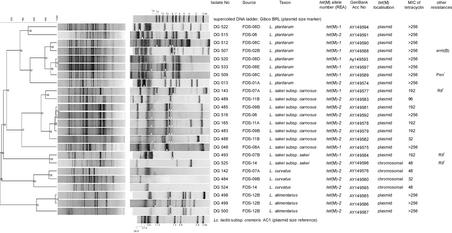
With (GTG)5-PCR fingerprinting, the 94 Tcr lactic acid bacterial isolates were identified as Lactobacillus plantarum (n = 31), L. sakei subsp. carnosus (n = 46), L. sakei subsp. sakei (n = 5), L. curvatus (n = 7), and L. alimentarius (n = 5). Only one species was isolated from the majority of the positive samples (64%), whereas in all other cases no more than two species were identified. In some cases, different species compositions were obtained in different batches of the same sausage type, e.g., batches FDS-07A and -07B and FDS-08A and -08C. Numerical analysis of the (GTG)5-PCR fingerprints showed that isolates originating from the same batch often displayed identical banding patterns (results not shown). Based on pattern similarity, the set of isolates was reduced to one isolate per unique (GTG)5-PCR fingerprint type, which resulted in a collection of 24 Tcr Lactobacillus isolates (Fig. 1).
FIG. 1.
Composite figure of (GTG)5-PCR fingerprints and inverted plasmid profiles of the 24 Tcr Lactobacillus isolates from FDS end products. The dendrogram was constructed after cluster analysis of the digitized (GTG)5-PCR fingerprints with the unweighted pair group method with arithmetic averages, with correlation levels expressed as a percentage of the Pearson correlation coefficient. Cophenetic correlations (shown on each branch of the dendrogram) indicate how faithfully the dendrogram represents the similarity matrix. A small triangle or circle indicates the place where the tet(M) or erm(B) probe, respectively, hybridized on the Southern blot of the plasmid DNA. Lactococcus lactis subsp. cremoris strain AC1 was used as a plasmid size marker (24). The tet(M)-1 and tet(M)-2 allele types correspond to the tet(M) genes found in Neisseria meningitidis (X75073) and Staphylococcus aureus MRSA 101 (M21136), respectively, based on restriction enzyme analysis (REA) with AccI and ScaI. Numerical designations indicate the fermented dry sausage type, and the letters A to D indicate different batches (type and batch are defined in Materials and Methods). tet(M) localization was determined by Southern blot analysis. The MIC of tetracycline was determined by Etest. Chr., chromosomal band.
Phenotypic characterization of resistance.
Etests revealed that the MIC of tetracycline ranged between 32 and >256 μg/ml (upper limit of test), and the MIC for 50% of strains was >256 μg/ml (Fig. 1). Next to the high-level tetracycline resistance found among the 24 selected isolates, some strains showed an additional phenotypic resistance to erythromycin (n = 1), rifampin (n = 4), and penicillin (n = 1) in disk diffusion susceptibility testing.
Detection, characterization, and localization of resistance genes.
Total genomic DNA preparations from all 24 Tcr Lactobacillus isolates were subjected to PCR amplification with the RPP set of primers, with class-specific primers for tet(M), tet(O), tet(S), tet(K), and tet(L) and with primers for detection of int genes of the Tn916/Tn1545 family of transposons. In all 24 isolates, only tet(M) was detected, and no int genes were found. Restriction enzyme analysis of the tet(M) PCR product (74% of the open reading frame) with AccI and ScaI revealed two different tet(M) allele types, tet(M)-1 and tet(M)-2 (Fig. 1). Most batches contained strains belonging to one allele type, except for batches FDS-02-B, FDS-07-A, and FDS-14, which contained strains displaying both allele types (Fig. 1).
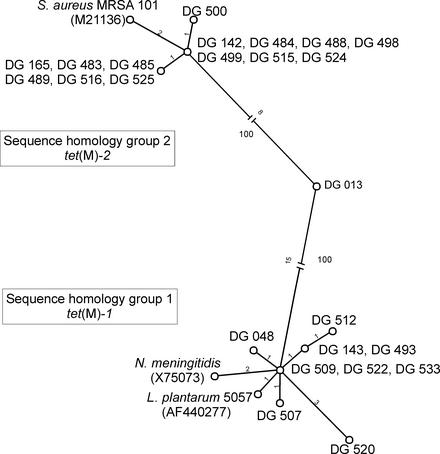
In order to characterize the tet(M) genes more profoundly, the open reading frame was partially (74%) sequenced from approximately position 280 (DI primer) to position 1700 (TetM-R primer). Sequence alignments revealed two sequence homology groups and one mosaic gene (Fig. 2). Between the homology groups, a difference of at least 25 bases was found. A first group comprised nine isolates in which the sequenced part of the tet(M) gene showed at most five base differences with the tet(M) gene of Neisseria meningitidis (GenBank accession no. X75073) (12). This group corresponds to tet(M)-1 found by restriction enzyme analysis. A second group of 14 isolates with a maximum of three base differences with the tet(M) gene of Staphylococcus aureus MRSA 101 (GenBank accession no. M21136) (23) corresponded to tet(M)-2 defined by restriction enzyme analysis.
