Abstract
Six diverse prokaryotic and five eukaryotic genomes were compared to deduce whether the protein synthesis termination signal has common determinants within and across both kingdoms. Four of the six prokaryotic and all of the eukaryotic genomes investigated demonstrated a similar pattern of nucleotide bias both 5′ and 3′ of the stop codon. A preferred core signal of 4 nt was evident, encompassing the stop codon and the following nucleotide. Codons decoded by hyper-modified tRNAs were over-represented in the region 5′ to the stop codon in genes from both kingdoms. The origin of the 3′ bias was more variable particularly among the prokaryotic organisms. In both kingdoms, genes with the highest expression index exhibited a strong bias but genes with the lowest expression showed none. Absence of bias in parasitic prokaryotes may reflect an absence of pressure to evolve more efficient translation. Experiments were undertaken to determine if a correlation existed between bias in signal abundance and termination efficiency. In Escherichia coli signal abundance correlated with termination efficiency for UAA and UGA stop codons, but not in mammalian cells. Termination signals that were highly inefficient could be made more efficient by increasing the concentration of the cognate decoding release factor.
INTRODUCTION
Termination of protein synthesis involves the decoding of a stop signal through an interaction between RNA (rRNA and mRNA) and proteins [release factors (RFs)] that facilitates the hydrolytic release of the nascent polypeptide chain from the peptidyl-transferase centre of the ribosome (1–5). Despite possessing some common features in their translation termination mechanisms, prokaryotes and eukaryotes display important differences. Bacteria possess two Class I decoding RFs (RF1 and RF2) with overlapping codon specificity, while eukaryotes possess only one decoding factor, eRF1. Each prokaryotic factor responds to UAA, whereas UAG is decoded only by RF1 and UGA is decoded only by RF2 (6). In contrast, eRF1 has an omnipotent decoding capacity and promotes completed polypeptide release in response to any of the three stop codons (7,8). This suggests that specificity for polypeptide release mediated by RFs may have evolved independently after the separation of these phylogenetic domains 2.7 billion years ago (9), and the fact that the RFs from the two kingdoms possess virtually no sequence or structural homology reinforces this view. If this were indeed the case, the appearance of prokaryotic RFs and the eukaryotic RFs would represent a fascinating example of parallel evolution (9). An interesting question arising from these observations is whether the termination signals in the mRNA are conserved between the two kingdoms.
The consequences of stop codons being recognized directly by protein factors, rather than a tRNA, as in polypeptide elongation, means that the signal for translation termination could extend beyond the 3 nt specified in the genetic code (10). In pro- and eukaryotes initial evidence that supports this concept has come both from bioinformatic and experimental studies. Bioinformatic analysis of nucleotide frequency around termination codons in bacteria, mainly derived from analyses of Escherichia coli, has highlighted that there is bias in the occurrence of specific codons 5′ and nucleotides 3′ of the stop codon (11–17).
A series of studies, in E.coli, have tested experimentally whether termination efficiency is correlated with changes in the nucleotide sequences 5′ and 3′ of the stop codon. In the limited number of sequences tested, the decoding of stop codons by RFs in competition with suppressor tRNAs was shown to be affected by the nucleotide sequence 3′ of UAG (18,19), UGA (20,21) and UAA (22) stop codons. The effect on termination efficiency of the nucleotide immediately 3′ of the stop codon could have been mediated through near cognate tRNA or RF decoding or both, as it has been shown with a zero-length crosslink reagent that the stop codon and 3 nt immediately 3′ of the stop codon are in close contact with the cognate decoding factor in termination complexes on E.coli ribosomes (23,24). Sequences 5′ and 3′of the UGA stop codon have been shown to interact in a cooperative manner to affect bacterial termination (25). In addition, identity of the last two amino acids of the nascent polypeptide has been demonstrated in vivo to affect termination efficiency in bacteria at UGA (26,27) and UAG (28,29) stop codons. The identity of the P site tRNA was also shown to influence termination efficiency (28).
Initial studies with a small subset of eukaryotic genes have also revealed bias in the occurrence of nucleotides 5′ and 3′ of stop codons. This led to the proposal that, as in prokaryotes, the base following the stop codon was important for termination efficiency, with eRF1 recognizing a tetra-nucleotide sequence containing limited redundancy, and not simply one of three tri-nucleotide stop codons (30,31). Subsequent studies of gene sequences in eukaryotes (32,33) and specific studies in yeast (34), plants (35) and mammals (15,36) have revealed a similar bias in nucleotide occurrence in the position following the stop codon. The eukaryotic decoding release factor, eRF1, requires a stop codon with an extra nucleotide to facilitate termination in vitro (37), and eRF1 has been shown through site-directed crosslink studies to be in contact with the triplet stop codon (38) although the study did not investigate the nucleotides following, as had the earlier equivalent studies in bacteria (23,24).
The translation termination efficiency of a limited set of selected eukaryotic sequences has been investigated experimentally in both yeast and mammalian cells (34,39–42). These studies have revealed that the nucleotide sequences both 5′ and 3′ of the stop codon can modulate termination efficiency.
The bioinformatic and experimental studies on the nature of the translation termination signal, undertaken in a limited number of both pro- and eukaryotic organisms, suggest that the signal extends 5′ and 3′ of the simple triplet codon. Now that a considerable number of genome sequences have been completed, it is possible to undertake a comprehensive comparison of translational termination contexts both within and between organisms of the same and of a different kingdom. In the current work we analysed six representative prokaryotic and five representative eukaryotic genomes in detail, and compared characteristics of the termination signals in their genes. The relationship between the occurrence of the translation termination signals and their functional efficiencies were tested experimentally, and both common and unique features of the mechanism have been highlighted.
MATERIALS AND METHODS
Bioinformatic analysis
Bioinformatic analyses were performed on gene sequences [open reading frames (ORFs)] entered into the TransTerm database (43). Gene sequences were excluded from the database if they did not contain a valid stop codon or if they contained an ORF before the stop codon of <100 nt. Files containing non-redundant cDNA sequences were selected for analysis from the following species: Prokaryo-tic, E.coli K12 (4199 sequences analysed, GC content of analysed sequences 47%), Bacillus subtilis (4095, 39%), Mycobacterium tuberculosis H37Rv (4148, 65%), Mycoplasma genitalium (481, 29%), Rickettsia prowazekii strain Madrid E (834, 25%) and Chlamydophila pneumoniae (1108, 35%); Eukaryotic, Saccharomyces cerevisiae (6258, 31%), Caenorhabditis elegans (15 329, 30%), Drosophila melanogaster (14 027, 38%), Arabidopsis thaliana (14 172, 33%) and Homo sapiens (16 778, 48%).
