Abstract
A comparative analysis was performed with 25 isolates of astroviruses (AstVs) detected in sewage sources and 22 concurrently identified clinical AstV isolates from the Tshwane (Pretoria) Metropolitan Area in South Africa. The samples and specimens were screened for AstVs by using an enzyme immunoassay and/or a reverse transcriptase PCR (RT-PCR) for the highly conserved untranslated region (3′ end) of the genome. The RT-PCR results were confirmed by oligonucleotide probe dot blot hybridization. Viable viruses were propagated in cell cultures for amplification when a minimal specimen was available or indeterminate sequences were obtained. AstV strains were characterized by RT-PCR and partial sequence analysis of the capsid region. The presence of multiple human AstV (HAstV) types in a single sewage sample complicated identification of individual strains, and additional type-specific RT-PCR and sequence analyses of the capsid region were required for characterization. Amplification and characterization of one genotype from a sample, therefore, did not preclude the possibility that a sample harbored additional different genotypes. Genotype and sequence information obtained from AstVs in wastewater samples were compared to information obtained from AstV strains from human stools. HAstV type 1 (HAstV-1), as well as HAstV-3, -5, -6, and -8, were identified among the clinical isolates, and HAstV-1, -2, -3, -4, -5, -7, and -8 were identified among the environmental samples. Phylogenetic analysis demonstrated that HAstV-1, -3, -5, and -8, which were present in human stool and sewage samples, clustered together, indicating that these viruses are closely related. The concurrent presence of identical HAstV strains in wastewater samples and in hospitalized patients suggests that AstVs present in the environment pose a potential risk to communities in which fecally contaminated water is used for recreational and domestic purposes.
Human astroviruses (HAstVs) cause human diarrhea (4) and have been identified as the second most important cause of viral infantile diarrhea in selected areas of South Africa (30, 57) and in other regions of the world (5, 32). HAstV infection has been reported for all age groups, and the young, elderly, and immunocompromised are at the greatest risk (12, 15). Astrovirus (AstV) disease is usually less severe than the disease caused by rotaviruses, and AstVs may also cause asymptomatic infections (8, 28). Transmission of HAstV infections occurs via the fecal-oral route (8, 15). Although contaminated food (47, 63) and water (11) have been associated with outbreaks of HAstV-associated gastroenteritis, the risk attributable to food and water contamination in the transmission of HAstVs has not been fully elucidated yet (15, 16). AstVs also have been associated with scours in young animals, such as calves, lambs, pigs, cats, dogs, and mink (8, 24, 25), as well as with a fatal hepatitis in ducklings (17), with hemorrhagic enteric syndrome in turkeys (23), and with acute intestinal nephritis in chickens (20). To date, eight HAstV serotypes (HAstV type 1 [HAstV-1] to HAstV-8) have been described (33, 37, 61, 65). Certainly two, and possibly three, serotypes of bovine AstVs (37) and one serotype of porcine AstV (38) have been recognized.
AstVs, which are classified in the distinct family Astroviridae, have a single-stranded polyadenylated positive-sense RNA genome that is approximately 6.8 to 7.2 kb long and contains three open reading frames (ORFs), designated ORF1a, ORF1b, and ORF2 (37). ORF2 is located at the 3′ end of the genome and encodes the capsid protein precursor (7). This protein has a well-conserved amino terminus (7). Nucleotide sequence analysis of a limited region of ORF2 has facilitated phylogenetic comparisons of HAstVs (45), and there is a good correlation between antigenic and genomic types (3, 45). Partial and complete sequence data are available for a limited number of animal (21, 65) and turkey (23) AstV isolates. Although the capsid proteins of AstVs infecting different hosts are reportedly highly divergent, similarities among HAstV, feline AstV, and porcine AstV capsid sequences suggest that zoonoses involving pigs, cats, and humans could occur (21). However, AstV infection appears to be species specific (32), and to date no interspecies transmission has been documented (21).
Surface waters in both rural and urban areas are affected by fecal contamination from human and animal sources (14, 18). The occurrence of AstVs in water sources (9, 29, 31, 40, 51, 60) and the occurrence of AstVs in sludge biosolids (10) have been reported, but the clinical significance and epidemiological impact of environmental AstV strains is unknown (60). Until recently, there have been no reports of antigenic or molecular characterization of environmental AstV isolates, but in a recent publication the authors described using restriction fragment length polymorphism (RFLP) to genotype such isolates (52). The aim of this study was to detect, by using type-common reverse transcriptase PCR (RT-PCR), and to characterize, by partially sequencing the 3′ end of the ORF2 capsid gene, AstV strains obtained from water and sewage samples and to compare these strains to AstV strains obtained from the stools of humans in the same geographic region. The comparative data obtained should provide valuable information concerning the possible sources of human infection or concerning sources of fecal contamination of surface waters in communities in which these water sources are used for domestic and recreational purposes.
MATERIALS AND METHODS
Sewage and water samples.
Three or four sewage samples (1 to 2 liters each) were collected from April 1999 to October 2000 at three sewage treatment plants serving residential areas of the Tshwane (Pretoria) Metropolitan Area, Gauteng, South Africa (Table 1). Three concurrent surface water samples (1 liter each) were collected from surface flows downstream from two of the sewage treatment plants (namely, the Daspoort and Baviaanspoort sewage works).
TABLE 1.
Detection and characterization of AstVs in sewage samples obtained from three sewage treatment plants and two urban streams downstream of the sewage treatment plants in the Tshwane (Pretoria) Metropolitan Area, South Africa, between April 1999 and October 2000
| Sampling site | Sampling date (mo/day/yr) | Sample | RT-PCRa
|
Genotype(s) | |
|---|---|---|---|---|---|
| Polyacrylamide gel electrophoresis | Oligonucleotide probe hybridization | ||||
| Sewage treatment plants | |||||
| Daspoort (west inflow) | 4/19/99 | DW1 | +b | − | Untypeable |
| 4/28/99 | DW2 | + | + | HAstV−1, −3, −4, −7, −2c | |
| 5/24/99 | DW3 | + | − | HAstV−1, −3, −4d | |
| 10/19/00 | DW4 | + | + | HAstV−1, −7e | |
| Daspoort (east inflow) | 4/19/99 | DE1 | + | + | Untypeable |
| 5/24/99 | DE2 | + | + | HAstV−1, −3, −5, −7d | |
| 7/26/99 | DE3 | + | + | HAstV−1f | |
| 10/19/00 | DE4 | + | + | HAstV−7, −2, −8g | |
| Baviaanspoort | 4/19/99 | B1 | + | + | Untypeable |
| 4/28/99 | B2 | + | + | HAstV−1, −2f | |
| 10/19/00 | B3 | + | + | HAstV−1h | |
| Zeekoegat | 4/28/99 | Z1 | + | + | HAstV−2h |
| 5/24/99 | Z2 | + | + | HAstV−1h | |
| 9/19/99 | Z3 | + | + | HAstV−3h | |
| 10/19/00 | Z4 | + | + | HAstV−1h | |
| Streams | |||||
| Pienaars River (downstream to Baviaanspoort) | 4/28/99 | R1 | − | − | |
| 5/24/99 | R2 | − | − | ||
| 8/02/99 | R3 | − | − | ||
| Apies River (downstream to Daspoort) | 4/19/99 | A1 | + | − | Untypeable |
| 5/24/99 | A2 | − | − | ||
| 7/26/99 | A3 | − | − | ||
RT-PCR was performed with type-common primers Mon2 and Mon67.
