Abstract
We have determined the NMR solution structures of the quadruplexes formed by d(TGLGLT) and d(TL4T), where L denotes LNA (locked nucleic acid) modified G-residues. Both structures are tetrameric, parallel and right-handed and the native global fold of the corresponding DNA quadruplex is retained upon introduction of the LNA nucleotides. However, local structural alterations are observed owing to the locked LNA sugars. In particular, a distinct change in the sugar–phosphate backbone is observed at the G2pL3 and L2pL3 base steps and sequence dependent changes in the twist between tetrads are also seen. Both the LNA modified quadruplexes have raised thermostability as compared to the DNA quadruplex. The quadruplex-forming capability of d(TGLGLT) is of particular interest as it expands the design flexibility for stable parallel LNA quadruplexes and shows that LNA nucleotides can be mixed with DNA or other modified nucleic acids. As such, LNA-based quadruplexes can be decorated by a variety of chemical modifications. Such LNA quadruplex scaffolds might find applications in the developing field of nanobiotechnology.
INTRODUCTION
Telomeric DNA is found at the ends of the chromosomes and contain continuous repeats of guanine (G)-rich sequences that possess the ability to form G-quadruplex structures by forming stacked guanine tetrads with cyclic Hoogsteen hydrogen bonding (Figure 1) (1). These non-coding DNA repeats are of kilobase length and protect the ends of the chromosomes from nuclease degradation, recombination and end-to-end fusion (2,3). Whether G-quadruplex structure is formed in vivo is a matter of debate but an indirect proof for the, at least transient, existence of in vivo quadruplex structures is the identification of proteins that recognize this structural motif (4,5).
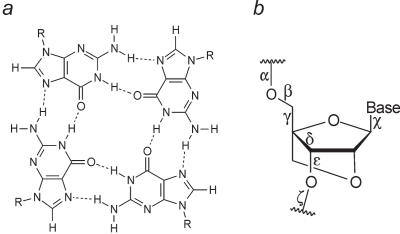
Figure 1.
(a) The base pairing in a Hoogsteen-paired G-tetrad. (b) The chemical structure of LNA with torsion angles labelled.
Quadruplex structures may be the target for therapeutic strategies. Telomerase is an enzyme which partakes in the maintenance process of the telomeres by recognition of the single-stranded or double-stranded form of the telomeric DNA. Recently, the quadruplex structures present at the ends of the telomeres have been stabilized by small molecule drugs thereby destabilizing telomere maintenance in tumour cells owing to less efficient telomerase binding (6,7). In addition, certain quadruplex molecules possess therapeutic potential themselves. Several folded quadruplexes can target nucleolin, a nucleolar protein involved in multiple aspects of cell growth, thereby inhibiting cell proliferation in some human tumour cell lines (8). A tetrameric quadruplex is able to target HIV integrase, (9) the enzyme responsible for the insertion of viral DNA into the host genome, and the HIV envelope-mediated cell fusion is inhibited by the parallel tetrameric quadruplex formed by T2G4T2 (10). This quadruplex drug is modified with phosphorothioate nucleotide linkages, which shows that the use of nucleic acid analogues may be useful in drug therapy with quadruplex molecules.
G-quadruplex structures have been studied intensively by X-ray crystallography and NMR spectroscopy (11,12). These studies show that G-quadruplexes are able to fold in different motifs, with the four strands being either parallel or anti-parallel with respect to each other. In the parallel conformation, guanosines have anti glycosidic angles as observed for the quadruplex formed by d(TG4T) (13,14). In the anti-parallel folding motif, the glycosidic angles for the guanosines are alternating between anti and syn conformations. An anti-parallel folding motif is often found when several guanine stretches are embedded in the same oligonucleotides as observed for the dimeric quadruplex formed by d(G4T4G4), (15,16) although parallel folds have been observed as well, e.g. for the quadruplex structure from the human telomeric DNA (17). The only observed anti-parallel four-stranded DNA quadruplexes containing only a single guanine stretch are those derived from d(TAG2) and d(T2AG2). In these structures, the tetrads are stabilized by capping TAA triads (18).
LNA (locked nucleic acid) is a ribonuclotide analogue that contains a 2′-O,4′-C methylene bridge.(19) Owing to this bridge, LNA is locked in a C3′-endo (N-type) sugar conformation, with a large puckering amplitude of ∼60° (Figure 1) (20). LNA nucleotides substantially increase the thermostability when incorporated into both DNA duplexes and triplexes and LNA has the potential for applications in drug design owing to its beneficial properties, such as non-toxicity, biostability and ease of synthesis. A further promising application is in the developing field of nanobiotechnology, where LNA, and functionalized LNA analogues, may find applications (21).
Recently, it was demonstrated that LNA modifications stabilize the tetrameric quadruplex of TG3T when it is fully modified (22). Also fully modified RNA and 2′-O-methyl-RNA quadruplexes are more stable than their unmodified d(TG4T) counterpart (23). Thus it appears that nucleotides with N-type sugar puckers stabilize parallel quadruplexes. For use both in therapeutics and nanobiotechnology, it may be useful to mix LNA nucleotides with either DNA or other nucleic acid analogues to tune pharmacokinetics or physicochemical properties. Thus we decided to investigate a quadruplex-forming sequence consisting of alternating DNA and LNA nucleotides.
In this study we have studied two sequences derived from the Oxytricha nova telomeric sequence, d(TG4T), modified with LNA. One with two guanines substituted at G3 and G5, TGLGLT and the other with all guanine nucleotides substituted with LNA guanine nucleotides, TL4T, (i.e. we denote an LNA guanine nucleotide by ‘L’). With the design of these oligonucleotides, we can study both the overall and the local structural implications of incorporation of N-type LNA nucleotides.
We determined the structures of the LNA modified quadruplexes using NMR spectroscopy and NOE-derived distance bounds in a simulated annealing (SA) scheme. In our analysis, we use the known NMR and crystal structures of the unmodified quadruplex as references. Although the global structures of the modified quadruplexes resembles the unmodified quadruplexes, both being parallel tetrameric quadruplexes, local structural alterations are observed owing to the locked LNA sugars. In particular, a distinct change in the sugar–phosphate backbone is observed at the G2pL3 and L2pL3 base steps and sequence dependent changes in the twist between tetrads are also seen.
MATERIALS AND METHODS
NMR sample preparation
d(TGLGLT) and d(TL4T) oligonucleotides were synthesized using standard phosphoramidite chemistry and purified by ultracentrifugation. The quadruplexes were annealed by heating the samples at 80°C followed by cooling to room temperature overnight. The buffer used consisted of 0.1 M KCl at pH 6. For experiments carried out in D2O, the solid quadruplexes were lyophilized three times from D2O and redissolved in 99.96% D2O (Cambridge Isotope Laboratories). A mixture of 90% H2O and 10% D2O was used for experiments examining exchangeable protons. The final concentrations were 0.75 and 0.25 mM for TGLGLT and TL4T, respectively in 500 µl.