FIG. 2.
Single most parsimonious tree (unrooted) for tet(M) gene relationships of the 24 Tcr lactic acid bacterial isolates and two reference strains, Neisseria meningitidis (X75073) and Staphylococcus aureus MRSA 101 (M21136). The recently published tet(M) gene of L. plantarum 5057 (AF440277) was included (8). The numbers of nucleotide substitutions are indicated on each branch. The bootstrap percentages (500 replicates) are indicated for the separation between the two homology groups.
The tet(M) gene of isolate DG 13 exhibited a mosaic structure, combining partial sequences of the two foregoing homology groups. The sequence up to position 1508 of the open reading frame displayed one base difference with the tet(M) gene of Neisseria meningitidis and was identical to the tet(M) gene of Staphylococcus aureus MRSA 101 in the remaining part. Restriction enzyme analysis classified the tet(M) gene of this strain as a tet(M)-2. The one isolate expressing phenotypic erythromycin resistance (DG 507) was shown to contain the erm(B) gene by PCR and partial sequencing of the PCR product.
Plasmid profiling of the 24 Tcr Lactobacillus isolates showed that 23 isolates (excluding DG 524) contained at least one and usually more than one plasmid (Fig. 1). By Southern blotting and hybridization with a tet(M)- and/or erm(B)-specific probe, the tet(M) genes of 20 isolates and the erm(B) gene in isolate DG 507 could be localized on a plasmid. Most of these R-plasmids had a size of approximately 10 kb, and in a few cases the R-plasmid was larger than 25 kb. Most plasmid profiles showed more than one band (up to three) that hybridized with the tet(M) probe (Fig. 1), due to hybridization with (low concentrations of) open circular and/or linear plasmid forms. For the remaining four Tcr Lactobacillus isolates that did not carry the tet(M) gene on a plasmid (DG 142, DG 484, DG 524, and DG 525), Southern blots of EcoRI-digested DNA hybridized to the tet(M) probe, with fragments of between 8 and 12 kb (results not shown).
DISCUSSION
To our knowledge, this is the first detailed molecular study of antibiotic resistance genes in Lactobacillus species isolated from fermented dry sausages. The presence of antibiotic-resistant Lactobacillus species has been documented in wine, cheese (17, 34), poultry, calf, swine (11, 21, 33, 36), pig feces (4, 10, 29), healthy human feces (18), and maize silage (8). In the first phase of the current study, a total of 94 tetracycline-resistant lactic acid bacterial isolates recovered from 14 batches representing nine different fermented dry sausage types were identified by (GTG)5-PCR fingerprinting. All strains could be identified as Lactobacillus species commonly associated with fermented meat products, i.e., L. plantarum, L. sakei subsp. carnosus, L. sakei subsp. sakei, L. curvatus, and L. alimentarius (16).
In four of the batches investigated, isolates belonged to two different Lactobacillus species, and in five batches more than one (GTG)5-PCR fingerprint type per species was found (results not shown). Different batches of one fermented dry sausage type were not found to contain identical (GTG)5-PCR fingerprints, indicating that the source of Tcr lactobacilli is variable. Interestingly, isolates DG 165, DG 485, and DG 516, originating from three different fermented dry sausage types (FDS-11B, FDS-09B, and FDS-06, respectively), displayed identical DNA fingerprints and harbored identical R-plasmids and tet(M) genes, suggesting a possible common source of Tcr Lactobacillus contamination. In order not to select multiple isogenic strains, one strain each of the 24 unique (GTG)5-PCR fingerprint types was selected.
