Abstract
Peptides derived from hydrolysis of αS1-casein(f1-9) [αS1-CN(f1-9)] and β-CN(f193-209) with cell extracts of Lactobacillus helveticus CNRZ32 and single-peptidase mutants (ΔpepC, ΔpepE, ΔpepN, ΔpepO, and ΔpepX) were isolated by using reverse-phase high-performance liquid chromatography and were characterized by mass spectrometry. The peptides identified suggest that there was activity of an endopeptidase, distinct from previously identified endopeptidases (PepE and PepO), with specificity for peptide bonds C terminal to Pro residues. Identification of hydrolysis products derived from a carboxyl-blocked form of β-CN(f193-209) confirmed that the peptides were derived from the activity of an endopeptidase.
Lactobacillus helveticus belongs to a diverse group of organisms known as the lactic acid bacteria (LAB), which are defined as organisms that produce lactic acid as a major product of carbohydrate fermentation. L. helveticus has multiple amino acid auxotrophies and thus is dependent on transport of amino acids and/or transport and hydrolysis of exogenous peptides to satisfy these nutritional requirements. In amino acid-containing defined media, L. helveticus CNRZ32 can grow without Ala, Asn, Cys, Gln, Gly, and Ser when these amino acids are absent individually (8). The fermentation of Bos taurus milk is a system that is commonly used to study the proteolytic system and physiology of L. helveticus; it provides a relatively consistent environment and a well-characterized set of proteins as a starting point, and it has adaptive significance for dairy-related LAB. Since all of the peptidases of L. helveticus that have been identified are believed to be intracellular, acquisition of amino acids is also likely to be dependent on the activity of at least one extracellular proteinase capable of hydrolyzing caseins (CNs) into transportable peptides (14). Therefore, obtaining amino acids through hydrolysis of CNs (the preferentially hydrolyzed milk proteins) requires a complex proteolytic system comprised of a proteinase(s), an endopeptidase(s), an aminopeptidase(s), a tripeptidase(s), a dipeptidase(s), and peptide transport systems (7, 14, 21).
Interest in the proteolytic system of L. helveticus CNRZ32 is related to this organism's ability to reduce bitterness and accelerate cheese flavor development when it is used as an adjunct culture in Gouda cheese production (1, 2). L. helveticus CNRZ32 has been shown to efficiently hydrolyze CN, and a comparison of its peptidolytic activity with the peptidolytic activities of L. helveticus ATCC 10797 and Lactobacillus delbrueckii subsp. bulgaricus ATCC 12278 demonstrated that L. helveticus CNRZ32 has higher general aminopeptidase and dipeptidase activities (13). Both αS1-CN(f1-9) and β-CN(f193-209), as well as other related hydrophobic peptide derivatives, are known to accumulate and have been associated with bitter defects in ripened cheeses (3, 4, 10, 12, 15, 17). The reduction in the bitterness of cheese is believed to be the result of preferential hydrolysis of low-molecular-weight hydrophobic peptides known to cause bitterness rather than the result of a lack of formation of bitter peptides from high-molecular-weight nonbitter CN-derived peptides (4, 11, 15, 16, 17).
The L. helveticus peptidases investigated in this study represent three different classes of enzymes. Broad-specificity aminopeptidases (PepC and PepN) remove the N-terminal amino acids from a peptide (X↓Y-Z…), and the specificity is dependent on the peptide length and the terminal amino acid residues. X-prolyl dipeptidyl aminopeptidase (PepX) has specificity for removal of proline-containing dipeptides (X-Pro↓Y…) from the N terminus of peptides. Endopeptidases (PepE and PepO) hydrolyze internal peptide bonds (…U-V-W↓X-Y-Z…) independent of the N-terminal amino acid residue but potentially with specificity for one or both residues flanking the hydrolyzed peptide bond. In this paper we describe the physiological availability and identification of hydrolysis products from the CN-derived peptides αS1-CN(f1-9) and β-CN(f193-209), which were obtained by using L. helveticus and several peptidase deletion mutants constructed as described previously (6, 8). Additionally, evaluation of the peptide hydrolysis profiles indicated that a previously undetected endopeptidase was present.