+, detected; −, not detected.
The HAstV−1, −3, −4, and −7 genotypes were determined by using amplicons derived directly from the sample by type-specific RT-PCR, and the HAstV-2 genotype was determined by using amplicons derived directly from the sample by group-specific RT-PCR.
The HAstV genotypes were determined by using amplicons derived directly from the sample by type-specific RT-PCR.
The HAstV-1 genotype was determined by using amplicons derived directly from the sample by group-specific RT-PCR, and the HAstV-7 genotype was determined by using amplicons derived directly from the sample by type-specific RT-PCR.
AstV genotypes were determined by using amplicons derived from infected CaCo-2 cell cultures by group-specific RT-PCR.
The HAstV-7 genotype was determined by using amplicons derived directly from the sample by type-specific RT-PCR, and the HAstV-2 and −8 genotypes were determined by using amplicons derived by group-specific RT-PCR from two different types of cell cultures inoculated with the same sample.
The HAstV genotype was determined by using amplicons derived directly from the sample by group-specific RT-PCR.
The sewage and water samples were clarified by centrifugation (Beckman GS-6R centrifuge) for 30 min at 3,000 × g. Each resultant pellet was resuspended in supernatant fluid (10 ml) and clarified by addition of chloroform (10%, vol/vol; Merck, Darmstadt, Germany) and further centrifugation (Beckman GS-6R centrifuge) for 10 min at 3,000 × g. The supernatants from the first and second clarification procedures were pooled, and AstVs were recovered from each supernatant in 10 ml (final volume) of phosphate-buffered saline (pH 7.4) (Sigma Chemical Co., St. Louis, Mo.) by using the polyethylene glycol-sodium chloride precipitation technique described by Minor (34) for the concentration of picornaviruses. The viral suspension was concentrated further to 2 ml by ultrafiltration by using a Biomax-100K NMWL membrane (Ultrafree 15 centrifugal filter device; Millipore Corporation, Bedford, Mass.). The final concentrate was divided into aliquots and stored at −20°C.
Clinical specimens.
Stool specimens from pediatric patients (age, <5 years) who presented with clinical symptoms of gastroenteritis at two tertiary referral hospitals in the Tshwane Metropolitan Area were submitted for routine diagnosis of gastroenteritis viruses. HAstVs were detected by enzyme immunoassay (EIA) (IDEIA Astrovirus; Dako Ltd., Ely, United Kingdom) in 32 of 1,303 (2.5%) stool specimens referred from January 1998 to October 2000 for analysis. An additional three HAstVs were detected by EIA retrospectively in 1% (3 of 356) of stool samples referred between January 1996 and December 1997 which had been stored at 4°C. Stool specimens and suspensions of stool specimens (10% in phosphate-buffered saline [Sigma]) were stored at 4°C.
Cell culture amplification.
To enhance detection by RT-PCR or to clarify sequencing data for certain isolates, AstVs in concentrates of selected water samples and stool suspensions were amplified by propagation in cell cultures. Sample concentrates and stool suspensions were treated with penicillin (50 μg/ml), streptomycin (50 μg/ml), and neomycin (100 μg/ml) (100× PSN antibiotic mixture; GIBCO BRL Life Technologies, Paisley, Scotland) and with 100 U of nystatin (GIBCO BRL) per ml and inoculated onto monolayers of human hepatoma cell line PLC/PRF/5 (ATCC CRL 8024) (passages 81 to 85) and human colonic carcinoma cell line CaCo-2 (ATCC HTB 37) (passages 35 to 58 and 178 to 198). Cells were grown in 25-cm2 cell culture flasks, as described previously (59), and were incubated for 7 days at 37°C and harvested; an aliquot was blind passaged, and this was followed by incubation for an additional 7 days at 37°C. Cell culture extracts (120 μl) from the initial harvest and after blind passage were assayed for AstV RNA by RT-PCR.
Detection of AstVs by RT-PCR.
Aliquots of the sludge samples, stool suspensions, and cell culture extracts were pretreated with an equal volume of 1,1,2-trichloro-trifluoroethane (Sigma) prior to extraction of total RNA from 120 μl of each treated sample with TRIZOL reagent (GIBCO BRL) used according to the manufacturer's instructions. The extracted RNA was resuspended in 25 μl (final volume) of sterile nuclease-free water (Promega Corp., Madison, Wis.) and stored at −70°C. For each extraction procedure, nuclease-free water was included as a negative control. The primers and probe were synthesized by Sigma-Genosys Ltd., Pampisford, United Kingdom. RT-PCR was performed by using 5 μl of RNA extract and type-common primers Mon2 and Mon67 (35). The amplicon was confirmed to be an AstV amplicon by performing an oligonucleotide probe hybridization assay as described previously (31, 60). For further characterization, a region at the 3′ end of the ORF2 capsid gene (nucleotides [nt] 6513 to 6781, HAstV-1 [accession no. L23513]) of all confirmed AstV-positive samples was amplified by using primers Mon2 and prBEG (54). These primers detect all HAstVs except HAstV-4. The reaction mixture and the conditions for the RT-PCR when these primers were used were essentially the same as those used when the Mon2-Mon67 primer pair was used (60), except that the 1× PCR buffer contained 10 mM Tris-HCl (pH 9.0), 50 mM KCl, 0.1% Triton X-100, and 1.5 mM MgCl2. Clinical isolates that were undetectable with primers Mon2 and prBEG were subjected to an HAstV-4 type-specific RT-PCR (64) or a type-common RT-PCR with primers Mon348 and Mon340, which amplified a region of ORF1a (3). Environmental isolates that were confirmed to be AstV positive by the RT-PCR-oligonucleotide probe hybridization assay but were undetectable or produced an uninterpretable sequence when primers Mon2 and prBEG were used were subjected to HAstV-1 to HAstV-7 type-specific RT-PCRs as described by Walter et al. (64). Cell culture extracts of HAstV-1 to HAstV-7 Oxford reference strains were used as positive controls.