Ultraviolet (UV) melting curves
The thermal stability of d(TGLGLT), d(TL4T) and d(TG4T) was determined spectrophotometrically with a PerkinElmer Lambda 35 spectrophotometer equipped with a thermoregulated Peltier element. The samples were prepared by dissolving 23 µM of each oligonucleotide in 1 ml containing 1 mM KCl (pH 7.7) and subsequently, the quadruplexes were annealed by heating to 90°C followed by slow cooling to 25°C. The melting profiles were recorded at 295 nm from 25 to 90°C at a heating and cooling rate of 0.1°C/min and the melting temperatures (Tm values) were obtained as the maxima of the first derivative of the melting curves.
NMR experiments
NMR experiments were performed Varian INOVA spectrometers operating at either 500 or 800 MHz at 25°C. The spectrometer operating at 800 MHz was equipped with a 3 mm HCN-triple resonance probe and for experiments at this field strength, the oligonucleotide concentrations were 3.0 and 1.0 mM, respectively.
For the structure determination, NOESY spectra in D2O were acquired at 800 MHz with mixing times of 60, 120, 200 and 300 ms and a NOESY spectrum in H2O was acquired at 5°C at 500 MHz with a mixing time of 250 ms for TGLGLT. The spectra in D2O were acquired using 4096 complex points in t2 and a spectral width of 6000 Hz. A total of 465 t1-experiments, each with 32 scans and a dwell-time of 3.141 s between scans, were recorded using the States phase cycling scheme. The residual signal from HOD was removed by low-power presaturation. The spectrum in H2O was acquired with the WATERGATE NOESY pulse sequence using 4096 complex points in t2 and a spectral width of 6600 Hz. A total of 600 t1-experiments, each with 32 scans and a dwell-time of 2.99 s between scans, were recorded using the States phase cycling scheme. For TL4T, two NOESY spectra in D2O were acquired at 800 MHz with mixing times of 100 and 300 ms and a NOESY spectrum in H2O was acquired at 5°C at 500 MHz with a mixing time of 250 ms. The spectra in D2O were acquired using 4096 complex points in t2 and a spectral width of 6000 Hz. A total of 322 t1-experiments, each with 80 scans and a dwell-time of 2.244 s between scans were recorded. The spectrum in H2O was acquired using 4096 complex points in t2 and a spectral width of 7500 Hz. A total of 345 t1-experiments, each with 96 scans and a dwell-time of 2.173 s between scans, were recorded using the States phase cycling scheme. In addition to the NOESY spectra, inversion recovery experiments were recorded at 800 MHz to extract T1 relaxation rates for both quadruplexes.
For assignment purposes and to estimate scalar coupling constants, DQF-COSY, TOCSY and 1H-31P HSQC experiments were acquired. The acquired data were processed using FELIX (version 97.2, MSI, San Diego, CA). All NOESY spectra were apodized by skewed sinebell squared functions in F1 and F2, linear predicted to double the number of points in F1, baseline corrected by the FLATT procedure in F1, (24) and by the automatic baseline correction procedure as implemented in FELIX in F2. Full experimental details are included in Supplementary Table S3.
Structure calculations
NOESY cross peak intensities were measured in the NOESY spectra for TGLGLT and TL4T using the Gaussian fit procedure in Sparky (25). The intensities were corrected for saturation effects using T1 relaxation times obtained from inversion recovery experiments and were subsequently transformed to distance restraints by calibrating against known distances using the method of Wijmenga et al. (26). In this variation of the isolated spin-pair approximation, spin diffusion is accounted for in an average manner. The final upper and lower distance bounds used in the structure determination were set to ±15% for restraints derived for the non-exchangeable protons and ±20% for the exchangeable protons.
In the analysis, NOEs were interpreted as intra-strand or inter-strand restraints by comparison of a theoretically calculated generalized volume contribution (GVC). This GVC was calculated using home written software based on the method of Güntert and co-workers (27). Calculation of the GVC requires a structure model and an iterative structure determination approach was adopted. For the first iteration, the structure model was a suitably modified copy of the structure of r(UG4U), in successive iterations models from the previous iteration were used. By relaxing the demand for the fraction between the GVC for inter- and intra-strand restraints (and for any multiple restraints in general) being close to unity, successively more and more NOE distance restraints could be included in the structure calculations. In the final iteration, totals of 175 and 104 NOE distance restraints were included in the structure calculations for TGLGLT and TL4T, respectively. The distributions of NOE restraints are included in Table 1. The average widths of the distance restraints were 1.44 and 1.24 Å and the average lengths 4.47 and 3.74 Å for TGLGLT and TL4T, respectively. All distance restraints were incorporated into the AMBER potential energy by a standard flat-well pseudo potential.
Table 1.
Number and distribution of restraints in calculations and structure statistics of the 20 structures calculateda
| TGLGLT | TL4T | |
|---|---|---|
| Distance restraints (per strand) | ||
| Intra-residue NOE | 95 | 63 |
| Inter-residue (intra-strand) NOE | 63 | 34 |
| Inter-strand NOE | 17 | 7 |
| Subtotal | 175 | 104 |
| Hydrogen bonding | 8 | 8 |
| Planarity | 8 | 8 |
| Total number of restraints (per strand) | 191 | 120 |
| Violations of experimental restraints | ||
| Mean number of NOE violations > 0.2 Å | 2.1 | 4.9 |
| Mean NOE violation (Å) | 0.022 | 0.029 |
| Maximum NOE violation (Å) | 0.24 | 0.46 |
| Mean violation of excluded restraintsb | 0.11 | 0.09 |
| Atomic r.m.s. deviations | ||
| Overallc | 0.11 ± 0.05 | 0.57 ± 0.16 |
| Cross validationd | 0.59 ± 0.16 | 1.09 ± 0.26 |
| Referencee | 0.44 ± 0.16 | 1.02 ± 0.26 |
aRestraints are deposited with the structures in the pdb databank.
bThe mean violation of the restraints excluded in the cross validation procedure (only non-terminal nucleotides included).
cAverage pair wise atomic r.m.s.d. for the 20 structures calculated using all restraints.
dFor cross validation, 20 new structures were calculated with 10% randomly removed NOE distance restraints for each one. The ‘r.m.s.d. of cross validation’ is the average pair wise atomic r.m.s.d. for these structures.
eThe ‘reference r.m.s.d.’ is the r.m.s.d. between the ensemble used for cross validation and a reference structure from the calculations where all restraints were included.
In both structure calculations, Hoogsteen base pairing restraints were included with target values taken from crystallographic data, (28) and loose planarity restraints on tetrads were included as well.