Codon adaptive index (CAI) values, calculated from published relative synonymous codon usage tables (44), were used to select subsets of genes with differential expression levels from the E.coli K12, S.cerevisiae and D.melanogaster genomes. Analysis of selected gene sequences was performed as described previously (36,45).
Media and bacterial strains
Cultures used for in vivo assays to measure termination signal efficiency in bacteria were grown in minimal media supplemented with all 19 l-amino acids and glycine added at recommended concentrations (46). Protein expression was induced from the Ptrc promoter with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). The following E.coli strains were used in this study: CDJ64 (Δ(lacproB) nalA rif sup9 valR thi), XAc (ara argE-am Δ(lacproB) nalA rif thi); XA101 (ara argE-am Δ(lacproB) nalA rif thi metB supD); XA105 (ara argE-oc Δ(lacproB) nalA rif thi metB supG) (47).
Constructs
Selected termination contexts identified from the bioinformatics studies were introduced into the 3A′ reporter system (26) for studies in bacteria, or into a dual luciferase reporter system adapted from Grentzmann et al., (48), for studies in reticulocyte lysates in vitro or in cultured mammalian cells (Table 2). Owing to the overlapping specificity of the bacterial Class I RFs, contexts were selected for each stop codon. Termination contexts were cloned using pairs of redundant oligonucleotides as described previously (49).
Prokaryotic translation termination assay
Termination contexts were tested in an assay that utilizes the IgG binding B domain from Staphylococcus aureus protein A as a reporter gene (49). E.coli cultures were grown to mid-log phase in the presence of 0.5 mM IPTG. The two or three domain reporter gene products (2A′ and/or 3A′) were purified by affinity chromatography on IgG Sepharose, followed by separation by SDS–PAGE. The efficiency of termination was determined by the ratio of protein arising from termination at the first stop codon (2A′) compared with total protein expressed from the reporter gene (2A′ + 3A′). The ratio was independent of the level of expression and optical density of the bacteria cell culture. E.coli were co-transformed, when appropriate with plasmids that contained wild-type RF2, RF2 T246S and RF1.
Eukaryotic translation termination assays
In vitro translation
The RiboMAX™ Large Scale RNA Production Systems—T7 (Promega) was used to synthesize mRNA for translation in Rabbit Reticulocyte Lysate (Promega). Dual luciferase reporter template DNA was prepared by restriction endonuclease digestion and was transcribed according to the manufacturer's instructions. Typically, 250–500 ng of RNA was incubated with nuclease-treated rabbit reticulocyte lysate supplemented with amino acids at 30°C for 90 min. Translation reactions were supplemented when appropriate with eRF1purified as described (7,50).
In vivo translation
COS-7 (51) cells were transiently transfected with a dual luciferase reporter plasmid using FuGENE 6™ reagent (Roche) according to the manufacturer's instructions. Samples of 2 × 104 cells per well were transfected with 500–1000 ng of plasmid DNA. Cells were harvested 48 h following transfection and lysed in 1× Passive Lysis Buffer (Gibco).
Dual luciferase assay
The Renilla reniformis luciferase (RLuc) 5′ of the test sequence was assayed using the conditions described by Matthews et al. (52) with the modifications of Srikantha et al. (53) and the Photinus pyralis luciferase (Luc+) 3′ of the test sequence was assayed using the conditions described by Tanguay and Gallie (54). Termination efficiencies were calculated from the ratio of Luc+ to RLuc activity in the test samples relative to the ratio of Luc+ to RLuc in control samples where the two reporters were in the same ORF.
RESULTS
Bias surrounds translational stop codons in genes from a range of prokaryotic and eukaryotic genomes
Genomes of representative pro- and eukaryotic organisms that varied in genome size, ORF number and GC content were selected for bioinformatic analysis. The prokaryotic species selected included free-living, pathogenic, non-pathogenic and parasitic strains. ORFs were analysed from the prokaryotes E.coli K12, B.subtilis, M.tuberculosis, M.genitalium, R.prowazekii and C.pneumoniae. In the case of eukaryotes, ORFs were analysed from S.cerevisiae, C.elegans, D.melanogaster, A.thaliana and H.sapiens.
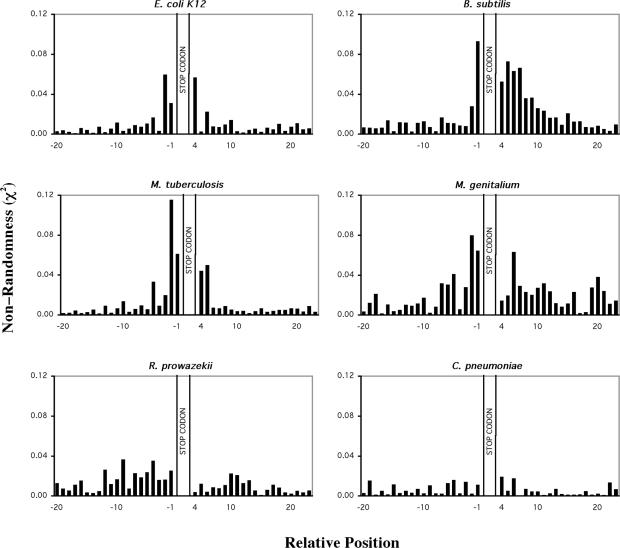
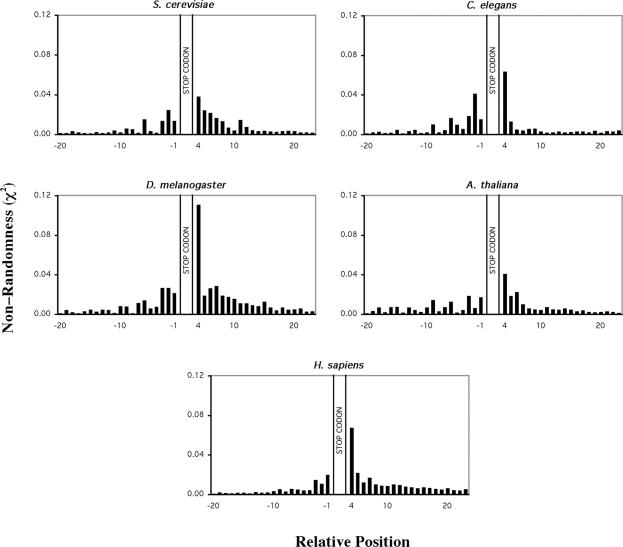
A database of sequences that included 100 bp 5′ and 3′ of ORF translational stop codons from the six prokaryotic and five eukaryotic genomes was compiled and analysed in detail. A frequency table detailing the occurrence of the 4 nt in each position for the region 100 bp 5′ and 3′ of the stop codon was constructed for all species. These frequency tables were used to determine the bias in occurrence of nucleotides in a region spanning 20 bases 5′, and 3′ to the stop codon (Figures 1 and 2). A clear bias in the occurrence of nucleotides both 5′ and 3′ of the stop codon was evident for all prokaryotic organisms, except for R.prowazekii and C.pneumoniae. The pattern of nucleotide bias observed in the prokaryotic species was characterized by an increase in nucleotide bias moving through the 5′ coding region towards the stop codon that peaked at the position immediately 3′ to the stop codon and declined through the non-coding region. This pattern of non-randomness is consistent with results from a previous analysis of a subset of E.coli K12 sequences (11). The pattern of 5′ and 3′ nt bias identified in the prokaryotic organisms was also evident in the eukaryotic genomes (Figure 2). However, among the eukaryotic genomes analysed there was less variation in the pattern of nucleotide bias compared with the prokaryotes examined.