Sequencing of RT-PCR amplicons.
DNA amplicons derived from the 3′ end of the ORF2 capsid gene or the 289-bp region of ORF1a were sequenced directly by the dideoxy chain termination method (56) by using a Sequenase version 2.0 PCR product sequencing kit (U.S. Biochemical Corp., Cleveland, Ohio) according to the manufacturer's instructions. The sequencing reaction mixtures were electrophoresed on 8% polyacrylamide-6 M urea gels in 1× Tris-borate-EDTA buffer. The gels were vacuum dried and exposed to X-ray film (Hyperfilm-βmax; Amersham) for 12 h at room temperature.
Sequence analysis and genotyping.
Nucleotide sequences were entered into a database in PC/Gene (version 6.85; IntelliGenetics Inc., Geneva, Switzerland). Basic sequence manipulation and verification were performed by using OMIGA (version 2.0; Accelrys, Madison, Wis.). ClustalX (62) was used to create multiple alignments of the amino acid sequences of selected isolates and reference strains. Nucleic acid sequences were added and aligned with GeneDoc (version 2.3) by using the corresponding amino acid alignment as a template, which resulted in a consensus length of 208 nt for the 3′ end of ORF2 (42). Pairwise comparisons of nucleotide sequences of all reference types with the selected isolates were performed by using GeneDoc (version 2.3) for preliminary genotype assignment and for identification of clusters of strains with 99 to 100% homology. Only one representative strain from each cluster was included in the phylogenetic analysis. The sequences of representative isolates were compared with AstV sequences present in GenBank by using the BLAST-N program (version 2.l.1) (1, 2) to search for the most similar strain available. The nucleotide sequence alignment was assessed by likelihood-mapping (43, 58) utilizing TREE-PUZZLE 5.0 (http://www.tree-puzzle.de) for tree-likeliness of the data and the proper sequence composition for phylogenetic analysis. Phylogenetic trees were constructed from the nucleic acid sequence alignments by using the maximum-likelihood algorithm of the program DNAML of PHYLIP (version 3.52c) running in a UNIX environment (13). We performed the analysis rooted (with HAstV-4 as a root) and unrooted. In the analysis the global rearrangement option was invoked, and the order of the sequence input was randomized 10 times. Phylograms generated in DNAML were visualized by using the TREEVIEW package (version 1.5) (48) and were further edited with Micrografx Designer (version 6.0a).
Definition of strains.
Although the definitions for strains and subtypes of AstVs are not clear, we arbitrarily considered an isolate with <95% nucleotide homology and a distance of >0.05 as determined by phylogenetic analysis at the 3′ end of ORF2 (consensus length, 208 nt) compared to the reference strain a different strain.
Nucleotide sequence accession numbers.
The accession numbers for the previously published HAstV capsid gene sequences that were used in the pairwise comparisons and phylogenetic analyses and included reference strains with complete capsid sequences are as follows: HAstV-1, L23513; HAstV-2, L13745; HAstV-3, AF117209; HAstV-4, Z33883; HAstV-5, U15136; HAstV-6, Z46658; HAstV-7, AF248738; and HAstV-8, Z66541.
The nucleotide sequence data for the clinical and environmental AstV isolates described here have been deposited in the EMBL/GenBank database under the following accession numbers: T3/SA/DW2_P3/1999, AY094090; T7/SA/DW2_P7/1999, AY094091; T4/DW3_P4/1999, AY094092; T5/SA/DE2_T5/1999, AY094089; T1/SA/DE3_C/1999, AY094082; T8/SA/DE4_P/2000, AY094083; T2/SA/DE4_S/2000, AY094084; T1/SA/B2_64/1999, AY094080; T2/SA/B2_61/1999, AY094079; T1/SA/B3/2000, AY094081; T2/SA/Z1/1999, AY094085; T1/SA/Z2/1999, AY094086; T3/SA/Z3/1999, AY094087; T1/SA/Z4/2000, AY094088; T8/SA/4759/1998, AY093649; T3/SA/5200/1998, AY093650; T5/SA/6899/1998, AY093651; T1/SA/7110/1998, AY093652; T6/SA/126729/1998, AY093653; T1/SA/7052/1999, AY093654; and T1/SA/26025/1999, AY093655.
RESULTS
AstV genotypes in water and sewage samples and clinical specimens.
AstVs were detected directly by the HAstV type-common RT-PCR in all of the human stool specimens previously identified by EIA, in all 15 sewage samples, and in one of six (17%) stream water samples. The presence of AstV amplicons was confirmed by an oligonucleotide probe hybridization assay of the RT-PCR products from 100% of the human stool specimens, from 13 of 15 (87%) of the sewage samples, and from none of the stream water samples. Of the 35 AstV isolates from human stool specimens, 22 (63%) could be characterized directly after amplification from the stool specimens by sequencing of the Mon2-prBEG amplicon (296 to 324 nt, depending on the HAstV type) from the 3′ end of ORF2. Two additional isolates were confirmed to be HAstV isolates by sequencing of a 246-bp region of ORF1a. After storage at 4°C for an extended period, AstVs could no longer be amplified by RT-PCR from the remaining 11 stool specimens by using type-common primers Mon2 plus prBEG and Mon348 plus Mon340 or type-specific primers. Seven environmental AstV isolates, from seven separate sewage samples, were amplified and characterized directly from the sewage samples by sequencing of the Mon2-prBEG amplicon. An additional five isolates, originating from three sewage samples, could be characterized only after isolation in cell culture. One of the sewage samples (sample B2) yielded two different HAstV types from separate flasks of CaCo-2 cell cultures with different numbers of passages, while another sample (sample DE4) yielded two different HAstV genotypes after amplification with two different cell culture types (i.e., CaCo-2 and PLC/PRF/5) (Table 1). Thirteen isolates, from five of the sewage samples, were typed by sequencing amplicons from amplicons generated by type-specific RT-PCR directly from the sewage samples. Indeterminate sequences were obtained from amplicons derived from sewage samples DW1, DE1, and B1 and their cell culture derivatives.