An SA protocol was utilized to obtain the structures of the quadruplexes. Initially the starting structure was restrained energy minimized before being subjected to 200 ps of molecular dynamics in time-steps of 1 fs: 20 ps of warming from 300 to 2000 K, 60 ps at 2000 K and finally cooling from 2000 to 300 K over 120 ps. Finally, a further restrained energy minimization was carried out. The force constants employed were 30 kcal/(mol Å2) for NOE restraints, 40 kcal/(mol Å2) for base pairing restraints and 20 kcal/(mol rad2) for base planarity restraints. During the SA scheme, the restraint weights were gradually raised from 0.5 to 2, kept at this weight and finally slowly decreased to 1 again. A distance dependent dielectric constant, ɛ = 4r, was used and the non-bonded cut-off was 16 Å in calculations.
Protein data bank accession codes
Coordinates and restraints employed in calculations have been deposited in the Protein Data Bank (accession codes TGLGLT: 2chj and TL4T: 2chk).
RESULTS
Thermal stability of TGLGLT and TL4T
We performed UV denaturation studies for TGLGLT, TL4T and TG4T for comparison. For TG4T the absorbance at 295 nm decreases from 0.36 to 0.28 as the temperature is raised from 25 to 90°C. This indicates a melting transition for the unmodified quadruplex with the melting temperature being 82°C. On the contrary, for the two modified quadruplexes, the absorbance remained constant during the entire temperature range sampled. This may indicate that these quadruplexes have melting temperatures above 90°C. As not all quadruplexes show a UV transition upon melting, we used NMR spectroscopy to verify the UV melting data (29). Samples with the same concentration as used for the UV melting studies (23 µM) were heated for 2 h at 80°C and subsequently 1D 1H-NMR spectra were acquired at 80°C. No broadening of imino resonances was observed for the TGLGLT and TL4T samples, indicating that no melting of the quadruplexes is initiated at this concentration at 80°C. For the TG4T sample, the imino resonances were significantly broadened, indicating that this quadruplex is starting to melt at 80°C.
Spectral analysis: assignment
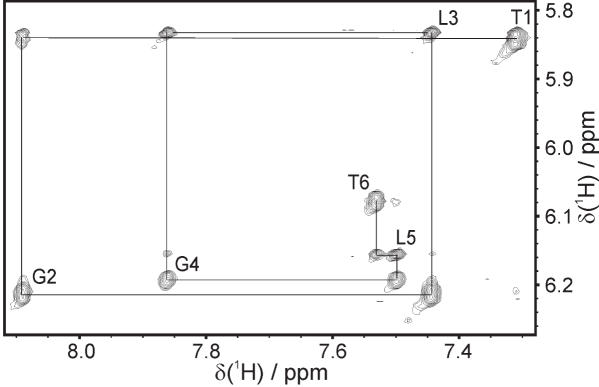
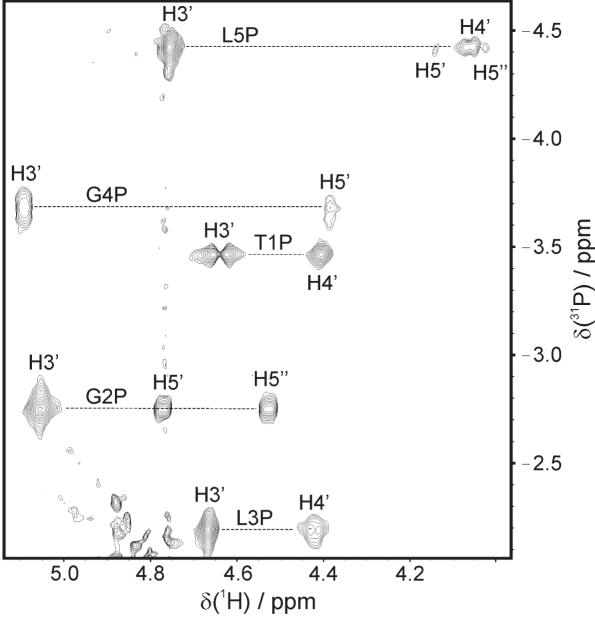
The 1H-NMR spectra recorded in 90:10% H2O/D2O of both TGLGLT and TL4T show four sharp resonances in the region around 11 p.p.m. (Supplementary Figure S1). This is indicative of Hoogsteen base paired guanines and quadruplex formation (11). The 1H-NMR spectra recorded in D2O show six signals from the six aromatic protons. Thus both molecules have six spectroscopic distinct residues and hence possess 4-fold symmetry. The 2D NOESY spectra show NOE contacts between H1 and H8 protons located on neighbouring strands but at the same end of the molecule, i.e. G(i)/L(i):H1 has an NOE with the symmetry-related G(i)/L(i):H8 (Supplementary Figure S1). This proves the formation of tetrads and shows that both molecules are parallel-stranded, as an anti-parallel strand orientation would give NOEs between protons located in opposite ends of the strands. All intra-residue aromatic-H1′ NOEs are weak so all nucleobases have anti glycosidic conformations and the inter-residue aromatic-H1′ connectivities are characteristic of a right-handed helix conformation (Figure 2). Accordingly, resonances could be assigned using standard methods (30,31). The sugar protons were assigned using COSY and TOCSY spectra except for the LNA nucleotides which have either small scalar coupling constants for the vicinal proton pairs or isolated spin systems. Consequently, the distance information from the NOESY spectra was used to assign the sugar protons in LNA nucleotides. For the TGLGLT molecule, the sample was more concentrated and good 31P-1H HSQC spectra could be obtained. Thus, the assignments for this molecule could be confirmed unambiguously through bond via H3′(i)-P(i)-H4′/H5′/H5′′(i + 1) connectivities (Figure 3). The chemical shifts of the two molecules are given in Supplementary Tables S1 and S2.
Figure 2.
The aromatic-H1′ region of a 300 ms NOESY spectrum of TGLGLT. The sequential assignment is shown and intra-nucleotide cross peaks are labelled.
Figure 3.
31P decoupled 1H-31P HSQC spectrum of TGLGLT. Inter-nucleotide P-H3′ and intra-nucleotide P-H4′/H5′/H5′′ cross peaks are indicated. The resonance of L3P is folded from −5.16 p.p.m.
In addition to the four signals from the guanine imino protons, the spectra of the modified oligonucleotides show two slightly broader lines upfield relative to the guanine imino protons. For TL4T these lines are only observed at pH 6 and 5°C and for TGLGLT only at either pH 7 and 5°C or pH 6 and 25°C. These lines are from the imino protons of the terminal thymines, which consequently are protected from exchange with the solvent through hydrogen bonding. The terminal thymine bases stack, at least transiently, on their neighbouring G-tetrads as judged from NOE contacts to guanine imino protons. These NOE contacts were employed for specific assignment of the thymine imino protons.