Figure 1.
Bias in nucleotides around the translation termination codons of six selected prokaryotic genomes. Non-randomness, χ2 values were calculated for 20 nt 5′ and 3′ of the stop codon. The stop codon is located at nucleotide positions +1 to +3. χ2 values were calculated for all genes E.coli K12 (4199 genes), B.subtilis (4095 genes), M.tuberculosis (4148 genes), M.genitalium (481 genes), R.prowazekii (834 genes), C.pneumoniae (1108 genes). Predicted nucleotide frequencies used in the calculation were derived from 99 nt 5′ and 3′ to the stop codon. Coding region nucleotide frequencies were corrected for triplet periodicity by determining the nucleotide frequency at each of the three positions within the triplet codon (1, 2 or 3).
Figure 2.
Bias in nucleotides around the translation termination codons of five selected eukaryotic genomes. Non-randomness, χ2 values were calculated for 20 nt 5′ and 3′ of the stop codon. The stop codon is located at nucleotide positions +1 to +3. χ2-Values were calculated for all genes S.cerevisiae (6258 genes), C.elegans (15 329 genes), D.melanogaster (14 027 genes), A.thaliana (14 172 genes) and H.sapiens (16 778 genes). Predicted nucleotide frequencies used in the calculation were derived from 99 nt 5′ and 3′ to the stop codon. Coding region nucleotide frequencies were corrected for triplet periodicity by determining the nucleotide frequency at each of the three positions within the triplet codon (1, 2 or 3).
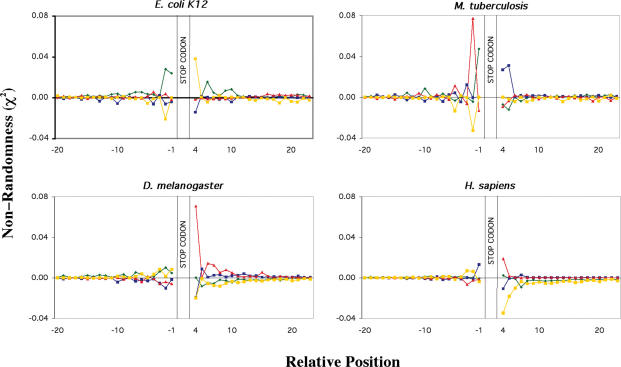
The datasets generated were further analysed to determine which particular nucleotides contributed to the bias observed in the sequences around the stop codons (Figure 3). In both prokaryotic and eukaryotic species, nucleotide bias at each position was caused by specific over- or under-representation of any one or more of the 4 nt. As expected, those species that exhibited the greatest overall bias also possessed the greatest deviations in occurrence of individual nucleotides. The pattern of individual nucleotide bias differed among the species, especially 3′ to the stop codon. This variation is illustrated with data from two of the prokaryotic genomes shown in Figure 3. E.coli K12 sequences exhibited an over-representation of T in the base following the stop codon and a under-representation of C, compared with an over-representation of C in this position and a under-representation of G or A in M.tuberculosis. A further variation was seen with B.subtilis in that A was preferred in this position (data not shown). In contrast, the eukaryotic genomes all exhibited an over-representation of purines (A or G) and an under-representation of pyrimidines (C or T) in the nucleotide position following the stop codon. There were differences, however, with both D.melanogaster and H.sapiens exhibiting over-representation of G (Figure 3), whereas in C.elegans A was predominant, and in A.thaliana and S.cerevisiae genes both G and A were over-represented (data not shown). In all cases the pyrimidines C or T were preferentially avoided in the position following the stop codon. This difference in bias indicates a difference in the preference for specific ‘tetra-nucleotide’ core stop signals between the two kingdoms. The pattern of individual nucleotide bias also differed in the 5′ region, but in both cases the bias went against the naturally occurring bias of G, non-G, R evident in coding regions.
Figure 3.