The distribution of the genotypes of the 24 HAstVs from clinical specimens characterized was as follows: HAstV-1, 63%; HAstV-3, 13%; HAstV-5, 8%; HAstV-6, 8%; and HAstV-8, 8%. The distribution of the genotypes of the 24 AstV isolates from the sewage samples was as follows: HAstV-1, 36%; HAstV-2, 16%; HAstV-3, 16%; HAstV-4, 8%; HAstV-5, 4%; HAstV-7, 16%; and HAstV-8, 4% (Table 1). Seasonal prevalence was not apparent in this small sample set.
Sequence analysis of South African AstVs.
The multiple alignment included sequences with a consensus length of 208 nt (after the primer sequences were removed) for all South African isolates. Pairwise comparisons revealed groups of South African isolates with 99 to 100% identity. The groups of isolates and representative strains of each group are summarized in Table 2.
TABLE 2.
Summary of the characterized South African HAstV strains from clinical and sewage sources with high levels of identity to the representative isolates included in the phylogenetic analysis
| Representative isolate (n = 21)a | Strain(s) with 99 to 100% nucleotide identity to the representative isolate (n = 25) |
|---|---|
| T3/SA/Z3/1999 | T3/SA/7169/1996, T3/SA/113768/1998, T3/SA/DE2_T3/1999, T3/SA/DW2_P3/1999 |
| T3/SA/5200/1998 | NDb |
| T3/SA/DW3_P3/1999 | ND |
| T7/SA/DW2_P7/1999 | T7/SA/DE2_T7/1999, T7/SA/DE4_T7/2000, T7/SA/DW4_T7/2000 |
| T6/SA/126729/1998 | T6/SA/5236/1998 |
| T8/SA/4759/1998 | ND |
| T8/SA/DE4_P/2000 | ND |
| T5/SA/DE2_T5/1999 | ND |
| T5/SA/6899/1998 | T5/SA/126585/1998 |
| T1/SA/DE3_C/1999 | T1/SA/DE2_T1/1999 |
| T1/SA/26025/1999 | T1/SA/3144/1997, T1/SA/124893/1998, T1/SA/4642/1999, T1/SA/6802/1999, T1/SA/9559/1999, T1/SA/22320/1999, T1/SA/25786/1999, T1/SA/5114/2000, T1/SA/DW3_P1/1999, T1/SA/DW4/2000 |
| T1/SA/Z4/2000 | T1/SA/29903/1999, T1/SA/3621/2000 |
| T1/SA/7110/1998 | ND |
| T1/SA/B3/2000 | ND |
| T1/SA/Z2/1999 | ND |
| T1/SA/7052/1999 | ND |
| T1/SA/B2_64/1999 | ND |
| T2/SA/Z1/1999 | ND |
| T2/SA/B2_61/1999 | ND |
| T2/SA/DE4_S/2000 | T2/SA/DW2_S/1999, T2/SA/DW2_P/1999 |
| T4/SA/DW3_P4/1999 | T4/SA/DW2_P4/1999 |
Type assignments were based on comparison with the Oxford reference strain.
ND, none detected.
To prepare for phylogenetic analysis, the multiple alignments of the nucleotide sequences were tested by likelihood mapping. The data had a tree-like structure, and all sequences were in the proper sequence composition range. The phylogenetic analysis was performed in two stages. First, all South African strains were included in an unrooted tree. Pairwise analysis and the phylogenetic tree revealed common branch points for the majority of the South African strains within types; therefore, 27 strains with 99 to 100% homology were withheld from the phylogenetic analysis to avoid repeats. Reference strains (HAstV-1 to -8) and representatives of the 21 distinctive South African strains were included in the final phylogenetic analysis. The HAstV-4 strain was less related to the other reference strains and was therefore used as a root for the analysis.
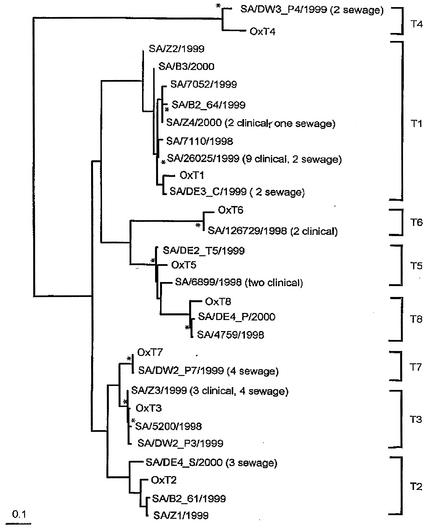
The analysis (Fig. 1) showed that clusters of types with HAstV-1 to -8 reference strains separated with confidence (distances, 0.09 to 0.62; P < 0.05). HAstV-3 and -7 and HAstV-5 and -8 were very closely related in this hypervariable region, with 90 and 82% pairwise identity and distances of 0.09 and 0.124, respectively. The HAstV-4 strain was significantly different from the other HAstV strains. All of the characterized South African isolates could be assigned to a type. The calculated intragenotypic distances suggest that the HAstV-1, -2, -4, -5, and -8 South African isolates represent new strains of the corresponding genotypes (for HAstV-1, 90 to 94% identity and distances of 0.05 to 0.13; for HAstV-2, 88 to 94% identity and distances of 0.10 to 0.13; for HAstV-4, 88% identity and distance of 0.12; for HAstV-5, 91 to 94% identity and distances of 0.05 to 0.10; and for HAstV-8, 94% identity and distance of 0.06). South African strains of HAstV-3, -6, and -7 appear not to be new strains or subtypes (for HAstV-3, 99% identity and distance of 0.01; for HAstV-6, 96% identity and distance of 0.04; and for HAstV-7, 99% identity and distance of 0.005).
FIG. 1.
Maximum-likelihood phylogenetic tree based on a 208-nt region of the 3′ end of ORF2, showing the relationships of representatives of the South African environmental and clinical HAstV isolates and the prototypes of HAstV-1 to -8. The branch points of the tree (rooted) had a confidence level of P < 0.05. Scale bar = 0.1 nucleotide substitution per site. Asterisks indicate nonsignificant branch points (P > 0.05).