Spectral analysis: sugar conformations
Deoxyribose sugars have the possibility of repuckering between N- and S-type conformations in solution, and the populations of each sugar pucker can be determined by analysis of intra-residue sugar proton cross peaks in COSY spectra (32). However, extensive resonance overlap precluded quantitative determination of sugar pucker parameters due to the limited number of well resolved cross peaks. The absence of an H3′–H4′ COSY cross peak for G2 in the mixed quadruplex, TGLGLT, indicates a small coupling constant for this proton pair and hence an almost pure S-type conformation for G2. On the contrary, G4 displays an intense H3′–H4′ COSY cross peak (3JH3′H4′ ≈ 10 Hz) and consequently possesses an N-type sugar pucker. The sugar puckers thus indiated for G2 and G4 were corroborated by the intensities of the NOE cross peaks between H3′ and H8 of each nucleotide. T1 is predominantly S-type as gauged by the weak H2′′–H3′ and H3′–H4′ COSY cross peaks, whereas T6 has more equal populations of S- and N-type sugars as indicated by the presence of both H1′–H2′ and H2′′–H3′ COSY cross peaks.
Spectral analysis: backbone torsion angles
We gauged semi quantitative coupling constants between phosphorus and H3′, H4′, H5′ and H5′′ sugar protons from 31P-1H HSQC spectra. In LNA nucleotides, no H4′ proton is present which spectroscopically simplifies 31P-1H spectra as passive 1H-1H couplings are reduced in number for H3′, H5′ and H5′′. For TL4T, the low sample concentration made it difficult to obtain 31P-1H correlation spectra with a decent signal-to-noise ratio and only 31P(i)-H3′(i) correlations (of similar intensity) were observed for the four guanines.
In the undecoupled 1H-31P HSQC spectrum of TGLGLT, we estimated 3JH3′P ≈ 4 Hz for L5 (Supplementary Figure S2), this value corresponds to an ɛ angle in the trans domain or in the −gauche domain according to the Karplus relationship (26). As all H3′-P cross peaks in the decoupled HSQC spectrum are of comparable intensity, ɛ values are estimated in the (high) trans or −gauche domains for all the nucleotides. Except for L3, all cross peaks between H5′ or H5′′ and P were very weak or absent in the decoupled HSQC spectrum, corresponding to small couplings (3JH5′-P < 3 Hz and 3JH5′′-P < 3 Hz). This is only possible with β in the trans domain. For the G2pL3 step, we found 3JH5′-P ≈ 7 Hz and 3JH5′′-P ≈ 10 Hz. Applying the Karplus relationship, this gives β ≈ 100° or −100° for L3. For G2, G4 and T6, cross peaks between H4′ and the preceding phosphorus atoms are observed (LNA nucleotides have no H4′). Non-vanishing JH4′-P coupling constants imply (β, γ) = (trans, +gauche) as the H4′-P fragment then assumes a W-conformation. A +gauche conformation of the γ angle is also consistent with the weak H4′–H5′ and H4′–H5′′ COSY cross peaks observed.
Structure calculations
Ensembles of 20 structures were calculated for the each of the modified quadruplexes, TL4T and TGLGLT, following the protocol described in the Materials and Methods section. Structural statistics for the two ensembles are shown in Table 1. Both ensembles have low r.m.s.d. values showing that the NMR restraints define the structures with good precision. Because of favourable spectral dispersion and higher sample concentration, more NOE restraints were determined for the TGLGLT structure. Consequently, the precision of this structure is higher than that of the TL4T structure with particularly the geometry of the sugar–phosphate backbone more well-defined in the mixed quadruplex. Even though the number of restraints is higher for the mixed quadruplex, there are fewer and smaller restraint violations for this structure. Each of the structures was cross validated by performing 20 calculations for each where 10% of the NOE restraints were randomly removed in every calculation (Table 1). Successful cross validation shows that the structures are not dependent on just few NOE restraints. The r.m.s.d. values from the cross validation probably give the most realistic measure of the precision of the structures (i.e. 0.59 Å for TGLGLT and 1.09 Å for TL4T).
In the cross validation procedure, we back-calculated the distances for the NOE restraints which were left out of the calculations. The average violation for these restraints is ≈0.1 Å for each structure. In addition, the backbone conformations in the structures are in agreement with the data derived from 1H-31P HSQC and COSY spectra even though no explicit restraints were included in the calculations to impose these data (see below). These points show that besides being precise, the structures are also accurate.
Description of the structures
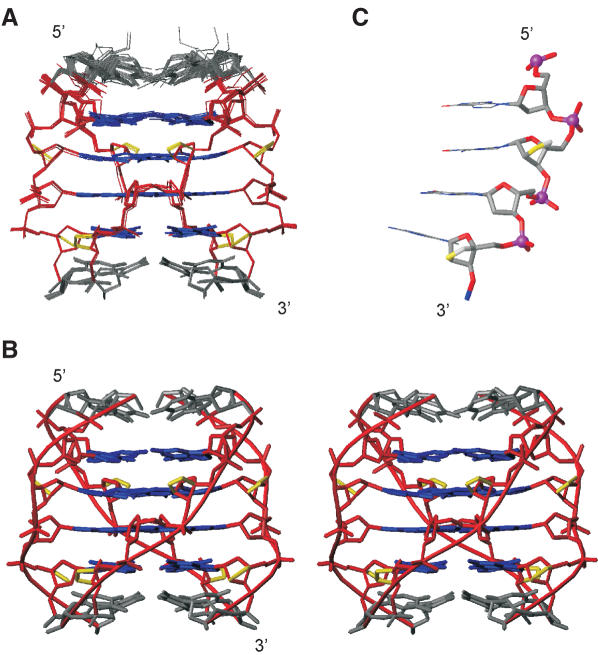
Both TL4T and TGLGLT form parallel right-handed quadruplexes with 4-fold symmetry and planar tetrads perpendicular to the helix axis. The four grooves are wide, and the oxymethylene bridges of the LNA nucleotides are located between neighbouring strands in the centre of the grooves. Overall, the two structures are similar albeit with local differences, namely in the sugar–phosphate backbone geometry and the geometry of the terminal thymines (see below). The TGLGLT structure is shown in Figure 4 and the TL4T structure in Supplementary Figure S3.
Figure 4.
(A) Overlay of the 20 structures calculated for the TGLGLT quadruplex. The LNA oxymethylene bridges are shown in yellow. The sugar–phosphate backbone, the guanines and the thymines are coloured red, blue and grey, respectively. For clarity, hydrogen atoms are not shown. (B) A stereo view of a representative TGLGLT structure. (C) View of the GLGL part of the TGLGLT quadruplex highlighting the sugar–phosphate backbone geometry. Atoms are coloured according to element type, carbon: grey, oxygen: red, LNA oxygen: yellow, nitrogen: blue and phosphorus: magenta.