Growth in amino acid-containing defined medium and defined medium supplemented with peptides.
To examine the effects of peptidase mutations on growth in media requiring hydrolysis of exogenous peptides, growth of L. helveticus and growth of the peptidase deletion mutants (Table 1) were determined by using defined media prepared with either αS1-CN(f1-9) or β-CN(f193-209) as the sole source of several essential amino acids. Growth in a medium not requiring hydrolysis of exogenous peptides to obtain amino acids was determined by using defined medium (8). The defined medium was not a minimal medium and contained all free amino acids (including the nonessential amino acids Ala, Asn, Gln, Gly, Cys, and Ser) as the sole nitrogen sources.
TABLE 1.
Bacterial strains
| Strain | Relevant feature(s) | Source or reference |
|---|---|---|
| CNRZ32 | Wild type; auxotrophic for all amino acids except Ala, Asn, Cys, Gln, Gly, and Ser | Laboratory strain |
| JLS241 | ΔpepC derivative of CNRZ32 | 8 |
| JLS242 | ΔpepN derivative of CNRZ32 | 8 |
| JLS243 | ΔpepX derivative of CNRZ32 | 8 |
| JLS233 | ΔpepE derivative of CNRZ32 | Fenster and Steelea |
| JLS232 | ΔpepO derivative of CNRZ32 | 6 |
K. M. Fenster and J. L. Steele, unpublished data.
The defined medium supplemented with αS1-CN(f1-9) was prepared with all of the components of complete defined medium except the essential amino acids present in this peptide (Arg, His, Ile, Lys, and Pro) (Table 1). Likewise, the defined medium supplemented with β-CN(f193-209) was prepared with all of the components of complete defined medium except the essential amino acids present in this peptide (Arg, Ile, Leu, Phe, Pro, Tyr, and Val). Inocula were prepared from cultures grown in MRS at 42°C to the late exponential phase. Cells were washed and resuspended in 0.85% NaCl to the original volume, defined medium was inoculated with ∼106 cells/ml, and the cultures were incubated at 42°C for 18 h.
There were no discernible differences for any of the strains between growth in amino acid-containing defined medium and growth in peptide-supplemented media (for all strains the optical density at 600 nm was ∼2.4 to 2.8 and the final pH was ∼3.6). The lack of growth deficiencies for the single-peptidase-deletion mutant strains (ΔpepC, ΔpepE, ΔpepN, ΔpepO, or ΔpepX) in defined peptide-containing media indicates that the target peptidases are not normally involved in hydrolysis of the peptides and/or that each essential amino acid is liberated efficiently via an alternative hydrolysis pathway in the absence of a given peptidase.
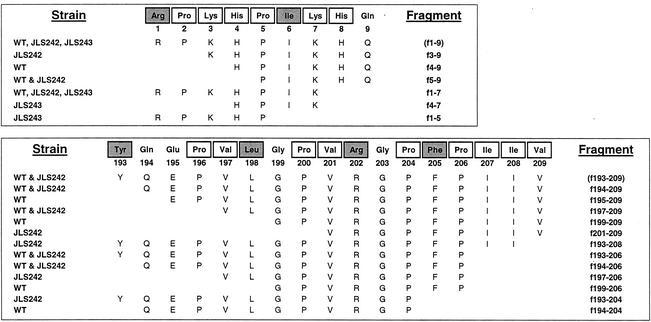
Complete hydrolysis of the peptide substrates was not necessarily required in order for L. helveticus to grow in the defined peptide media used. Some of the amino acids required by L. helveticus occur only once in αS1-CN(f1-9) (Arg and Ile) and β-CN(f193-209) (Tyr, Leu, Arg, and Phe) and therefore must be liberated for growth in the defined media (Fig. 1). However, there are two residues of each of the remaining essential amino acids per peptide in αS1-CN(f1-9). In β-CN(f193-209), there are multiple residues per peptide for Pro (four), Val (three), and Ile (two).
FIG. 1.