HAstV-1, -3, -5, and -8 were detected among the clinical samples and environmental isolates. HAstV-1 isolates comprised 22 (48%) of the 46 isolates characterized by sequence analysis of the 3′ end of ORF2. The phylogenetic analysis included eight representative strains, as previously described. The nucleotide identity among the South African HAstV-1 isolates was 88 to 100%, compared to 89 to 94% identity to the prototype strain. Two isolates recovered from wastewater sources formed a distinct subtype (cluster T1a) but were more closely related to the Oxford reference strain (distance, 0.05) than the other South African strains were. Another cluster (cluster T1b), which was distinct from cluster T1a (distance, 0.04 to 0.6), was also observed. Cluster T1b included multiple closely related environmental strains and strains from human stool samples represented by SA/Z4/2000, SA/B2_64/1999, SA/7110/1998, SA/7052/1999, and SA/26025/1999. The other two strains (SA/B3/2000 and SA/Z2/1999) had unique sequences and were identified only once.
The environmental and clinical HAstV-3 isolates showed high levels of nucleotide sequence identity (≥98%) to each other and to the prototype strain and clustered together in a single genotypic cluster, cluster T3 (Table 2; Fig. 1). The HAstV-5 environmental isolate exhibited a higher level of nucleotide identity (95%) to the prototype strain than the clinical isolates exhibited (nucleotide identity, 91%). The HAstV-5 environmental isolate and clinical isolates exhibited only 93% nucleotide identity (distance, 0.07); as a result, we considered them unique strains of HAstV-5 (Fig. 1).
Two HAstV-8 strains, one from a clinical specimen obtained in 1998 and the other from a sewage sample collected in 2000, were analyzed. These two strains are closely related to each other (98% nucleotide sequence identity) but are distinct from the prototype strain (93 to 94% nucleotide sequence identity) (Fig. 1). A nucleotide identity of 100% was recorded for the two clinical HAstV-6 strains, and they exhibited 96% nucleotide identity to the Oxford reference strain. No HAstV-6 strains were detected among the environmental isolates.
HAstV-2, -4, and -7 isolates were detected only among the environmental isolates. The levels of nucleotide identity among the HAstV-2 isolates were 88 to 98%, but when these viruses were compared to the prototype strain, the levels of identity were lower (88 to 94%); this resulted in clustering of the South African isolates separate from the prototype strain within HAstV-2 (Fig. 1). The South African HAstV-2 isolates were members of two unique clusters, represented by SA/B2_61/1999 and SA/Z1/1999 in one group and by SA/DE4_S/2000 in the other group (distance, 0.013) (Fig. 1). The HAstV-4 isolates, both from the same sewage works, showed 100% nucleotide sequence homology to each other but 88% nucleotide sequence homology to the prototype strain. The HAstV-7 isolates showed high levels of nucleotide sequence identity (≥98%) to each other and to the prototype strain and grouped together in a single cluster (Fig. 1).
The South African HAstV strains from clinical and sewage specimens were compared to HAstV strains from different geographic locations from the same time period (data not shown). The analysis showed that the South African environmental and clinical strains in clusters T1b and T8 cluster together and are distinct from strains isolated at the same time from different geographical locations.
DISCUSSION
In this study an RT-PCR-oligonucleotide probe hybridization assay followed by partial sequencing of the C terminus of ORF2 was successfully used to detect and characterize clinical and environmental AstV isolates from the Tshwane Metropolitan Area in South Africa. The RT-PCR-oligonucleotide probe hybridization assay has previously been shown to be a valuable tool for detection of HAstVs in stool specimens (30) and, in conjunction with cell culture, for detection of infectious HAstVs in water from different sources (31, 60). In this investigation prior amplification in cell culture was also shown to facilitate detection and characterization of multiple HAstV types in a single sample (Table 1). PCR is a procedure that is widely used to detect and genotype viruses (9, 27, 49, 50, 66) and parasites (53) from clinical and environmental sources. Sequence analysis of the 3′ region of HAstV ORF2 provides type information, as well as enough diversity to provide additional strain information. There are, however, no data on the ability of RT-PCR amplification of ORF2 to detect and characterize mixed populations of HAstV genotypes. For this study, single HAstV genotypes obtained from the clinical specimens and obtained directly from six of the sewage samples were amplified by RT-PCR by using type-common primers Mon2 and prBEG, and they were characterized by sequencing a 208-nt region of ORF2. However, sequence analysis with type-common primers Mon2 and prBEG for a number of the sewage samples resulted in indeterminate or untypeable sequences (Table 1). We then asked if these strains were truly unique strains or represented a mixed population of strains that underwent the sequencing reaction simultaneously. Subsequent RT-PCR amplification of the same samples with HAstV-1 to -7 type-specific primers, which also amplified the 3′ end of ORF2, resulted in identification of multiple genotypes in at least five of the sewage samples (Table 1). In one of the sewage samples, DW4, HAstV-1 was identified by RT-PCR by using the type-common primers Mon2 and prBEG, while HAstV-7 was subsequently detected in the same specimen by using type-specific primers. As has been reported previously for Cryptosporidium parvum (53), we showed that amplification and characterization of a single genotype from a clinical specimen or water sample do not preclude the possibility that multiple genotypes are present. A similar finding was reported for human caliciviruses; in this case cloning of PCR products and sequencing of several individual clones resulted in identification of multiple genotypes in a single sewage sample (27).
Among the clinical isolates characterized by sequence analysis of the 3′ end of ORF2, HAstV-1 (64%) was the most frequent type identified, and HAstV-3 (14%) and HAstV-5 (9%) were less common. This finding is similar to findings obtained in other regions of the world (19, 22, 26, 39, 44, 45, 46, 49, 55). The occurrence of HAstV-6 and -8 in 9 and 5% of the specimens, respectively, is important as these types reportedly are seldom detected (15, 37). HAstV-8, however, appears to be more common on the African continent (36, 41, 61) and in Barcelona, Spain (19). The absence of HAstV-2 in the South African clinical specimens is noteworthy as this serotype was identified as the predominant type in other parts of the world, including in a periurban community of Mexico City (64). The distribution of HAstV genotypes in the South African environmental isolates is similar to that observed for the clinical isolates in that HAstV-1 was the predominant type identified (36% of the isolates). The difference was the occurrence of HAstV-2, -4, and -7 in the sewage samples, which comprised 16, 8, and 16% of the environmental isolates, respectively, while none of these types were detected in the clinical specimens. Further research is therefore warranted to ascertain whether these types are possibly more resistant to environmental degradation or whether human infection by these types is not as severe as human infection by the other types and thus does not require medical attention. Finally, is there a difference in the reservoir and/or mode of transmission between HAstV types?