The helix parameters were calculated for the two ensembles with Curves 5.1 (33,34). As the molecule is symmetric and the geometry of the G-quartets programmed by the planar hydrogen bond arrangement, the pertinent helix parameters are rise and twist (Table 2). For the three tetrad steps largely similar mean values were obtained for the two structures (rise ≈ 3.3 Å and twist ≈ 23–25°). Locally, the variation of the twist is pronounced in the TGLGLT quadruplex, with a value of 31° for the LpG step and values of ≈21° for the two GpL steps. The grooves of the LNA modified quadruplexes are similar (6.8 and 7.1 Å for TGLGLT and TL4T, respectively) and in both cases the grooves are narrowing towards the 3′ end (Table 2).
Table 2.
Sugar conformations, groove widths and selected helix parameters for the LNA quadruplexesa
| Nucleotide | TGLGLT | TL4T | ||||
|---|---|---|---|---|---|---|
| number | Pseudorotation angle (°) | Sugar pucker | Puckering amplitude (°) | Pseudorotation angle (°) | Sugar pucker | Puckering amplitude (°) |
| 1 | 143 | S-type | 39 | 148 | S-type | 40 |
| 2 | 156 | S-type | 43 | 20 | N-type | 58 |
| 3 | 23 | N-type | 62 | 28 | N-type | 59 |
| 4 | 25 | N-type | 23 | 15 | N-type | 58 |
| 5 | 23 | N-type | 59 | 5 | N-type | 56 |
| 6 | 64 | 28 | 148, −123b | 21 | ||
| Nucleotide step | Rise (Å) | Twist (°) | Groove width (Å) | Rise (Å) | Twist (°) | Groove width (Å) |
|---|---|---|---|---|---|---|
| 2–3 | 3.7 | 21 | 7.0 | 3.5 | 22 | 8.4 |
| 3–4 | 3.2 | 31 | 7.0 | 3.5 | 23 | 6.4 |
| 4–5 | 3.0 | 22 | 6.4 | 3.2 | 28 | 6.4 |
| Mean | 3.3 | 25 | 6.8 | 3.4 | 24 | 7.1 |
aThe groove width is measured as the shortest inter-strand phosphorus distances minus 5.8 Å, which is the combined Van der Waals radii of two phosphate groups.
bTwo different conformations are observed.
There are some differences in the conformations of thymines at the ends of the two LNA quadruplexes. In the mixed quadruplex, a T-tetrad is formed at the 5′-terminus (Supplementary Figure S4). At the 3′-terminus, the thymine H3 proton hydrogen bonds to N7 of a guanine in the fourth G-tetrad (Supplementary Figure S4). In TL4T, both unstructured thymines and T:T base pairs are observed in different structures in the ensemble.
The sugar puckers were measured for both structures (Table 2). In agreement with the observations from COSY spectra, G2 has an S-type pucker whereas G4 possesses an N-type pucker in TGLGLT. In addition, T1 is in the expected S-type conformation and T6 has a pseudorotation angle in-between the values of S- and N-type conformations, therefore indicating a pseudorotational equilibrium. Thus all residues are in accordance with the spectral data even though no explicit sugar pucker restraints were included in the calculations.
Next, the backbone torsion angles were analysed for the two structural ensembles (Table 3). All glycosidic angles are in anti conformations, except for G2 in TGLGLT, which is in the far syn range (χ = −82°). The most frequent conformation found for the guanines in both structures is (α, γ) = (−gauche, +gauche), β = trans and (ɛ, ζ) = (trans, −gauche), these values follow the standard genus for RNA and DNA duplexes (28). The observed conformation for ɛ, ζ, α, β is the BI conformation, which is the lowest energy conformation in a DNA nucleotide segment (35). In the mixed quadruplex, a distinctly different backbone geometry is observed in the G2pL3 step. For G2, (ɛ, ζ) = (−86°, 171°) and for L3 (α, γ) = (53°, −168°) and β = −114° (Figure 4). As noted above, the backbone geometry of the fully modified quadruplex, TL4T is rather poorly determined with our NMR data. However, some structural heterogeneity is observed in the L2pL3 step for this quadruplex as well (Table 3).
Table 3.
Backbone torsion angle ranges and glycosidic angle ranges for the LNA quadruplexesa
| Base step | δ | ɛ | ζ | α | β | γ | χ |
|---|---|---|---|---|---|---|---|
| TGLGLT | |||||||
| G2pL3 | t | g− | t | g+ | g− | t | −82° |
| Other steps | g+ | t | g− | g− | t | g+ | anti |
| TL4T | |||||||
| T1pL2 | t | t (g+) | g− (g+) | t (g+) | t | g+ (t) | anti |
| L2pL3 | g+ | t | g− | g+ (g−) | g+ (t) | t | anti |
| L3pL4 | g+ | t | g− | g− | t | g+ | anti |
| L4pL5 | g+ | t | g− | g− | t | g+ | anti |
| L5pT6 | t | g− (t) | g+ (g−) | g− (g+) | t (g+) | g− (g+) | anti |
aIf more than one rotamer is populated in the structure ensembles, the minor rotamers are included in brackets.
The backbone conformations identified in the two structures are in good agreement with the experimental NMR data. It is particularly gratifying that the atypical G2pL3 conformation is validated as no explicit backbone restraints were included in the structure calculations. In this step β = −114°, which fits perfectly with the observation of large JH5′P and JH5′′P coupling constants (see above).
DISCUSSION
Stability of TGLGLT and TL4T
It is known that LNA modifications stabilize duplex and triplex DNA (19). In a previous report, a fully modified LNA quadruplex, d(TG3T), was stabilized relative to the DNA quadruplex by 20°C (22). Here, we show that partly modified LNA strands can stabilize a parallel quadruplex as well, i.e. d(TGLGLT) is stabilized relative to the all DNA quadruplex. This opens for design flexibility and for the prospect of combining LNA nucleotides with other modified nucleic acids in functionalized stable quadruplex scaffolds with possible applications in nanobiotechnology.
Design of mixed LNA/DNA quadruplex
In anti-parallel quadruplex topologies, the glycosidic angles of the guanosines alternate between syn and anti conformations (15,16). In LNA guanosines, the syn conformation is prohibited due to a steric clash between H3′ and N3. We designed the mixed DNA/LNA quadruplex sequence, d(TGLGLT), so that the alternating pattern of glycosidic angles and thus an anti-parallel fold would be possible. However, the NMR data collectively show that the mixed quadruplex has a parallel fold identical to the unmodified and fully LNA modified quadruplexes. One possible manner of forcing an anti-parallel fold in a tetrameric quadruplex would be to combine anti LNA nucleotides with guanosine analogues forced to adopt a syn conformation, e.g. by introduction of bulky groups at the C8 position of the nucleobase (36).
Overall appearance of the structures
Overall the appearances of our LNA modified quadruplex structures are similar to each other and to the corresponding unmodified DNA and RNA quadruplexes (14,31,37). Thus, LNA nucleotides can be incorporated in parallel quadruplexes without major distortions of the structure.