Bias in individual nucleotides around the translational termination codons of two representative prokaryotic and eukaryotic genomes. Non-randomness of individual nucleotides, χ2 values were calculated for 20 bases 5′ and 3′ of the stop codon. The stop codon is located at nucleotide positions +1 to +3. χ2-Values were calculated for all genes E.coli K12 (4199 genes), M.tuberculosis (4148 genes), D.melanogaster (14 027 genes) and H.sapiens (16 778 genes). Individual nucleotides are represented by a green diamond A, dark blue square C, red triangle G and yellow square T. Predicted nucleotide frequencies used in the calculation were derived from 99 nt 5′ and 3′ to the stop codon. Coding region nucleotide frequencies were corrected for triplet periodicity by determining the nucleotide frequency at each of the three positions within the triplet codon (1, 2 or 3)
Preferred core termination signals: the stop codon and the nucleotide following
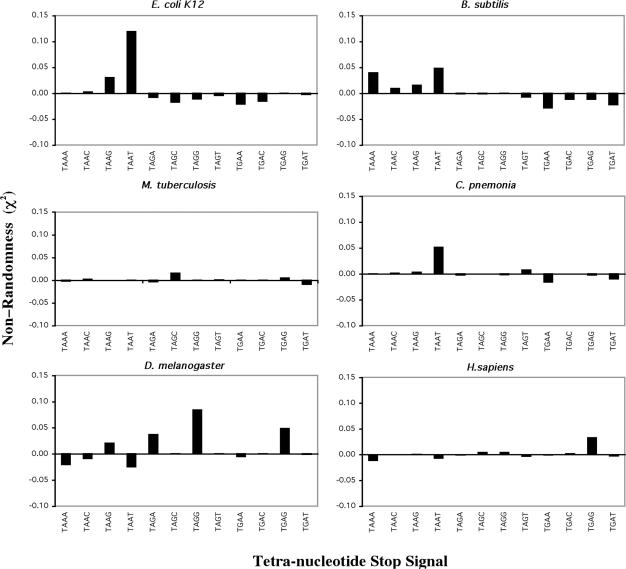
Previous bioinformatic analysis (30) predicted a core termination signal that was comprised of the stop codon and the nucleotide following, forming a tetra-nucleotide stop signal. Comparison of the genomes used in this study revealed that there was distinct bias in the abundance of specific tetra-nucleotide stop signals. Generally, one tetra-nucleotide signal was preferred out of the 12 possible T A/G A/G N signals (excluding TGGN). The identity of the tetra-nucleotide signal differed between the two kingdoms, as illustrated in Figure 4. The genomes of E.coli K12 and M.genitalium (data not shown), exhibited a strong preference for TAAT. The genome of C.pneumoniae that exhibited little overall nucleotide bias (Figure 1) showed a strong bias for the TAAT tetra-nucleotide signal. R.prowazekii exhibited a slight bias for the same TAAT tetra-nucleotide signal (data not shown). The B.subtilis genome showed a preference for both the TAAA and TAAT signal, consistent with strong over representation of A in the position immediately following the stop codon in ORFs from this genome. In contrast, M.tuberculosis showed a preference for the TAGC signal, which reflected the very high GC content (65%) and the dominance of C in the fourth position in this organism. All five eukaryotic genomes showed preferred tetra-nucleotide signals of a stop codon followed by a purine (G or A). This is illustrated in D.melanogaster TAGG>TGAG>TAGA> TAAG and in H.sapiens TGAG (Figure 4). The other three genomes fitted this pattern although it was not as marked with A.thaliana genes (data not shown).
Figure 4.
Bias in tetra-nucleotide translational stop signal in selected prokaryotic and eukaryotic genomes. Non-randomness of tetra-nucleotide sequences, χ2 values were calculated for the +1 to +4 nt for all genes E.coli K12 (4199 genes), B.subtilis (4095 genes), M.genitalium (481 genes), R.prowazekii (834 genes), D.melanogaster (14 027 genes) and H.sapiens (16 778 genes). Predicted codon frequencies used in the calculation were derived from 99 nt 3′ to the stop codon.
Nucleotide bias surrounds stop codons in highly expressed but not lowly expressed genes of prokaryotes and eukaryotes
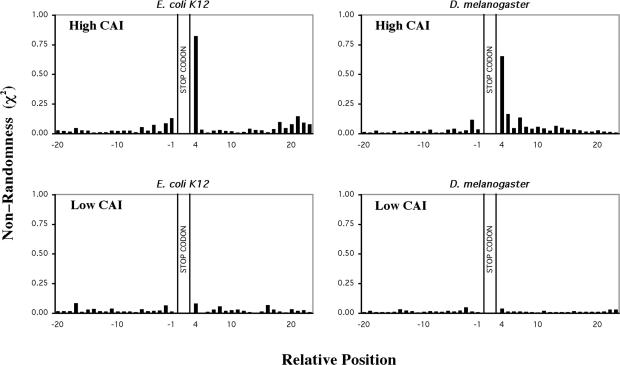
To investigate whether the observed bias in nucleotide sequence identified around the stop codons correlated with translation efficiency, subsets of the most highly and most lowly expressed genes were selected for further analysis. CAI values were used to identify subset of genes with differential expression levels from the E.coli K12, S.cerevisiae and D.melanogaster genomes. The striking finding of this analysis was that the most highly expressed genes (highest 5% CAI) exhibited marked nucleotide bias in the sequences surrounding the stop codon, whereas those with low expression (lowest 5% CAI) showed insignificant bias. This is illustrated in Figure 5 for both E.coli K12 and D.melanogaster sequences. Between the highest and lowest subsets the patterns were intermediate (data not shown). The pattern of nucleotide bias observed in the analysis of the highest CAI genes was similar to that determined for the whole genome, except that the magnitude of the bias was greater. This was common to both prokaryotic and eukaryotic organisms. The finding implied that translational efficiency is correlated to over- or under-representation of nucleotides in certain positions. When only the core tetra-nucleotide signal was considered, the same pattern of bias as observed for the whole genome analysis was identified in the highly expressed datasets, with no preference visible in the lowest CAI dataset (data not shown). These findings suggested that a preferred tetra-nucleotide core termination signal(s) might correlate with efficient translation termination in vivo.
Figure 5.
Bias in nucleotides around the translational termination codons of high and low CAI genes from E.coli K12 and D.melanogaster. Non-randomness, χ2-values were calculated for 20 nt 5′ and 3′ of the stop codon. The stop codon is located at nucleotide to ‘positions +1 to +3’. χ2-Values were calculated for the highest 5% (upper panels) and lowest 5% (lower panels) of genes ranked according to their CAI value E.coli (214 genes) D.melonogaster (701 genes). Predicted nucleotide frequencies used in the calculation were derived from 99 nt 5′ and 3′ to the stop codon. Coding region nucleotide frequencies were corrected for triplet periodicity by determining the nucleotide frequency at each of the three positions within the triplet codon (1, 2 or 3).
Nucleotide bias 5′ of the stop codon reflects codons decoded by hyper-modified tRNAs
In the prokaryotic species the pattern of nucleotide bias 5′ of the stop codon was characterized by a repeating bias in the penultimate and C-terminal codon. This repeating bias resulted in the over-representation of A or G in the second position and A in the third positions of the codon, and under-representation of T or C in the second position (Figure 3). In contrast, eukaryotic genomes exhibited bias predominantly in the last codon with an over-representation of T and C in the first and third positions of the codon respectively, and under-representation of G and C in the first and second positions, respectively (Figure 3). The pattern observed was not influenced by the intrinsic sense codon bias (G, non-G, R) as the analysis method independently determined the nucleotide frequency at each of the three positions within the triplet codon. The observed bias affected the subset of codons utilized in the position 5′ of the stop codon. This, in turn, alters both the subset of tRNAs involved in decoding and the encoded amino acids.