Our results clearly indicate that the HAstVs detected in both clinical and environmental samples are closely related and probably represent identical strains (Table 2; Fig. 1). Further analysis showed that the closely related environmental and clinical isolates were distinct from other HAstV strains detected during the same time period (1997 to 2000) in other geographical locations. This suggests that fecally polluted water could be a reservoir for human infection. In addition, the presence of different strains in the same community indicates that multiple strains and multiple genotypes circulate concurrently. Phylogenetic analysis demonstrated that the South African strains aligned with the corresponding reference strains but were sufficiently different that they represent new strains or subtypes (P < 0.05). Two South African HAstV-1 subtypes were detected; one of these subtypes includes both clinical and environmental isolates (distance, 0.05; P < 0.01). Two separate subtypes of HAstV-5 and -2 were identified. South African HAstV-8 strains formed a new subtype that included environmental and clinical subtypes. South African HAstV-4 strains also clustered in a distinct subtype. The nucleotide sequences of HAstV-3 and -7 isolates from the South African clinical and environmental sources, as well as from other geographic regions, appeared to be highly conserved, exhibiting 98 to 100% identity. This is similar to what was observed for hepatitis A virus (50).
The AstVs detected by RT-PCR-oligonucleotide probe hybridization in the sewage samples were characterized as HAstVs (namely, HAstV-1, -2, -3, -4, -5, -7, and -8) (Table 1). This suggests that the integrated RT-PCR-oligonucleotide probe hybridization assay used in this and previous studies (31, 60) for the detection of AstVs in water and sewage samples selects for AstVs of human origin. As cross-species infection in vitro appears to occur only after prior adaptation of an AstV isolate in a cell culture of the species of origin (6), the amplification of AstVs from water and sewage specimens in cell lines of human origin (namely, PLC/PRF/5 and CaCo-2) should further enhance the selection and detection of viruses of human origin. Additional RT-PCRs performed with primers specific for animal AstVs and/or cell cultures of animal origin would therefore be required to detect AstVs of animal origin in water and sewage samples. The role of zoonotic infection of AstVs is not currently understood; consequently, the possible risk of infection of humans by animal AstVs in water sources needs further clarification. The type of HAstVs found in sewage is a reflection of the clinical epidemiology of HAstVs (52). Therefore, the presence of HAstVs in the environment could pose a potential health risk to persons using contaminated water for domestic or recreational purposes. Although quantitative RT-PCR and larger surface water sample volumes would be required to obtain conclusive evidence, the absence of AstVs in the surface waters downstream of the sewage works in which multiple genotypes of HAstVs were detected suggests that these viruses were removed effectively by the sewage treatment process. This study provides valuable new data on the molecular epidemiology of HAstVs circulating in the communities in the Tshwane Metropolitan Area of South Africa and in southern Africa and provides a feasible alternative to RFLP analysis (52) for characterization of HAstVs detected in water sources. However RT-PCR with analysis of the sequence in the capsid region provides more information for characterization of environmental isolates than RFLP analysis provides as RFLP analysis is limited by the amount of the RT-PCR product and requires secondary amplification by internal primers.
Acknowledgments
This work was supported by grants from the Water Research Commission and the National Research Foundation, South Africa. S. Nadan was supported by grant 99/19 from the Poliomyelitis Research Foundation, South Africa, and by a grant holder-linked bursary from the National Research Foundation. This study was supported in part by grant NIAD RO1 AI45872-01.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belliot, G., H. Laveran, and S. S. Monroe. 1997. Detection and genetic differentiation of human astroviruses: phylogenetic grouping varies by coding region. Arch. Virol. 142:1323-1334. [DOI] [PubMed] [Google Scholar]
- 4.Bern, C., and R. I. Glass. 1994. Impact of diarrheal diseases worldwide, p. 1-26. In A. Z. Kapikian (ed.), Viral infections of the gastrointestinal tract, 2nd ed. Marcel Dekker, Inc. New York, N.Y.
- 5.Bon, F., P. Fascia, M. Dauvergne, D. Tenenbaum, H. Planson, A. M. Petion, P. Pothier, and E. Kohli. 1999. Prevalence of group A rotavirus, human calicivirus, astrovirus, and adenovirus type 40 and 41 infections among children with acute gastroenteritis in Dijon, France. J. Clin. Microbiol. 37:3055-3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brinker, J. P., N. R. Blacklow, and J. E. Herrmann. 2000. Human astrovirus isolation and propagation in multiple cell lines. Arch. Virol. 145:1847-1856. [DOI] [PubMed] [Google Scholar]
- 7.Carter, M. J., and M. M. Willcocks. 1996. The molecular biology of astroviruses. Arch. Virol. Suppl. 12:277-285. [DOI] [PubMed] [Google Scholar]
- 8.Caul, E. O. 1996. Viral gastroenteritis: small round structured viruses, caliciviruses and astroviruses. Part II. The epidemiological perspective. J. Clin. Pathol. 49:959-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chapron, C. D., N. A. Ballester, J. H. Fontaine, C. N. Frades, and A. B. Margolin. 2000. Detection of astroviruses, enteroviruses, and adenovirus types 40 and 41 in surface waters collected and evaluated by the information collection rule and an integrated cell culture-nested PCR procedure. Appl. Environ. Microbiol. 66:2520-2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chapron, C. D., N. A. Ballester, and A. B. Margolin. 2000. The detection of astrovirus in sludge biosolids using an integrated cell culture nested PCR technique. J. Appl. Microbiol. 89:11-15. [DOI] [PubMed] [Google Scholar]
- 11.Cubitt, W. D. 1991. A review of the epidemiology and diagnosis of waterborne viral infections. Water Sci. Technol. 24:197-203. [Google Scholar]
- 12.Cubitt, W. D., D. K. Mitchell, M. J. Carter, M. M. Willcocks, and H. Holzel. 1999. Application of electronmicroscopy, enzyme immunoassay, and RT-PCR to monitor an outbreak of astrovirus type 1 in a paediatric bone marrow transplant unit. J. Med. Virol. 57:313-321. [DOI] [PubMed] [Google Scholar]
- 13.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), 3.5c ed. Department of Genetics, University of Washington, Seattle.