If one assumes that the tetrad geometry is prescribed, rise and twist are the two helix parameters pertinent for description of the global structure of a quadruplex. In a symmetric parallel quadruplex with the tetrads perpendicular to the helix axis, rise matches the distance between tetrads. The common value for the LNA and DNA quadruplexes, ≈3.3 Å, agrees with the sum of the Van der Waals radii for atoms in the tetrads (Table 2). Accordingly, incorporation of LNA nucleotides does not stretch the quadruplex architecture and preserves the native length of the molecule. The molecule can then adjust the twist between tetrads so as to optimize stacking interactions and coordination of metal ions. This is done by fine tuning the backbone torsion angles to obtain the desired geometry. The LNA quadruplexes studied are slightly unwound compared to the DNA quadruplex as judged by average twist values of 25° and 24° for TGLGLT and TL4T, respectively. The twist for the DNA quadruplex is 30° and 29° in NMR and X-ray structures, respectively (Table 2) (14,31). Notably, in the previous report of the TLL3TL structure, the average twist is 28.5° (22). This change in twist is associated with variations in the imino chemical shifts of the central nucleotides, which is 11.48 p.p.m. in TLL3TL and 11.15 and 11.18 p.p.m. for the two central nucleotides in TL4T. The change in twist between the L3 and L4 quadruplexes could be a reflection upon that in the L3 structure there would be room for only two cations in the quadruplex core whereas there is room for three ions in the L4 structure. In the TGLGLT structure there is a large difference in the twist between TpL and LpT tetrads (Table 2 and Supplementary Figure S5). This difference could again be a consequence of cation binding in the quadruplex core or because of the restraints imposed in the sugar–phosphate backbone by the locked nucleotides which will restrain the molecule's freedom to optimize tetrad stacking.
The unwinding of the LNA quadruplexes causes a great widening of their grooves as compared to their DNA counterpart (≈7 Å as compared to 2.5 Å) (Table 2). The wider grooves in the LNA quadruplexes lead to a larger distance between the negatively charged phosphate groups in neighbouring strands. This may contribute to the enhanced thermostability of the LNA quadruplexes. In the crystal structure of d(TG4T), a spine of hydration is observed in each of the grooves (14). In the LNA quadruplexes, the O2′ oxygen of the LNA nucleotides provide an additional anchoring point for hydration, however, with the much wider grooves, the hydration has to be altered as compared to the DNA quadruplex. An alternative hydration pattern might also play a role in the change in the twist between tetrads in the LNA and DNA structures.
In the mixed quadruplex, a T-tetrad is formed at the 5′-terminus. This is similar to the U-tetrad observed in the RNA quadruplex structure, (37) albeit the geometry of the tetrads differs. In the LNA quadruplex, the hydrogen bonding is between the thymine imino proton and the O5′ oxygen of a neighbouring sugar. In the TL4T structure ensemble, some structures have T:T base pairs while other ones do not. However, the conformation of the terminal thymidines should not be over interpreted as no explicit experimental data are available to confirm these structural findings.
Sugar–phosphate backbone geometry
The sugar puckers obtained in the structures are in agreement with the analysis of COSY spectra. This validates the structures as no explicit restraints were included to drive sugar puckers into the right conformation. In the mixed quadruplex, G2 is S-type and G4 is N-type. In duplex a context, we have observed that LNA nucleotides perturb neighbouring sugars, particularly 3′-flanking ones, towards N-type puckers (38). Here, in a quadruplex context, an identical trend is seen.
The sugar–phosphate backbone of DNA structures is malleable and, to some extent, the exact conformation is dictated by the restraints imposed and by the nucleobases' participation in stacking and hydrogen bonding. In general, it is troublesome to determine the backbone geometry using NMR spectroscopy due to spectral overlap of H4′, H5′ and H5′′ resonances. In the case of the TGLGLT quadruplex, the 4-fold symmetry and favourable spectral dispersion induced by the modified nucleotides allow us to determine a rather high number of NOEs pertaining to backbone atoms and consequently, the geometry of the backbone is well-defined. In addition, the backbone conformation was validated by the NMR data derived from 1H-31P correlation spectra, even though no explicit backbone torsion angle restraints were included in our calculations to impose these data. For the TL4T structure, the backbone is less well-defined and in most backbone steps more rotamers than one are observed in the structure ensemble (Table 3).
If considering the guanosine–guanosine steps, the backbone angles fall in the standard duplex genus, i.e. (α, γ) = (−gauche, +gauche), β = trans, (ɛ, ζ) = (trans, −gauche), except for the G2pL3 and L2pL3 steps in TGLGLT and TL4T, respectively. In the G2pL3 step of TGLGLT, we have for G2 (ɛ, ζ) = (−86°, 171°) and for L3 (α, β, γ) = (53°, −114°, −168°). There is a strong correlation between ɛ and ζ in duplex DNA X-ray structures (39,40). The (ɛ, ζ) correlations found in our structures adapt perfectly to that relation. The (ɛ, ζ) = (−gauche, trans) conformation leads to a downfield shift of the 31P resonance, (41) which we indeed observe as the G2pL3 phosphorus resonance is shifted 0.9 p.p.m. downfield as compared to the G4pL5 resonance.
A further anomaly in the G2pL3 step is the glycosidic angle of G2 which is −82° and hence just on the border of the syn domain. In the RNA quadruplex structure, r(UG4U), the glycosidic angle of G2 is shifted to χ = −122° (37). Furthermore, the backbone of the G2pG3 step is also irregular in this structure and similar to the G2pL3 step in our structure. Moreover, the phosphorus resonance is shifted downfield and G2 has an S-type sugar pucker, which is unusual for a ribose sugar in a regular structure. The change in glycosidic angle for G2 and downfield resonance shift of the G2pG3 phosphorus resonance are not observed in the DNA quadruplex and appear to be features peculiar to quadruplexes with nucleotides having N-type sugar puckers. In the TGLGLT and r(UG4U) quadruplex structures, the N-type nucleotides are accommodated by adjustment of the 5′-terminal tetrad. Presumably, this tetrad is more prone to adjustment as it only stacks with another tetrad at one of its faces. The adjustment in tetrad geometry is accomplished by changing the glycosidic angle of G2 and the backbone torsion angles of the G2pL3 step. These observations suggest that designing the most stable parallel LNA quadruplex could be done by substituting every guanosine, except the 5′-terminal one, with LNA guanosines.
In the TL4T structure, the L2pL3 step is again irregular (Table 3). However, as L2 is locked in an N-type sugar conformation (with the glycosidic angle in the high anti domain), the G2pL3 geometry of TGLGLT cannot be adopted. For TL4T as well, it appears that the molecule responds to the incorporation of LNA nucleotides by changes in the backbone at the 5′ end of the tetrad stretch. In the recent NMR study of TLL3TL, the backbone torsion angles were restrained to standard A-type helical values by artificial restraints during calculations and thus we are not able to make comparison with the backbone geometry of this quadruplex.