Given the fact that the Tcr Lactobacillus isolates were recovered from nine different fermented dry sausage types and contained 24 (GTG)5-PCR fingerprint types representing five different Lactobacillus species, it was somewhat surprising that in all isolates, of the five tet genes tested, only tet(M) was detected. According to current insights, tet(M) is the most widely distributed tet gene, being detected in at least eight gram-negative and 18 gram-positive genera, including the lactic acid bacterial genera Enterococcus, Streptococcus, and Bifidobacterium (6). It was suggested that the origin of tet(M) is most probably the tetracycline-producing species of Streptomyces and that its integration into mobile genetic elements (plasmids and transposons) has led to its widespread distribution (6, 25).
At the moment of discovery, only tet(O) and tet(Q) have been reported in members of the genus Lactobacillus (6), but recently a tet(M) gene was also found in an L. plantarum strain (8). Partial sequencing revealed that the tet(M) genes in the Tcr Lactobacillus isolates belonged to two homology groups and one individual. The two homology groups correspond to sequences that were published before; group I corresponds with the tet(M) found in Neisseria meningitidis (12), and group II with tet(M) of Staphylococcus aureus MRSA 101 (23). Isolate DG 13 represents a new allelic variation, showing high partial similarities with both the N. meningitidis and S. aureus tet(M) genes. This group may have arisen from homologous recombination and corresponds to the mosaic structures exhibited by the tet(M) gene, as previously described (25). The tet(M) genes found in these Lactobacillus isolates differ from those found in other lactic acid bacteria, including Enterococcus faecalis (X56353, M85225, X92947, and X04388), with a base difference ranging between 12 and 115 bases, and Streptococcus pneumoniae (X90939), with a base difference ranging between 69 and 86 bases (results not shown).
The tet(M) genes of N. meningitidis and S. aureus MRSA 101 are located on a plasmid and on the chromosome, respectively (12, 23). Within the set of Tcr Lactobacillus isolates, the tet(M) genes of sequence homology group I and isolate DG 13 were found exclusively on plasmids (n = 10), whereas for sequence homology group II, tet(M) genes were localized on the chromosome (n = 4) or on a plasmid (n = 10). R-plasmids encoding tetracycline, chloramphenicol, gentamicin, and macrolide-lincosamide-streptogramin B resistance have been reported previously in L. reuteri (4, 21, 33, 36), L. fermentum (10, 18), L. acidophilus (36), and L. plantarum (1, 8) isolated from raw meat, feces, and maize silage. Most of these R-plasmids were smaller than 10 kb. The tetracycline MIC was significantly lower for the Tcr Lactobacillus isolates with a chromosomal tet gene in this study than for those with plasmid-encoded tetracycline resistance, 32 to 48 μg/ml and >192 μg/ml, respectively, with the exception of two L. sakei subsp. carnosus isolates (DG 488 and DG 489). More research is needed to investigate to what extent this marked difference in phenotypic resistance levels is linked to, e.g., the location of the tet(M) gene or the copy number of the R-plasmid.
In conclusion, the results of the current study indicate that Lactobacillus species from fermented meat products can harbor acquired tetracycline resistance encoded by a tet(M) gene, which is usually located on a plasmid and displays very high genotypic similarities with tet(M) genes previously reported in two pathogenic species. Further research may focus on the diversity and transferability of these Lactobacillus plasmids into other commensal bacteria and on the source of Tcr lactobacilli in the production of fermented dry sausages.
Acknowledgments
This research was carried out with financial support from the Institute for the Encouragement of Scientific and Technological Research in the Industry (IWT). The Fund for Scientific Research—Flanders (Belgium) (F.W.O.-Vlaanderen) is acknowledged by J.S. for support in the framework of contract G0.0309.01 and for the postdoctoral fellowship of G.H.
We thank Geertrui Rasschaert and Hanne Schoeters for their contribution to the practical work.