Bioinformatic analysis revealed that there was bias in the amino acids encoded by the last two codons 5′ of the stop codon. Nevertheless it was smaller than the individual nucleotide bias observed previously in both pro- and eukaryotic databases (data not shown). Isaksson and co-workers had shown strong evidence for a correlation between efficiency of termination at a relatively weak UGA stop signal (UGAA) and the hydrophilic/hydrophobic character of the last two encoded amino acids where there isoelectric points were around pl 6 (26,29). However, we were unable to identify a common pattern of over- or under-representation of specific amino acids in the region 5′ of the stop codon when we analysed the three stop codons across the different genomes (data not shown), either for all genes or the high CAI set.
Translation efficiency in both prokaryotes and eukaryotes is known to correlate with tRNA abundance (55,56). It was possible that the bias observed in nucleotide usage in the region 5′ of the stop codon was correlated with a preference for codons decoded by specific tRNAs. Additional analysis revealed that there was a bias for codons decoded by specific iso-accepting tRNAs (Table 1). Specifically, the last two C-terminal amino acids were preferentially decoded by tRNAs that contained hyper-modified bases, at positions 32, 34 and 37 of the anticodon stem–loop (ASL). For example, a single modified species of tRNALys (mnm5s2U at position 34) decodes preferred AAA and AAG codons in the E.coli K12 genome. In addition, the codon decoded by the hyper-modified is over-represented whereas that decoded by the non-modified is not (Table 1). Similar trends are also indicated in the other examples shown in Table 1. This observation suggested that the specific tRNA selected by the last codon position could affect termination in a manner that is independent of the identity of the encoded amino acid. This observation was common for the genomes from both kingdoms implying that the effect of hyper-modified tRNAs on termination was mediated through a similar mechanism. The same pattern of codon usage observed in the complete datasets was identified in the highly expressed datasets indicating that the role of the hyper-modified tRNAs might be correlated with efficient translation termination. We are currently examining whether the structural characteristics of the tRNA at the ribosomal P site affect the binding site of the decoding release factor.
Table 1.
Bias of last sense codon recruits specific iso-accepting tRNAs to the ribosome
| Genome | Codon | tRNA | Bias χ2 | tRNA modification position | ||
|---|---|---|---|---|---|---|
| 32 | 34 | 37 | ||||
| E.coli K12 | AAA | Lys1 | +0.027 | C | mnm5s2U | t6A |
| AAG | Lys1 | +0.029 | C | mnm5s2U | t6A | |
| CAA | Gln2 | +0.017 | Um | mnm5s2U | m2A | |
| CAG | Gln1 | − | Um | C | m2A | |
| B.subtilis | AAA | Lys1 | +0.045 | unkC | cmnm5s2U | ms2t6A |
| AAG | Lys1 | +0.006 | unkC | cmnm5s2U | ms2t6A | |
| CAA | Gln2 | +0.008 | Um | mnm5s2U | m2A | |
| CAG | Gln1 | − | Um | C | m2A | |
| ATA | Ile2 | +0.009 | C | k2C | m6A | |
| ATC | Ile1 | −0.010 | C | G | t6A | |
| ATT | Ile1 | −0.008 | C | G | t6A | |
| S.cerevisiae | AAA | Lys2 | +0.030 | C | cmnm5Um | t6A |
| AAG | Lys1 | − | C | U | t6A | |
| ATA | Ile2 | +0.011 | C | I | t6A | |
| ATC | Ile1 | − | U | G | t6A | |
| ATT | Ile1 | − | U | G | t6A | |
| D.melanogaster | AAA | Lys2 | +0.021 | C | cmnm5Um | t6A |
| AAG | Lys1 | − | C | C | t6A | |
Bias (χ2) values were calculated for the last codon before the stop codon signal as described in Materials and Methods. Bias is indicated when >±0.005, where bias was <±0.005 it is indicated as a ‘minus’. χ2-Values were calculated for all genes E.coli K12 (4199 genes), B.subtilis (4095 genes), S.cerevisiae (6258 genes) and D.melanogaster (14 027 genes). Codons exhibiting the greatest bias are presented. Predicted nucleotide frequencies used in the calculation were derived from 33 codons 5′ to the stop codon. Iso-accepting species of tRNA are indicated with their respective modifications. tRNA modifications are from Sprinzl and Vassilenko (75). Modification are: m2A; 2-methyladenosine, m6A; N6-methyladenosine, t6A; N6-threonylcarbamoyladenosine, ms2t6A; 2-methylthio-N6-threonyl carbamoyladenosine, unkC; unknown modification, k2C; lysidine [4-amino-2-(N6-lysino)-1-beta-d-ribofuranosyl pyrimidine], Um; 2′-O-methyluridine, mnm5s2U; 5-methylaminomethyl-2-thiouridine, cmnm5Um; 5-carboxymethylaminomethyl- 2′-O-methyluridine, cmnm5s2U; 5-carboxymethylaminomethyl-2-thiouridine.
Abundance of a termination signal in the genome correlates with termination efficiency in prokaryotes but not eukaryotes
The comparative bioinformatic studies of the 11 genomes described above implied that a correlation might exist between specific sequences 5′ and 3′ of the stop codon and the efficiency of the translation termination signal. Experiments were undertaken to determine if the bias in nucleotides observed in the bioinformatics analysis allowed prediction of relative functional termination efficiencies of specific signals.
Translation termination signals comprised of 6 nt 5′ and 3 nt 3′ of the stop codon, that occurred more often (labelled abundant) and less often (labelled rare) were selected for experimental analysis. For each stop codon (UAA, UGA and UAG) an abundant (Ab) signal (e.g. 5′Ab UAA Ab3′) and a rare (Ra) signal (5′Ra UAA Ra3′) were selected. Owing to the overlapping specificity of the bacterial Class I RFs, unique contexts were selected for each stop codon so that UAG signals would reflect RF1 decoding, UGA signals RF2 decoding and UAA signals would reflect either or both. These signals represent extremes of occurrence in the pro- and eukaryotic genomes. In the case of the prokaryotes, selection was based on the E.coli K12 genome analysis since the signals could be tested in E.coli, and, in the case of eukaryotes, because of their more conserved features, selection was based on characteristics from all five genomes for testing in vitro, or in cultured mammalian cells. Sequences selected for analysis are listed in Table 2. In each case, hybrid signals were also constructed containing the abundant and rare 5′ and 3′ contexts together. These formed a total of 24 specific contexts that were tested for termination efficiency.