Peptide sequences of αS1-CN(f1-9) and β-CN(f193-209) and fragments derived from hydrolysis with CFEs. The residues that are enclosed in boxes are essential amino acids for L. helveticus. The shaded residues are essential amino acids that occur only once in the peptide and therefore must be liberated for growth of L. helveticus when the peptide is supplied as the sole substrate. The peptides correspond to unique values obtained for RP-HPLC fractions from mass spectrometry data. The strain column indicates from which CFE reaction(s) a given peptide was identified. The peptide profile obtained from hydrolysis of αS1-CN(f1-9) by CFE from the wild type (WT) is also representative of the results of the reactions with the ΔpepC, ΔpepO, and ΔpepE strains. Similarly, the peptide profile obtained from hydrolysis of β-CN(f193-209) by CFE from the wild type is also representative of the results of the reactions with the ΔpepC, ΔpepO, ΔpepE, and ΔpepX strains.
Hydrolysis of peptides by deenergized whole cells.
To analyze the growth with the peptides derived from cell extracts (CFEs), the possibility that the substrates undergo initial hydrolysis by a cell envelope proteinase prior to transport of the resulting peptides was also investigated. Deenergized whole-cell suspensions were prepared and incubated with the peptides under the conditions used for CFE reactions, and the reaction supernatants were analyzed by reverse-phase high-performance liquid chromatography (RP-HPLC) (see below). We found no evidence of extracellular hydrolysis when we examined extended-time reactions (120 min) of the peptides with whole cells (data not shown). Although this result was not expected, it suggests that extracellular hydrolysis prior to transport is not necessary for these peptides. There is a precedent for other multiply amino acid-auxotrophic LAB to transport even larger peptides via the oligopeptide transport system (Opp) than reported for homologues in bacteria with fewer amino acid auxotrophies (19, 20). Kinetic analysis of Opp from Lactococcus indicates that peptides which are 4 to at least 18 residues long can be transported with little specificity for particular amino acid side chains (9). Therefore, it is possible that L. helveticus is capable of transporting αS1-CN(f1-9) and β-CN(f193-209) without prior hydrolysis.
Hydrolysis of αS1-CN(f1-9) and β-CN(f193-209) with CFEs of L. helveticus and peptidase mutants.
The peptides αS1-CN(f1-9) and β-CN(f193-209) were synthesized and purified at the Utah State University Biotechnology Center. CFEs were prepared as described previously (8). The protein concentrations in the CFEs and in bovine serum albumin standards were determined by using a bicinchoninic acid assay kit (Sigma, St. Louis, Mo.) (22).
Peptide hydrolysis reactions with L. helveticus CFEs were performed by using 50-μl (total volume) mixtures. Each sample contained 45 μl of CFE or an appropriate dilution (0.95 to 1.05 mg of protein per ml). The reactions were initiated by adding the peptide substrate, and the mixtures were incubated at 37°C. The initial substrate concentration in the reaction mixtures was 5.0 μg/μl for αS1-CN(f1-9), β-CN(f193-209), and β-CN(f193-209-n-butyl amide). An incubation time of 40 min was determined to result in hydrolysis products with relatively even distributions, and approximately 25 to 30% (as determined by peak area) of the original peptide substrates remained (data not shown). The reactions were stopped by adding 200 μl of 40% (vol/vol) acetonitrile. The sample stability was determined to be >12 h at room temperature.
Hydrolysis samples were separated and collected by using RP-HPLC as described in the accompanying paper (5). The masses of RP-HPLC-separated peptide fractions were determined by using a Perkin-Elmer API 365 triple quadrupole electrospray ionization mass spectrometer or a Bruker Reflex II instrument for matrix-assisted laser desorption ionization-time of flight mass spectrometry. Peptides that could not be positively identified on the basis of mass alone were analyzed by electrospray ionization mass spectrometry-mass spectrometry to measure fragments of the parent peptide. Electrospray ionization mass spectrometry was performed at the University of Wisconsin Biotechnology Center, and matrix-assisted laser desorption ionization-time of flight mass spectrometry was performed and the University of Wisconsin Chemistry Instrument Center.