REFERENCES
- 1.Ahn, C., D. Collins-Thompson, C. Duncan, and M. E. Stiles. 1992. Mobilization and location of the genetic determinant of chloramphenicol resistance from Lactobacillus plantarum caTC2R. Plasmid 27:169-176. [DOI] [PubMed] [Google Scholar]
- 2.Aminov, R. I., N. Garrigues-Jeanjean, and R. I. Mackie. 2001. Molecular ecology of tetracycline resistance: development and validation of primers for detection of tetracycline resistance genes encoding ribosomal protection proteins. Appl. Environ. Microbiol. 67:22-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anderson, D. G., and L. L. McKay. 1983. Simple and rapid method for isolating large plasmid DNA from lactic streptococci. Appl. Environ. Microbiol. 46:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Axelsson, L. T., S. Ahrné, M. C. Andersson, and S. R. Stahl. 1988. Identification and cloning of a plasmid-encoded erythromycin resistance determinant from Lactobacillus reuteri. Plasmid 20:171-174. [DOI] [PubMed] [Google Scholar]
- 4a.Bauer, A. W., W. M. M. Kirby, J. C. Sherris, and M. Turck. 1966. Antibiotic susceptibility by a standardized single disc method. Am. J. Clin. Pathol. 45:493-496. [PubMed] [Google Scholar]
- 5.Charpentier, E., G. Gerbaud, and P. Courvalin. 1993. Characterization of a new class of tetracycline-resistance gene tet(S) in Listeria monocytogenes BM4210. Gene 131:27-34. [DOI] [PubMed] [Google Scholar]
- 6.Chopra, I., and M. Roberts. 2001. Tetracycline antibiotics: mode of action, applications, molecular biology, and epidemiology of bacterial resistance. Microbiol. Mol. Biol. Rev. 65:232-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clermont, D., O. Chesneau, G. DeCespedes, and T. Horaud. 1997. New tetracycline resistance determinants coding for ribosomal protection in streptococci and nucleotide sequence of tet(T) isolated from Streptococcus pyogenes A498. Antimicrob. Agents Chemother. 41:112-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danielsen, M. 2002. Characterization of the tetracycline resistance plasmid pMD5057 from Lactobacillus plantarum 5057 reveals a composite structure. Plasmid 48:98-103. [DOI] [PubMed]
- 9.Doherty, N., K. Trzcinski, P. Pickerill, P. Zawadzki, and C. G. Dowson. 2000. Genetic diversity of the tet(M) gene in tetracycline-resistant clonal lineages of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44:2979-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fons, M., T. Hege, M. Ladire, P. Raibaud, R. Ducluzeau, and E. Maguin. 1997. Isolation and characterization of a plasmid from Lactobacillus fermentum conferring erythromycin resistance. Plasmid 37:199-203. [DOI] [PubMed] [Google Scholar]
- 11.Frei, A., D. Goldenberger, and M. Teuber. 2001. Antimicrobial susceptibility of intestinal bacteria from Swiss poultry flocks before the ban of antimicrobial growth promoters. Syst. Appl. Microbiol. 24:116-121. [DOI] [PubMed] [Google Scholar]
- 12.Gascoyne-Binzi, D. M., J. Heritage, P. M. Hawkey, and M. S. Sprott. 1994. Characterization of a tet(M)-carrying plasmid from Neisseria meningitidis. J. Antimicrob. Chemother. 34:1015-1023. [DOI] [PubMed] [Google Scholar]
- 13.Gevers, D., G. Huys, F. Devlieghere, M. Uyttendaele, J. Debevere, and J. Swings. 2000. Isolation and identification of tetracycline resistant lactic acid bacteria from pre-packed sliced meat products. Syst. Appl. Microbiol. 23:279-284. [DOI] [PubMed] [Google Scholar]
- 14.Gevers, D., G. Huys, and J. Swings. 2001. Applicability of rep-PCR fingerprinting for identification of Lactobacillus species. FEMS Microbiol. Lett. 205:31-36. [DOI] [PubMed] [Google Scholar]
- 15.Giraffa, G., and F. Sisto. 1997. Susceptibility to vancomycin of enterococci isolated from dairy products. Lett. Appl. Microbiol. 25:335-338. [DOI] [PubMed] [Google Scholar]
- 16.Hammes, W. P., A. Bantleon, and S. Min. 1990. Lactic acid bacteria in meat fermentation. FEMS Microbiol. Rev. 87:165-174. [Google Scholar]
- 17.Herrero, M., B. Mayo, B. González, and J. E. Suárez. 1996. Evaluation of technologically important traits in lactic acid bacteria isolated from spontaneous fermentations. J. Appl. Bacteriol. 81:565-570. [Google Scholar]
- 18.Ishiwa, H., and S. Iwata. 1980. Drug resistance plasmids in Lactobacillus fermentum. J. Gen. Appl. Microbiol. 26:71-74. [Google Scholar]