Table 2.
List of termination contexts investigated in this work
| 5′ Context | Stop Codon | 3′ Context | Abbreviation |
|---|---|---|---|
| Prokaryotic termination contexts | |||
| CAG AAG | UAA | UCU | Aba UAA Ab |
| CAG AAG | UAA | CUG | Ab UAA Rab |
| GAA ACC | UAA | UCU | Ra UAA Ab |
| GAA ACC | UAA | CUG | Ra UAA Ra |
| GUC GCG | UAG | GGG | Ab UAG Ab |
| GUC GCG | UAG | CAU | Ab UAG Ra |
| GAU GGA | UAG | GGG | Ra UAG Ab |
| GAU GGA | UAG | CAU | Ra UAG Ra |
| AAU UUC | UGA | UUU | Ab UGA Ab |
| AAU UUC | UGA | CUA | Ab UGA Ra |
| GAA ACU | UGA | UUU | Ra UGA Ab |
| GAA ACU | UGA | CUA | Ra UGA Ra |
| Eukaryotic termination contexts | |||
| AAG GUC | UAA | GCU | Ab UAA Ab |
| AAG GUC | UAA | UGU | Ab UAA Ra |
| UUC CCA | UAA | GCU | Ra UAA Ab |
| UUC CCA | UAA | UGU | Ra UAA Ra |
| AAG GUC | UAG | GCU | Ab UAG Ab |
| AAG GUC | UAG | UGU | Ab UAG Ra |
| UUC CCA | UAG | GCU | Ra UAG Ab |
| UUC CCA | UAG | UGU | Ra UAG Ra |
| AAG GUC | UGA | GCU | Ab UGA Ab |
| AAG GUC | UGA | UGU | Ab UGA Ra |
| UUC CCA | UGA | GCU | Ra UGA Ab |
| UUC CCA | UGA | UGU | Ra UGA Ra |
aAb = abundant.
bRa = rare.
Prokaryotic termination contexts
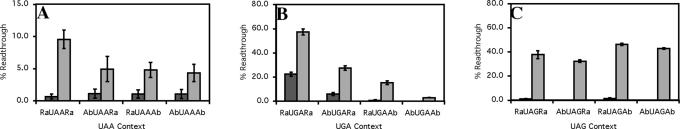
The termination efficiency of the prokaryote signals were assessed in both wild-type E.coli strains, carrying no suppressor tRNAs, and in stop codon-specific suppressor strains. The signals were placed between encoded sequences from the IgG binding B domains of the protein A from S.aureus (49). There was variation in the termination efficiency of the different signals, in wild-type strains in some instances that were more evident in the suppressor strains (Figure 6).
Figure 6.
Termination efficiency of the (A) UAA, (B) UGA and (C) UAG ‘abundant’ and ‘rare’ termination signals in E.coli. Termination efficiency was derived from the 3A′ protein synthesis termination assay (26). Readthrough (%) was calculated by comparison of readthrough protein (3A′) to total protein (3A′ + 2A′). Constructs were assayed in specific strains of E.coli XAc wild-type strain (dark grey), XA105 UAA suppressor strain (light grey), CDJ64 UGA suppressor strain (light grey) and XA101 UAG suppressor strain (light grey). The mean values from six experiments are presented. Error bars are ±SEM.
In the UAA signal set (Figure 6A), the percentage termination was high at >99% in the wild-type strain, indicating that both abundant and rare termination contexts that contained UAA were efficient in competition with near cognate tRNAs. Readthrough increased as expected in the presence of a suppressor tRNA to between 4 and 9% dependent on the sequence surrounding the stop codon. The sequences that promoted most efficient termination were indeed those most commonly used for translation termination in E.coli genes as identified by the bioinformatics analysis. The rare context was least able to counter competition from the suppressor tRNA. The hybrid contexts with one preferred element gave intermediate termination efficiencies but were more similar to the most abundant contexts.
With the UGA series (Figure 6B), there was also a hierarchy of termination efficiencies, although termination efficiency varied more widely than with the UAA contexts (Figure 6A). The rare sequence was quite inefficient exhibiting only ∼80% termination in wild-type strains where it competed relatively poorly with near cognate tRNAs. This inefficiency increased markedly in the suppressor strain to ∼40% termination. In contrast, the abundant context was highly efficient as a termination signal (>99% termination) in the wild-type strain, and in the suppressor strain (∼95% termination). The hybrid contexts were intermediate in their termination efficiencies. These data best reflected the predicted bioinformatic termination efficiency.
The efficiencies of the selected UAG stop codon contexts (Figure 6C) did not match the predicted termination efficiencies as well as for the UAA and UGA signals. In the wild-type strains, UAG sequences exhibited termination efficiencies comparable with those of the UAA signals. In contrast, in two different suppressor strains carrying different amber suppressor tRNAs, the 5′ termination contexts matched the predicted effect while the 3′ contexts did not. This may reflect other selective pressures in E.coli that have resulted in UAG being the most under-represented stop codon, with only 12% of genes terminating with UAG.
Eukaryotic termination contexts
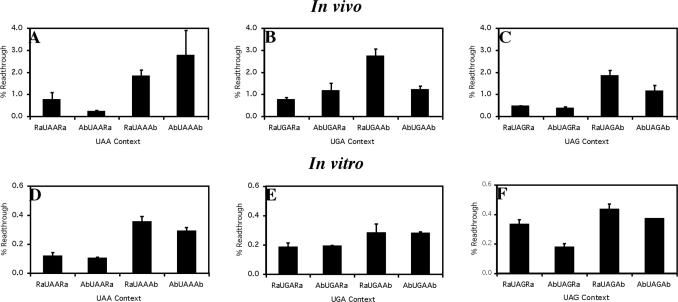
Sequences, identified as either abundant or rare from the analysis of the eukaryotic genomes (Table 2), were assessed for their termination efficiency in cultured COS-7 cells with a dual luciferase reporter gene assay that determines stop signal failure (Figure 7A–C). This sensitive assay allows effective comparisons of termination efficiencies even when the readthrough is <1%. In this reporter system a hierarchy of termination efficiencies were observed but, in contrast to the E.coli signals, no correlation between abundance and efficiency was identified. Indeed, the abundant UAA signal was the least efficient and, generally, the rare signals were among the most efficient. Given this significant difference in the selected sequences from prokaryotic and eukaryotic genomes, the study was then repeated in vitro in reticulocyte lysate. In the reticulocyte lysate assay, termination was several fold more efficient (Figure 7D–F) probably due to a lower concentration of near cognate competitor tRNAs, but there was a general concordance between the COS-7 and the in vitro data. Most significantly in both cases, signal abundance did not correlate with efficiency. Comparison of the results of the two eukaryotic assay systems highlighted that the absolute levels of readthrough of each signal may be specific to the assay system utilized. This precluded in our opinion any comparisons of the absolute termination efficiencies of pro- and eukaryotic signals.