The reactions of αS1-CN(f1-9) or β-CN(f193-209) with CFEs of the L. helveticus wild type and the peptidase deletion mutants were investigated to assess hydrolytic differences due to the absence of a given peptidase. The rates of depletion of αS1-CN(f1-9) and accumulation of the primary hydrolysis product, αS1-CN(f1-7), were indistinguishable for all six strains. However, the remaining peaks that accumulated after hydrolysis by ΔpepN or ΔpepX strains were different from each other and different from the peaks obtained with the wild type (as well as the peaks obtained with the ΔpepC, ΔpepO, and ΔpepE strains [Fig. 1]).
The chromatograms for reactions of β-CN(f193-209) with CFEs from wild-type, ΔpepC, ΔpepO, ΔpepE, and ΔpepX strains were indistinguishable in terms of the rate of hydrolysis and accumulation of the detectable peptides (Fig. 1). However, the chromatogram peak profiles resulting from hydrolysis with the ΔpepN strain were distinctly different. The most notable differences in the reaction with the ΔpepN strain were the decreased rate of hydrolysis of the initial substrate and the increased accumulation of peptide β-CN(f193-206).
Several other unique peptides accumulated in the absence of PepN activity. For the αS1-CN(f1-9) hydrolysis reactions, these peptides resulted from a decreased ability to liberate Lys-3 from αS1-CN(f3-9) (Fig. 1). For the hydrolysis of β-CN(f193-209), these peptides resulted from a decreased ability to liberate Tyr-193, Val-197, and Leu-198 from derived peptides. The results for liberation of Lys and Leu residues are consistent with the reported amino acid specificities of PepN for amino acid-ρ-nitroanilide substrates (7). The activities measured for liberation of Tyr and Val from dipeptide substrates are routinely relatively low (18). However, an evaluation of the activity of purified lactococcal PepN for a tryptic digest of β-CN also indicated that this peptidase is able to liberate Tyr from β-CN(f193-202) and Val from β-CN(f170-176), as well as peptides containing Glu at the N terminus (23).
The absence of PepX activity from CFEs resulted in accumulation of αS1-CN(f1-5) and αS1-CN(f4-7), two peptides that have an Xaa-Pro N terminus (Fig. 1). This is consistent with the known substrate specificity of PepX (7).
Surprisingly, no differences were detected between hydrolysis of either of the peptides with CFEs from the endopeptidase deletion mutants JLS232 (ΔpepO) and JLS233 (ΔpepE) and hydrolysis of the peptides with CFEs from the wild type. The chromatograms for hydrolysis of αS1-CN(f1-9) by all six strains were indistinguishable with respect to the rate of hydrolysis of αS1-CN(f1-9) and the accumulation of the primary product, αS1-CN(f1-7), indicating that none of the deleted peptidases is responsible for hydrolysis of the Lys-7-His-8 peptide bond. In addition, the fractions analyzed after hydrolysis of β-CN(f193-209) by all six strains contained several peptides with Pro-204 or Pro-206 at the C terminus (Fig. 1). The hydrolysis results indicate that either (i) PepE and PepO do not have significant specificity for αS1-CN(f1-9) or β-CN(f193-209) or derived peptides, (ii) PepE and PepO are not expressed significantly under the growth conditions used, or (iii) the peptides that are substrates for these enzymes did not accumulate at sufficient levels to be detected in our investigation. Since deletion of PepE and PepO did not affect the rate of formation of these peptides, these results indicated that the hydrolysis was due to an as-yet-unidentified endopeptidase and/or carboxypeptidase.
Hydrolysis of the carboxy terminus-protected peptide β-CN(f193-209-n-butyl amide).
To determine whether the carboxy-terminal hydrolysis of β-CN(f193-209) resulted from endopeptidase activity, hydrolysis products of the carboxyl-protected substrate β-CN(f193-209-n-butyl amide) were identified. β-CN(f193-209-n-butyl amide) was synthesized and purified at the University of Wisconsin Biotechnology Center. In order to reduce the extent of hydrolysis of peptides resulting from aminopeptidase activity, hydrolysis reactions with β-CN(f193-209) and β-CN(f193-209-n-butyl amide) were performed with CFE prepared from JLS242 (ΔpepN). The predominant peptide formed from hydrolysis of both β-CN(f193-209) and β-CN(f193-209-n-butyl amide) was identified as β-CN(f193-206), indicating that an endopeptidase has a role in formation of peptides with Pro-206 at the C terminus.