- 14.Fogarty, J., L. Thornton, C. Hayes, M. Laffoy, D. O'Flanagan, J. Devlin, and R. Corcoran. 1995. Illness in a community associated with an episode of water contamination with sewage. Epidemiol. Infect. 114:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glass, R. I., J. Noel, D. Mitchell, J. E. Herrmann, N. R. Blacklow, L. K. Pickering, P. Dennehy, G. Ruiz-Palacios, M. L. de Guerrero, and S. S. Monroe. 1996. The changing epidemiology of astrovirus-associated gastroenteritis: a review. Arch. Virol. Suppl. 12:287-300. [DOI] [PubMed] [Google Scholar]
- 16.Goodgame, R. W. 2001. Viral causes of diarrhea. Gastroenterol. Clin. N. Am. 30:779-795. [DOI] [PubMed] [Google Scholar]
- 17.Gough, R. R., M. S. Collins, E. Borland, and L. F. Keymer. 1984. Astrovirus-like particles associated with hepatitis in ducklings. Vet. Rec. 14:279.. [DOI] [PubMed] [Google Scholar]
- 18.Grabow, W. O. K. 1996. Waterborne diseases: update on water quality assessment and control. Water SA (Pretoria) 22:193-202. [Google Scholar]
- 19.Guix, S., S. Caballero, C. Villena, R. Bartolomé, C. Lattore, N. Rabella, M. Simó, A. Bosch, and R. M. Pintó. 2002. Molecular epidemiology of astrovirus infection in Barcelona, Spain. J. Clin. Microbiol. 40:133-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imada, T., S. Yamaguchi, M. Masaji, K. Tsukamoto, M. Kubo, and A. Morooka. 2000. Avian nephritis virus (ANV) as a new member of the family Astroviridae and construction of infectious ANV cDNA. J. Virol. 74:8487-8493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jonassen, C. M., T. Ø. Jonassen, Y. M. Saif, D. R. Snodgrass, H. Ushijima, M. Shimizu, and B. Grinde. 2001. Comparison of capsid sequences from human and animal astroviruses. J. Gen. Virol. 82:1061-1067. [DOI] [PubMed] [Google Scholar]
- 22.Kjeldsberg, E. 1994. Serotyping of human astrovirus strains by immunogold staining electron microscopy. J. Virol. Methods 50:137-144. [DOI] [PubMed] [Google Scholar]
- 23.Koci. M. D., B. S. Seal, and S. Schultz-Cherry. 2000. Molecular characterization of an avian astrovirus. J. Virol. 74:6173-6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurtz, J. B. 1994. Astroviruses, p. 569-580. In A. Z. Kapikian (ed.), Viral infections of the gastrointestinal tract, 2nd ed. Marcel Dekker, Inc., New York, N.Y.
- 25.Kurtz, J. B., and T. W. Lee. 1987. Astroviruses: human and animal. CIBA Found. Symp. 128:92-101. [DOI] [PubMed] [Google Scholar]
- 26.Lee, T. W., and J. B. Kurtz. 1994. Prevalence of human astrovirus serotypes in the Oxford region 1976-92, with evidence for two new serotypes. Epidemiol. Infect. 112:187-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lodder, W. J., J. Vinjé, R. van de Heide, A. M. de Rode Husman, E. J. T. M. Leenen, and M. P. G. Koopmans. 1999. Molecular detection of Norwalk-like caliciviruses in sewage. Appl. Environ. Microbiol. 65:5624-5627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Madeley, C. R., and B. P. Cosgrove. 1975. Viruses in infantile gastroenteritis. Lancet i:124. [DOI] [PubMed]
- 29.Marx, F. E., M. B. Taylor, and W. O. K. Grabow. 1995. Optimization of a PCR method for the detection of astrovirus type 1 in environmental samples. Water Sci. Technol. 31:359-362. [Google Scholar]
- 30.Marx, F. E., M. B. Taylor, and W. O. K. Grabow. 1998. The prevalence of human astrovirus and enteric adenovirus infection in South African patients with gastroenteritis. Southern Afr. J. Epidemiol. Infect. 13:5-9. [Google Scholar]
- 31.Marx, F. E., M. B. Taylor, and W. O. K. Grabow. 1998. The application of a reverse transcriptase-polymerase chain reaction oligonucleotide probe assay for the detection of human astroviruses in environmental water. Water Res. 32:2147-2153. [Google Scholar]
- 32.Matsui, S. M., and H. B. Greenberg. 1996. Astroviruses, p. 811-824. In B. N. Fields, D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, Pa.
- 33.Méndez-Toss, M., P. Romero-Guido, P., M. E. Munguía, E. Méndez, and C. F. Arias. 2000. Molecular analysis of a serotype 8 human astrovirus genome. J. Gen. Virol. 81:2891-2897. [DOI] [PubMed] [Google Scholar]
- 34.Minor, P. D. 1985. Growth, assay and purification of picornaviruses, p. 25-41. In B. W. J. Mahy (ed.), Virology: a practical approach. IRL Press Ltd., Oxford, United Kingdom.
- 35.Mitchell, D. K., S. S. Monroe, X. Jiang, D. O. Matson, R. I. Glass, and L. K. Pickering. 1995. Virologic features of an astrovirus diarrhea outbreak in a day care center revealed by reverse transcriptase-polymerase chain reaction. J. Infect. Dis. 172:1437-1444. [DOI] [PubMed] [Google Scholar]
- 36.Monceyron, C., B. Grinde, and T. Ø. Jonassen. 1997. Molecular characterisation of the 3′-end of the astrovirus genome. Arch. Virol. 142:699-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monroe, S. S. 1999. Astroviruses (Astroviridae), p. 104-108. In A. Granoff and R. G. Webster (ed.), Encyclopedia of virology, 2nd ed., vol. 1. Academic Press Ltd., London, United Kingdom.
- 38.Monroe, S. S., M. J. Carter, J. E. Herrmann, J. B. Kurtz, and S. M. Matsui. 2000. Family Astroviridae, p. 741-745. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Seventh report of the International Committee on Taxonomy of Viruses. Academic Press, San Diego, Calif.