- 19.Jensen, L. B., N. Frimodt-Moller, and F. M. Aarestrup. 1999. Presence of erm gene classes in Gram-positive bacteria of animal and human origin in Denmark. FEMS Microbiol. Lett. 170:151-158. [DOI] [PubMed] [Google Scholar]
- 20.Klein, G., A. Pack, and G. Reuter. 1998. Antibiotic resistance patterns of enterococci and occurrence of vancomycin-resistant enterococci in raw minced beef and pork in Germany. Appl. Environ. Microbiol. 64:1825-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin, C. F., Z. F. Fung, C. L. Wu, and T. C. Chung. 1996. Molecular characterization of a plasmid-borne (pTC82) chloramphenicol resistance determinant (cat-Tc) from Lactobacillus reuteri G4. Plasmid 36:116-124. [DOI] [PubMed] [Google Scholar]
- 22.Morse, S. A., S. R. Johnson, J. W. Biddle, and M. C. Roberts. 1986. High-level tetracycline resistance in Neisseria gonorrhoeae is result of acquisition of streptococcal tetM determinant. Antimicrob. Agents Chemother. 30:664-670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nesin, M., P. Svec, J. R. Lupski, G. N. Godson, B. Kreiswirth, J. Kornblum, and S. J. Projan. 1990. Cloning and nucleotide sequence of a chromosomally encoded tetracycline resistance determinant, tetA(M), from a pathogenic, methicillin-resistant strain of Staphylococcus aureus. Antimicrob. Agents Chemother. 34:2273-2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neve, H., A. Geis, and M. Teuber. 1984. Conjugal transfer and characterization of bacteriocin plasmids in group N (lactic acid) streptococci. J. Bacteriol. 157:833-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oggioni, M. R., C. G. Dowson, J. M. Smith, R. Provvedi, and G. Pozzi. 1996. The tetracycline resistance gene tet(M) exhibits mosaic structure. Plasmid 35:156-163. [DOI] [PubMed] [Google Scholar]
- 26.Pavia, M., C. G. A. Nobile, L. Salpietro, and I. F. Angelillo. 2000. Vancomycin resistance and antibiotic susceptibility of enterococci in raw meat. J. Food Prot. 63:912-915. [DOI] [PubMed] [Google Scholar]
- 27.Perreten, V., F. Schwarz, L. Cresta, M. Boeglin, G. Dasen, and M. Teuber. 1997. Antibiotic resistance spread in food. Nature 389:801-802. [DOI] [PubMed] [Google Scholar]
- 28.Quednau, M., S. Ahrné, A. C. Petersson, and G. Molin. 1998. Antibiotic-resistant strains of Enterococcus isolated from Swedish and Danish retailed chicken and pork. J. Appl. Microbiol. 84:1163-1170. [DOI] [PubMed] [Google Scholar]
- 29.Rinckel, L. A., and D. C. Savage. 1990. Characterization of plasmids and plasmid-borne macrolide resistance from Lactobacillus sp. strain 100-33. Plasmid 23:119-125. [DOI] [PubMed] [Google Scholar]
- 30.Salyers, A. A. 1995. Out of the ivory tower: bacterial gene transfer in the real world, p. 109-136. In A. A. Salyers (ed.), Antibiotic resistance transfer in the mammalian intestinal tract: implications for human health, food safety and biotechnology. Springer-Verlag, Berlin, Germany.
- 31.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 32.Sougakoff, W., B. Papadopoulou, P. Nordmann, and P. Courvalin. 1987. Nucleotide-sequence and distribution of gene tet(O) encoding tetracycline resistance in Campylobacter coli. FEMS Microbiol. Lett. 44:153-159. [Google Scholar]
- 33.Tannock, G. W., J. B. Luchansky, L. Miller, H. Connell, S. Thodeandersen, A. A. Mercer, and T. R. Kalenhammer. 1994. Molecular characterization of a plasmid-borne (pGT633) erythromycin resistance determinant (ermGT) from Lactobacillus reuteri 100-163. Plasmid 31:60-71. [DOI] [PubMed] [Google Scholar]
- 34.Teuber, M., L. Meile, and F. Schwarz. 1999. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie van Leeuwenhoek 76:115-137. [PubMed] [Google Scholar]
- 35.Teuber, M., and V. Perreten. 2000. Role of milk and meat products as vehicles for antibiotic-resistant bacteria. Acta Vet. Scand. Suppl. 93:75-87. [PubMed]
- 36.Vescovo, M., L. Morelli, and V. Bottazzi. 1982. Drug resistance plasmids in Lactobacillus acidophilus and Lactobacillus reuteri. Appl. Environ. Microbiol. 43:50-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vidal, C. A., and D. Collins-Thompson. 1987. Resistance and sensitivity of meat lactic acid bacteria to antibiotics. J. Food Prot. 50:737-740. [DOI] [PubMed] [Google Scholar]
- 38.Witte, W. 1998. Medical consequences of antibiotic use in agriculture. Science 279:996-997. [DOI] [PubMed] [Google Scholar]