Hydrolysis of peptides at a low pH and a high ionic strength.
To evaluate hydrolysis of peptides under the pH and ionic conditions associated with ripening Cheddar cheese, reactions of L. helveticus wild-type CFE with αS1-CN(f1-9) or β-CN(f193-209) were performed at pH 5.1 in 120 mM MES (morpholineethansulfonic acid) buffer-0.68 M NaCl (4% NaCl). Hydrolysis reactions with CFE were also performed at pH 6.5 (as described above) for direct chromatographic comparison. The CFEs were preincubated (5 min) in the buffers before substrate addition. Following addition of substrate, samples were incubated for 20, 40, and 60 min in order to evaluate hydrolysis over time. A comparative evaluation of the chromatograms for each type of reaction conditions (pH 6.5 versus pH 5.1, 0.68 M NaCl) for αS1-CN(f1-9) at a given reaction time revealed no differences in the hydrolysis of the initial substrate and the accumulation of the primary hydrolysis product, αS1-CN(f1-7) (data not shown). In subtle contrast, the chromatograms for reactions with β-CN(f193-209) indicated that there was a slight increase in the rate of hydrolysis of the initial substrate but no significant difference in the subsequent hydrolysis of the primary product, β-CN(f194-209) (data not shown). The activity of an unidentified endopeptidase at pH 5.1 and 0.68 M NaCl indicates that this enzyme may be important for initiating the hydrolysis of αS1-CN(f1-9), β-CN(f193-209), and perhaps other Pro-containing bitter peptides. We are currently pursuing the identification and characterization of other endopeptidases from L. helveticus CNRZ32 (5).
Acknowledgments
We thank Marie Strickland and the Utah State University Biotechnology Center for providing αS1-CN(f1-9) and β-CN(f193-209). Also, we thank Martha M. Vestling of the University of Wisconsin Chemistry Instrument Center and Amy Harms of the University of Wisconsin Biotechnology Center for providing mass spectrometry expertise.
This project was funded by Dairy Management, Inc. through the Center for Dairy Research and the College of Agricultural and Life Sciences at the University of Wisconsin-Madison.
REFERENCES
- 1.Bartels, H. J., M. E. Johnson, and N. F. Olson. 1987. Accelerated ripening of Gouda cheese. I. Effect of freeze-shocked Lactobacillus helveticus on proteolysis and flavor development. Milchwissenschaft 42:139-144. [Google Scholar]
- 2.Bartels, H. J., M. E. Johnson, and N. F. Olson. 1987. Accelerated ripening of Gouda cheese. I. Effect of heat-shocked thermophilic lactobacilli and streptococci on proteolysis and flavor development. Milchwissenschaft 42:83-88. [Google Scholar]
- 3.Broadbent, J. R., M. Barnes, C. Brennand, M. Strickland, K. Houck, M. E. Johnson, and J. L. Steele. 2002. Contribution of Lactococcus lactis cell envelope proteinase specificity to peptide accumulation and bitterness in reduced-fat Cheddar cheese. Appl. Environ. Microbiol. 68:1778-1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Broadbent, J. R., M. Strickland, B. C. Weimer, M. E. Johnson, and J. L. Steele. 1998. Peptide accumulation and bitterness in Cheddar cheese made using single-strain Lactococcus lactis starters with distinct proteinase specificities. J. Dairy Sci. 81:327-337. [Google Scholar]
- 5.Chen, Y.-S., J. E. Christensen, J. R. Broadbent, and J. L. Steele. 2003. Identification and characterization of Lactobacillus helveticus PepO2, an endopeptidase with post-proline specificity. Appl. Environ. Microbiol. 69:1276-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen, Y. S., and J. L. Steele. 1998. Genetic characterization and physiological role of endopeptidase O from Lactobacillus helveticus CNRZ32. Appl. Environ. Microbiol. 64:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Christensen, J. E., E. G. Dudley, J. A. Pederson, and J. L. Steele. 1999. Peptidases and amino acid catabolism in lactic acid bacteria. Antonie Leeuwenhoek. 76:217-246. [PubMed] [Google Scholar]
- 8.Christensen, J. E. 2000. Peptidases of Lactobacillus helveticus: role in physiology and casein hydrolysis. Ph.D. thesis. University of Wisconsin-Madison, Madison.