Figure 7.
Termination efficiency of the eukaryotic UAA, UGA and UAG ‘abundant’ and ‘rare’ termination signals in vitro and in vivo. Termination efficiency was derived from a dual luciferase protein synthesis termination assay in vitro (A–C) and in vivo using COS-7 cells (D–F). Readthrough (%) was calculated by comparison to a control construct (UGG). The mean values from three experiments are presented. Error bars are ±SEM.
Modulating the decoding RF concentration can strengthen the termination efficiency of inefficient and rare signals
What does an inefficient signal mean from the perspective of the mechanism of translation termination? One possibility is that inefficient signals are simply decoded more slowly by decoding RFs, so that near cognate (or cognate suppressor tRNAs) are more competitive. If these inefficient signals provided a poor template for binding of the RF into the active site of a termination competent ribosome, then increasing the concentration of the decoding factor might increase the termination signal efficiency through an increase in the productive binding rate of the RF to the stop signal. Contexts that supported a high productive binding rate of RF would not be expected to be influenced by increasing the concentration of RF as their binding to the stop signal contexts would not be rate limiting. This hypothesis was tested in both the prokaryotic and eukaryotic systems.
Over-expressing prokaryotic RFs
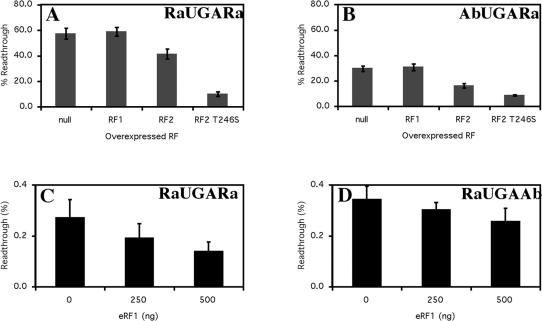
A rare context, 5′Ra UGA Ra3′, that was a poor termination signal and a hybrid context 5′Ab UGA Ra3′ that was more efficient were tested in E.coli co-transformed with a compatible plasmid over-expressing, by 3- to 5-fold, a variant of RF2 (RF2T246S) (57) or the wild-type RF2. Although tested, the other hybrid and the abundant construct gave termination efficiencies that were beyond the sensitivity of the assay that relies on the detection of protein products separated by SDS–PAGE.
Over-expression of RF2wt from an exogenous plasmid produces a factor of lower specific activity for peptide release than its endogenous counterpart, possibly because the factor is lacking its natural modification at residue 252 (58). The RF2 T246S variant does not exhibit diminished specific ctivity and therefore was utilized for this study. A plasmid encoding non-cognate RF1 and one that lacked an insert (null) were used as controls. Both the 5′Ra UGA Ra3′, and the hybrid context 5′Ab UGA Ra3′, became much more efficient when RF2T246S was over-expressed, as shown by a dramatic decrease in readthrough (Figure 8A and B). In the presence of RF2 T246S the poor 5′Ra UGA Ra3′ and the more efficient 5′Ab UGA Ra3′ signals exhibited comparable termination efficiencies which were similar to those observed for the abundant signals assessed in non-supplemented cells. It is of interest that the exogenously expressed RF2wt with the lowered specific activity was less effective at strengthening the efficiency of the rare signal. Expression from the vector expressing the non-cognate RF1 had no effect in each case.
Figure 8.
Effect of increasing E.coli RF1, RF2 and RF2-T246S and human eRF1 expression on termination efficiency. Termination efficiency of the prokaryotic 5′Ra UGA Ra3′ (A) and 5′Ab UGA Ra3′ (B) sequence contexts were measured with the 3A′ protein synthesis termination assay (26). Constructs were assayed in the specific E.coli UGA suppressor strain CDJ64. The mean values from three experiments are presented. Error bars are ±SEM. Termination efficiency of the eukaryotic 5′Ra UGA Ra3′ (C) and 5′Ra UGA Ab3′ (D) sequence contexts were measured with the dual luciferase protein synthesis termination assay in vitro. The assay was supplemented with 250 ng and 500 ng of eRF1 where indicated. Readthrough (%) was calculated by comparison to a control construct (UGG). Experiments were performed in triplicate with a single construct of each signal context. Mean values from three experiments are presented. Error bars are ±SEM.
Over-expressing eukaryotic RF
We have shown that mammalian cells transfected with an expression plasmid encoding eRF1 tightly controls the cellular concentration of the protein, limiting the increase to at most 2-fold (S.F. Mathews and W.P. Tate, unpublished data). For this reason, the equivalent eRF1 over-expression studies were carried out in vitro in reticulocyte lysates supplemented with exogenous eRF1 protein. (Figure 8C and D). As with the prokaryotic signals higher concentrations of eRF1 increased the termination efficiency of the selected sequences (1.5-fold for 5′Ra UGA Ra3′ compared with 1.25-fold for 5′Ra UGA Ab3′) and suggested in both cases the affinity of the decoding RF for its ribosomal-binding site was strongly influenced by the characteristics of the termination signal extending in both directions from the stop codon.
DISCUSSION
5′ and 3′ contexts of prokaryotic and eukaryotic stop signals
Bias in occurrence of nt 5′ and 3′ of stop codons was observed in four of six prokaryotic genomes. While the complete genomes of E.coli K12, B.subtilis, M.tuberculosis and M.genitalium showed clear deviation in nucleotide use around the stop codon, by comparison, the parasitic species R.prowazekii and C.pneumoniae did not. The lack of nucleotide bias in the R.prowazekii and C.pneumoniae species was mimicked by the lowly expressed genes identified from E.coli K12. Studies by Sharp et al. (59) have confirmed that the R.prowazekii and C.pneumoniae genomes have not undergone positive selection for factors that influence translation accuracy or efficiency. Indeed, the magnitude of nucleotide bias 5′ and 3′ of stop codons in the bacterial species investigated here showed positive correlation with the strength of selected codon usage bias (S) as described by Sharp et al. (59). These results suggest that the R.prowazekii and C.pneumoniae parasitic bacteria have evolved under conditions where there has been little selection for efficient termination as appears to have been the case for the other bacterial species. R.prowazekii and C.pneumoniae also contain fewer genes than the other bacteria investigated indicating that differential translation of genes may become more important in organisms with greater complexity in their transcriptomes.