- 39.Mustapha, H., E. A. Palombo, and R. F. Bishop. 2000. Epidemiology of astrovirus infection in young children hospitalized with acute gastroenteritis in Melbourne, Australia, over a period of four consecutive years, 1995 to 1998. J. Clin. Microbiol. 38:1058-1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myint, S., R. Manley, and D. Cubitt. 1994. Viruses in bathing waters. Lancet 343:1640-1641. [DOI] [PubMed] [Google Scholar]
- 41.Naficy, A. B., M. R. Rao, J. L. Holmes, R. Abu-Elyazeed, S. J. Savarino, T. F. Wierzba, R. W. Frenck, S. S. Monroe, R. I. Glass, and J. D. Clemens. 2000. Astrovirus diarrhea in Egyptian children. J. Infect. Dis. 182:685-690. [DOI] [PubMed] [Google Scholar]
- 42.Nicholas, K. B., H. B. J. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualisation of genetic variation. EMBNEW News 4:14. [Google Scholar]
- 43.Nieselt-Struwe, K., and A. von Haeseler. 2001. Quartet-mapping, a generalization of the likelihood-mapping procedure. Mol. Biol. Evol. 18:1204-1219. [DOI] [PubMed] [Google Scholar]
- 44.Noel, J., and D. Cubitt. 1994. Identification of astrovirus serotypes from children treated at the Hospitals for Sick Children. Epidemiol. Infect. 113:153-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noel, J. S., T. W. Lee, J. B. Kurtz, R. I. Glass, and S. S. Monroe. 1995. Typing of human astroviruses from clinical isolates by enzyme immunoassay and nucleotide sequencing. J. Clin. Microbiol. 33:797-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh, D., and E. Schreier. 2001. Molecular characterization of human astroviruses in Germany. Arch. Virol. 146:443-455. [DOI] [PubMed] [Google Scholar]
- 47.Oishi, I., K. Yamazaki, T. Kimoto, Y. Minekawa, E. Utagawa, S. Yamazaki, S. Inouye, G. S. Grohmann, S. S. Monroe, S. E. Stine, C. Carcamo, T. Ando, and R. I. Glass. 1994. A large outbreak of acute gastroenteritis associated with astrovirus among students and teachers in Osaka, Japan. J. Infect. Dis. 170:439-443. [DOI] [PubMed] [Google Scholar]
- 48.Page, R. 1996. TREEVIEW: an application to display phylogenetic trees on personal computers. Comput. Applic. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 49.Palombo, E. A., and R. F. Bishop. 1996. Annual incidence, serotype distribution, and genetic diversity of human astrovirus isolates from hospitalized children in Melbourne, Australia. J. Clin. Microbiol. 34:1750-1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pina, S., M. Buti, R. Jardí, P. Clemente-Casares, J. Jofre, and R. Girones. 2001. Genetic analysis of hepatitis A virus strains recovered from the environment and from patients with acute hepatitis. J. Gen. Virol. 82:2955-2963. [DOI] [PubMed] [Google Scholar]
- 51.Pintó, R. M., F. X. Abad, R. Gajardo, and A. Bosch. 1996. Detection of infectious astroviruses in water. Appl. Environ. Microbiol. 62:1811-1813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pintó, R. M., C. Villena, F. le Guyader, S. Guix, S. Calallero, M. Pommepuy, and A. Bosch. 2001. Astrovirus detection in wastewater. Water Sci. Technol. 43:73-77. [PubMed] [Google Scholar]
- 53.Reed, C., G. D. Sturbaum, P. J. Hoover, and C. R. Sterling. 2002. Cryptosporidium parvum mixed genotypes detected by PCR-restriction fragment length polymorphism analysis. Appl. Environ. Microbiol. 68:427-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Saito, K., H. Ushijima, O. Nishio, M. Oseto, H. Motohiro, Y. Ueda, M. Takagi, S. Nakaya, T. Ando, R. Glass, and K. Zaiman. 1995. Detection of astroviruses from stool samples in Japan using reverse transcription and polymerase chain reaction amplification. Microbiol. Immunol. 39:825-828. [DOI] [PubMed] [Google Scholar]
- 55.Sakamoto, T., H. Negishi, Q. H. Wang, S. Akihara, B. Kim, H. Nishimura, K. Kaneshi, Y. Nak, K. Sugita, T. Motohiro, T. Nishimura, and H. Ushijima. 2000. Molecular epidemiology of astroviruses in Japan from 1995 to 1998 by reverse transcription-polymerase chain reaction with serotype specific primers (1-8). J. Med. Virol. 61:326-331. [PubMed] [Google Scholar]
- 56.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Steele, A. D., H. R. Basetse, N. R. Blacklow, and J. E. Herrmann. 1998. Astrovirus infection in South Africa: a pilot study. Ann. Trop. Paediatr. 18:315-319. [DOI] [PubMed] [Google Scholar]
- 58.Strimmer, K., and A. von Haeseler. 1997. Likelihood-mapping: a simple method to visualize phylogenetic content of sequence alignment. Proc. Natl. Acad. Sci. USA 94:6815-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Taylor, M. B., W. O. K. Grabow, and W. D. Cubitt. 1997. Propagation of human astroviruses in the PLC/PRF/5 hepatoma cell line. J. Virol. Methods 67:13-18. [DOI] [PubMed] [Google Scholar]
- 60.Taylor, M. B., N. Cox, M. A. Vrey, and W. O. K. Grabow. 2001. The occurrence of hepatitis A and astroviruses in selected river and dam waters in South Africa. Water Res. 35:2653-2660. [DOI] [PubMed] [Google Scholar]
- 61.Taylor, M. B., J. Walter, T. Berke, W. D. Cubitt, D. K. Mitchell, and D. O. Matson. 2001. Characterisation of a South African human astrovirus as type 8 by antigenic and genetic analyses. J. Med. Virol. 64:256-261. [DOI] [PubMed] [Google Scholar]
- 62.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Walter, J. E., and D. K. Mitchell. 2000. Role of astroviruses in childhood diarrhea. Curr. Opin. Pediatr. 12:275-279. [DOI] [PubMed] [Google Scholar]
- 64.Walter, J. E., D. K. Mitchell, M. L. Guerrero, T. Berke, D. O. Matson, S. S. Monroe, L. K. Pickering, and G. Ruiz-Palacios. 2001. Molecular epidemiology of human astrovirus diarrhea among children from a periurban community of Mexico City. J. Infect. Dis. 183:681-686. [DOI] [PubMed] [Google Scholar]
- 65.Wang, Q.-H., J. Kakizawa, L.-Y. Wen, M. Shimizu, O. Nishio, Z.-Y. Fang, and H. Ushijima. 2001. Genetic analysis of the capsid region of astroviruses. J. Med. Virol. 64:245-255. [DOI] [PubMed] [Google Scholar]
- 66.Wolfaardt, M., C. L. Moe, and W. O. K. Grabow. 1995. Detection of small round structured viruses by enzymatic amplification. Water Sci. Technol. 31:375-382. [Google Scholar]