- 9.Detmers, F. J. M., E. R. S. Kunji, F. C. Lanfermeijer, B. Poolman, and W. N. Konings. 1998. Kinetics and specificity of peptide uptake by the oligopeptide transport system of Lactococcus lactis. Biochemistry 37:16671-16679. [DOI] [PubMed] [Google Scholar]
- 10.Exterkate, F. A., and A. C. Alting. 1995. The role of starter peptidases in the initial proteolytic events leading to amino acids in Gouda cheese. Int. Dairy J. 5:15-28. [Google Scholar]
- 11.Gomez, M. J., P. Gaya, M. Nuñez, and M. Medina. 1996. Debittering activity of peptidases from selected lactobacilli strains in model cheese. Milchwissenschaft 51:315-319. [Google Scholar]
- 12.Kaminogawa, S., T. R. Yan, N. Azuma, and K. Yamauchi. 1986. Identification of low molecular weight peptides in Gouda-type cheese and evidence for the formation of these peptides from 23 amino-terminal residues of alpha-s1-casein by proteinases of Streptococcus cremoris H61. J. Food Sci. 51:1253-1256, 1264.
- 13.Khalid, N. M., M. El Soda, and E. H. Marth. 1991. Peptide hydrolases of Lactobacillus helveticus and Lactobacillus delbrueckii ssp. bulgaricus. J. Dairy Sci. 74:29-45. [Google Scholar]
- 14.Kunji, E. R. S., I. Mierau, A. Hagting, B. Poolman, and W. N. Konings. 1996. The proteolytic systems of lactic acid bacteria. Antonie Leeuwenhoek 70:187-221. [DOI] [PubMed] [Google Scholar]
- 15.Lee, K. D., C. G. Lo, and J. J. Warthesen. 1996. Removal of bitterness from the bitter peptides extracted from Cheddar cheese with peptidases from Lactococcus lactis ssp. cremoris SK11. J. Dairy Sci. 79:1521-1528. [DOI] [PubMed] [Google Scholar]
- 16.Lemieux, L., and R. E. Simard. 1991. Bitter flavour in dairy products. I. A review of the factors likely to influence its development, mainly in cheese manufacture. Lait 71:599-636. [Google Scholar]
- 17.Lemieux, L., and R. E. Simard. 1991. Bitter flavour in dairy products. II. A review of bitter peptides from caseins: their formation, isolation and identification, structure masking and inhibition. Lait 72:335-382. [Google Scholar]
- 18.Niven, G. W., S. A. Holder, and P. Stroman. 1995. A study of the substrate specificity of aminopeptidase N from Lactococcus lactis subsp. cremoris Wg2. Appl. Microbiol. Biotechnol. 44:100-105. [DOI] [PubMed] [Google Scholar]
- 19.Payne, J. W. 1968. Oligopeptide transport in Escherichia coli. J. Biol. Chem. 243:3395-3403. [PubMed] [Google Scholar]
- 20.Perego, M., C. F. Higgins, S. R. Pearce, M. P. Gallagher, and J. A. Hoch. 1991. The oligopeptide transport system of Bacillus subtilis plays a role in the initiation of sporulation. Mol. Microbiol. 5:173-185. [DOI] [PubMed] [Google Scholar]
- 21.Pritchard, G. G., and T. Coolbear. 1993. The physiology and biochemistry of the proteolytic system in lactic acid bacteria. FEMS Microbiol. Rev. 12:179-206. [DOI] [PubMed] [Google Scholar]
- 22.Smith, P. K., R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson, and D. C. Klenk. 1985. Measurement of protein using bicinchoninic acid. Anal. Biochem. 150:76-85. [DOI] [PubMed] [Google Scholar]
- 23.Tan, P. S. T., T. A. J. M. Van Kessel, F. L. M. Van De Veerdonk, P. F. Zuurendonk, A. P. Bruins, and W. N. Konings. 1993. Degradation and debittering of a tryptic digest from beta-casein by aminopeptidase N from Lactococcus lactis subsp. cremoris WG2. Appl. Environ. Microbiol. 59:1430-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]