The non-standard backbone conformation in the first base pair step in the tetrad arrangement (G2pL3 and L2pL3, respectively) might be a consequence of accommodating the locked sugars in the backbone or be an attempt to optimize tetrad stacking interaction and/or cation binding in the core of the quadruplex.
Thermostability and geometry of LNA modified quadruplexes
A few reports have appeared with LNA modified quadruplexes of more complex nature than the simple tetrameric fold studied here. LNA modified versions of the (G4T4)3G4 sequence were investigated (42). LNA nucleotides were incorporated at both syn and anti positions in the native DNA quadruplex and in all cases caused destabilization of the quadruplex. When incorporated at syn positions, a change in quadruplex geometry followed as indicated by CD measurements. Recently, two LNA modified versions of the Thrombine binding aptamer was studied (43). When all nucleotides were LNA modified, quadruplex formation was abolished. With only all non-loop positions modified, a quadruplex was formed albeit the thermostability was reduced as compared to the native Thrombine binding aptamer. In both cases, syn positions in the native quadruplex were LNA modified. This renders the syn conformation unavailable for these residues and consequently the native fold-back conformation impossible. The LNA modified quadruplex probably adopts a parallel folding topology. So whether LNA modifications stabilize quadruplexes depends on the context of the molecule. The parallel tetrameric quadruplexes are stabilized but the more complex fold-back structures are destabilized. Thus it appears that the N-type LNA nucleotides are compatible with a parallel quadruplex fold but less suitable for quadruplex fold-back structures with alternating glycosidic angles.
In a duplex context, LNA nucleotides increase the thermostability and change the duplex towards an A-type (RNA-like) geometry (38). The parallel quadruplex geometry is locally similar to A-type duplex geometry. In addition, we have shown in the present study that the DNA nucleotide following an LNA nucleotide has changed its sugar pucker to N-type as is happening in duplex contexts (44). Accordingly, the local changes in geometry when LNA is incorporated into a tetrameric, parallel DNA quadruplex is very similar to the local changes when incorporating LNA into a DNA duplex and hence it is not surprising that parallel quadruplexes are stabilized by incorporation of LNA and RNA nucleotides.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
Supplementary Material
Acknowledgments
The authors thank The Instrument Centre for NMR Spectroscopy of Biological Macromolecules at The Carlsberg Laboratory, Copenhagen granted by The Danish Natural Science Research Council for providing spectrometer time at the 800 MHz spectrometer. The Danish National Research Foundation is thanked for financial support. Funding to pay the Open Access publication charges for this article was provided by the Danish National Research Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Cech T.R. Life at the end of the chromosome: telomeres and telomerase. Angew. Chem. Int. Ed. 2000;39:34–43. doi: 10.1002/(sici)1521-3773(20000103)39:1<34::aid-anie34>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 2.Blackburn E.H. Structure and function of telomeres. Nature. 1991;350:569–573. doi: 10.1038/350569a0. [DOI] [PubMed] [Google Scholar]
- 3.Zakian V.A. Telomeres: beginning to understand the end. Science. 1995;270:1601–1607. doi: 10.1126/science.270.5242.1601. [DOI] [PubMed] [Google Scholar]
- 4.Erlitzki R., Fry M. Sequence-specific binding protein of single-stranded and unimolecular quadruplex telomeric DNA from rat hepatocytes. J. Biol. Chem. 1997;272:15881–15890. doi: 10.1074/jbc.272.25.15881. [DOI] [PubMed] [Google Scholar]
- 5.Horvath M.P., Schultz S.C. DNA G-quartets in a 1.86 Å resolution structure of an Oxytricha nova telomeric protein–DNA complex. J. Mol. Biol. 2001;310:367–377. doi: 10.1006/jmbi.2001.4766. [DOI] [PubMed] [Google Scholar]
- 6.Neidle S., Parkinson G. Telomere maintenance as a target for anticancer drug discovery. Nature Rev. Drug Discov. 2002;1:383–393. doi: 10.1038/nrd793. [DOI] [PubMed] [Google Scholar]
- 7.Rezler E.M., Bearss D.J., Hurley L.H. Telomere inhibition and telomere disruption as processes for drug targeting. Annu. Rev. Pharmacol. Taxicol. 2003;43:359–379. doi: 10.1146/annurev.pharmtox.43.100901.135733. [DOI] [PubMed] [Google Scholar]
- 8.Bates P.J., Kahlon J.B., Thomas S.D., Trent J.O., Miller D.M. Antiproliferative activity of G-rich oligonucleotides correlates with protein binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 9.Koizumi M., Koga R., Hotoda H., Ohmine T., Furukawa H., Agatsuma T., Nishigaki T., Abe K., Kosaka T., Tsutsumi S., et al. Biologically active oligodeoxyribonucleotides part 11: the least phosphate-modification of quadruplex-forming hexadeoxyribonucleotide TGGGAG, bearing 3′- and 5′-end-modification, with anti-HIV-1 activity. Bioorg. Med. Chem. 1998;6:2469–2475. doi: 10.1016/s0968-0896(98)80021-7. [DOI] [PubMed] [Google Scholar]
- 10.Wyatt J.R., Vickers T.A., Roberson J.L., Buckheit R.W., Klimkait T., Debaets E., Davis P.W., Rayner B., Imbach J.L., Ecker D.J. Combinatorially selected guanosine-quartet structure is a potent inhibitor of human-immunodeficiency-virus envelope-mediated cell-fusion. Proc. Natl Acad. Sci. USA. 1994;91:1356–1360. doi: 10.1073/pnas.91.4.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keniry M.A. Quadruplex structures in nucleic acids. Biopolymers. 2001;56:123–146. doi: 10.1002/1097-0282(2000/2001)56:3<123::AID-BIP10010>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 12.Neidle S., Parkinson G.N. The structure of telomeric DNA. Curr. Opin. Struct. Biol. 2003;13:275–283. doi: 10.1016/s0959-440x(03)00072-1. [DOI] [PubMed] [Google Scholar]
- 13.Aboul-ela F., Murchie A.I.H., Norman D.G., Lilley D.M.J. Solution structure of a parallel-stranded tetraplex formed by d(TG4T) in the presence of sodium-ions by nuclear-magnetic-resonance spectroscopy. J. Mol. Biol. 1994;243:458–471. doi: 10.1006/jmbi.1994.1672. [DOI] [PubMed] [Google Scholar]
- 14.Phillips K., Dauter Z., Murchie A.I.H., Lilley D.M.J., Luisi B. The crystal structure of a parallel-stranded guanine tetraplex at 0.95 Å resolution. J. Mol. Biol. 1997;273:171–182. doi: 10.1006/jmbi.1997.1292. [DOI] [PubMed] [Google Scholar]
- 15.Schultze P., Hud N.V., Smith F.W.F.J. The effect of sodium, potassium and ammonium ions on the conformation of the dimeric quadruplex formed by the Oxytricha nova telomere repeat oligonucleotide d(G(4)T(4)G(4)) Nucleic Acids Res. 1999;27:3018–3028. doi: 10.1093/nar/27.15.3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haider S., Parkinson G.N., Neidle S. Crystal structure of the potassium form of an Oxytricha nova G-quadruplex. J. Mol. Biol. 2002;320:189–200. doi: 10.1016/S0022-2836(02)00428-X. [DOI] [PubMed] [Google Scholar]
- 17.Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 18.Kettani A., Bouaziz S., Wang W., Jones R.A., Patel D.J. Bombyx mori single repeat telomeric DNA sequence forms a G-quadruplex capped by base triads. Nature Struct. Biol. 1997;4:382–389. doi: 10.1038/nsb0597-382. [DOI] [PubMed] [Google Scholar]
- 19.Petersen M., Wengel J. LNA: a versatile tool for therapeutics and genomics. Trends Biotechnol. 2003;21:74–81. doi: 10.1016/S0167-7799(02)00038-0. [DOI] [PubMed] [Google Scholar]
- 20.Nielsen K.E., Rasmussen J., Kumar R., Wengel J., Jacobsen J.P., Petersen M. NMR studies of fully modified locked nucleic acid (LNA) hybrids: solution structure of an LNA:RNA hybrid and characterization of an LNA:DNA hybrid. Bioconjugate Chem. 2004;15:449–457. doi: 10.1021/bc034145h. [DOI] [PubMed] [Google Scholar]
- 21.Wengel J. Nucleic acid nanotechnology-towards Angstrom-scale engineering. Org. Biomol. Chem. 2004;2:277–280. doi: 10.1039/b313986g. [DOI] [PubMed] [Google Scholar]
- 22.Randazzo A., Esposito V., Ohlenschläger O., Ramachandran R., Mayol L. NMR solution structure of a parallel LNA quadruplex. Nucleic Acids Res. 2004;32:3083–3092. doi: 10.1093/nar/gkh629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saccà B., Lacroix L., Mergny J.-L. The effect of chemical modifications on the thermal stability of different G-quadruplex-forming oligonucleotides. Nucleic Acids Res. 2005;33:1182–1192. doi: 10.1093/nar/gki257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Güntert P., Wüthrich K. FLATT: a new procedure for high-quality baseline correction of multidimensional NMR spectra. J. Magn. Reson. 1992;96:403–407. [Google Scholar]
- 25.Goddard T.D., Kneller D.G. San Francisco: University of California; SPARKY 3. [Google Scholar]
- 26.Wijmenga S.S., Van Buuren B.N.M. The use of NMR methods for conformational studies of nucleic acids. Prog. Nucl. Magn. Reson. Spectrosc. 1998;32:287–387. [Google Scholar]
- 27.Herrmann T., Güntert P., Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 28.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag, NY; 1984. [Google Scholar]
- 29.Dapic V., Abdomerovic V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxynucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aboul-ela F., Murchie A.I.H., Lilley D.M.J. NMR study of parallel-stranded tetraplex formation by the hexadeoxynucleotide d(TG4T) Nature. 1992;360:280–282. doi: 10.1038/360280a0. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y., Patel D.J. Solution structure of a parallel-stranded G-quadruplex DNA. J. Mol. Biol. 1993;234:1171–1183. doi: 10.1006/jmbi.1993.1668. [DOI] [PubMed] [Google Scholar]
- 32.Van Wijk J., Huckriede B.D., Ippel J.H., Altona C. Furanose sugar conformations in DNA from NMR coupling constants. Meth. Enzymol. 1992;211:286–306. doi: 10.1016/0076-6879(92)11017-d. [DOI] [PubMed] [Google Scholar]
- 33.Lavery R., Sklenar H. The definition of generalized helicoidal parameters and of axis curvature of irregular nucleic acids. J. Biomol. Struc. Dyn. 1988;6:63–91. doi: 10.1080/07391102.1988.10506483. [DOI] [PubMed] [Google Scholar]
- 34.Lavery R., Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J. Biomol. Struc. Dyn. 1989;7:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 35.Foloppe N., MacKerell A.D. Jr. Contribution of the phosphodiester backbone and glycosyl linkage intrinsic torsional energetics to DNA structure and dynamics. J. Phys. Chem. B. 1999;103:10955–10964. [Google Scholar]
- 36.Petraccone L., Erra E., Esposito V., Randazzo A., Galeone A., Barone G., Giancole C. Biophysical properties of quadruple helices of modified human telomeric DNA. Biopolymers. 2005;77:75–85. doi: 10.1002/bip.20189. [DOI] [PubMed] [Google Scholar]
- 37.Cheong C., Moore P.B. Solution structure of an unusually stable RNA tetraplex containing G- and U-quartet structures. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 38.Petersen M., Bondensgaard K., Wengel J., Jacobsen J.P. Locked nucleic acid (LNA) recognition of RNA: NMR solution structures of LNA:RNA hybrids. J. Am. Chem. Soc. 2002;124:5974–5982. doi: 10.1021/ja012288d. [DOI] [PubMed] [Google Scholar]
- 39.Dickerson R.E., Drew H.R. Structure of a B-DNA dodecamer. J. Mol. Biol. 1981;149:761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- 40.Dickerson R.E. Base sequence and helix structure variation in B-DNA and A-DNA. J. Mol. Biol. 1983;166:419–441. doi: 10.1016/s0022-2836(83)80093-x. [DOI] [PubMed] [Google Scholar]
- 41.Gorenstein D.G. Conformation and dynamics of DNA and protein–DNA complexes by P-31 NMR. Chem. Rev. 1994;94:1315–1338. [Google Scholar]
- 42.Dominick P.K., Jarstfer M.B. A conformationally constrained nucleotide analogue controls the folding topology of a DNA G-quadruplex. J. Am. Chem. Soc. 2004;126:5050–5051. doi: 10.1021/ja039192z. [DOI] [PubMed] [Google Scholar]
- 43.Randazzo A., Esposito V., Ohlenschläger O., Ramachandran R., Virgilio A., Mayol L. Structural studies on LNA quadruplexes. Nucl. Nucl. Nucl. 2005;24:795–800. doi: 10.1081/ncn-200060279. [DOI] [PubMed] [Google Scholar]
- 44.Bondensgaard K., Petersen M., Singh S.K., Rajwanshi V.K., Kumar R., Wengel J., Jacobsen J.P. Structural studies of LNA:RNA duplexes by NMR: conformations and implications for RNase H activity. Chem. Eur. J. 2000;6:2687–2695. doi: 10.1002/1521-3765(20000804)6:15<2687::aid-chem2687>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.