This bioinformatic analysis revealed that the previously well documented bias both 5′ and 3′ of the stop codon of E.coli genes is widespread among bacterial species, although it is not found in every bacterium. Similarly, for eukaryotic genes a number of previous studies had indicated a bias in both 5′ and 3′ positions. The current study has reinforced not only that this is generally found within a diverse range of genomes, but also, and importantly, that the individual nucleotide patterns of bias are quite similar in contrast to the diversity seen with the prokaryotes. The comparison of the nucleotides over- and under-represented 5′ and 3′ of the stop codon between pro- and eukaryotes, however, indicated that similar mechanisms could be involved in efficient translation termination.
Significance of the bias in the 5′ context of the stop codon
Bias in the last two codons 5′ of the stop codon was coupled previously to a correlation between the physical and chemical properties of the last two encoded amino acids and their bias (26,27,29). Additionally, the termination efficiency of a UGAA core signal was linked with the hydrophilic/hydrophobic character of the amino acids encoded by the last two codons before the stop signal (26,29). In the current study where several genomes have been compared, we have confirmed the bias of the amino acids particularly at the last position, but while the properties seem important in specific instances, we have not been able to identify the effect as a dominant determinant of efficiency for termination at all stop signals. In some cases there exists a positive bias for a codon recognized by more than one iso-accepting tRNA species (Table 1), supporting a possible role for the amino acid itself rather than the tRNA, in these instances, influencing efficient termination.
The last two 5′ codons specifying amino acids are decoded by a subset of specific tRNAs. Our investigation of the coding region 5′ of the stop codon revealed that in both pro- and eukaryotic genomes there is an over-representation of codons decoded by hyper-modified tRNAs, especially tRNAs that contained modifications in the anticodon region. In bacteria, tRNA modifications have been shown to improve reading frame maintenance (60) and thermodynamics of ribosomal binding (61). One tRNA that is highly selected for decoding the 5′ codon is tRNALys (Table 1). Structures derived from Thermus thermophilus 30S crystals soaked with the anticodon stem–loop (ASL) of the have revealed that the hyper-modified nucleotides (t6A37 and mnm5U34) contribute to the free energy of their binding to the ribosomal A site. In this case, the modifications act in concert to stabilize the through pre-ordering the ASL into a structure suitable for presentation to the codon in the decoding centre (62). These two modifications allow to bind both AAA and AAG lysine codons. In bacterial genomes there is a positive bias for the AAA lysine codon before stop codons. The AAA codon is preferentially decoded by the modified indicating that a modified tRNA in the ribosomal P site might confer efficient translation termination. The over-representation of other codons in this final position that are also decoded by hyper-modified tRNAs in both pro- and eukaryotes, suggest that these tRNAs confer a similar effect to that observed for .
Class I RF recognition of the termination signal
Hyper-modified tRNAs may have an effect on translation termination efficiency in vivo by promoting similar types of interactions with the decoding site and mRNA or by inducing an A site conformation that promotes or stabilizes RF binding. These stabilizing interactions may be important for stop codon recognition as the decoding RFs are larger than the tRNAs that normally dock into the A site [RF2, (63–65); eRF1, (66)]. The P site tRNA provides one potential binding face for the decoding RF. This is well illustrated in bacteria with a study showing two glycine codons decoded by a specific iso-accepting tRNA species promote low efficiency of decoding at a UAG stop codon compared with other glycine codons decoded by a different tRNA, but this can be improved with a variant of the decoding factor, RF1, or a mutant form of a different tRNA, suggesting a functional interaction between the tRNA and the decoding factor (67).
Bias around the stop codons of genes was generally most prominent in the nucleotide following the stop codon (+4) with differences among prokaryotic genomes most marked in the 3 nt following the stop codon (+4 to +6). These 3 nt along with the stop codon have been shown with a zero-length crosslink reagent to be in close contact with the cognate decoding factor in termination complexes on E.coli ribosomes (23,24). It has been suggested from these studies that RFs mediate termination by interacting directly with stop codons, and the nucleotides +4 to +6 are positioned to form part of a recognition signal. Although no direct interaction between nucleotides beyond the stop codon and the eukaryotic factor, eRF1, have been reported to date, the studies of Chavatte et al. (38) suggest that eRF1 could also be in close enough proximity to form such an interaction. In this model, the rate of binding of the RF to the stop codon in the ribosomal A site would be influenced by the nucleotide sequence 3′ of the stop codon and, thereby, affect the rate of termination. The very recently published crystal structures of bacterial ribosomal termination complexes have sufficient detail to suggest that direct interaction with the stop codon by the RF is possible through an unstructured loop in its domain IV, and that the +4 base of the mRNA could form part of this interaction (68) as predicted (23).
The diversity in the preference for a particular 3′ nucleotide(s) reflected in the prokaryotic genomes may relate to divergence in the amino acid sequence of the Class I RF in the domain that interacts with the region 3′ of the stop. A comparative study of Class I RF sequences have revealed a correlation between several conserved residues and nucleotide bias at translation termination sites (69), although these residues are not universally conserved across bacteria. This suggests that there is a correlation between the divergence in Class I RFs and the variability in the pattern of over- and under-representation of nucleotides in the 3′ region of bacteria.
If the eukaryotic Class I RF arose independently of the prokaryotic equivalents, as their primary and tertiary structures suggest (9), then it is not surprising there are differences in the nucleotides that constitute the recognition signal between the factors from the two kingdoms. The eukaryotic genomes collectively exhibit their own distinct signature of a strong preference for purine nucleotides 3′ of the stop codon found in this study, but there is variation as to which is preferred among the different genomes. This subtle and distinct variability in the +4 to +6 nt of the stop signals among the eukaryotic genomes might reflect evolution of a single omnipotent eRF1, compared with the divergent prokaryotic Class I RFs. Simultaneously with our study, Liu (70) analysed six eukaryotic genomes for bias around stop codons, particularly highlighting the −2 and +4 nt as positions of strong and consistent bias that might form a possible extended signal. Additionally, from multiple alignments of eRF1 from 20 species, Liu (70) suggested 16 highly conserved amino acids that might be involved in its recognition.
Efficiency of extended termination signals
The bioinformatic analyses were used to identify translation termination signals that were abundant or rare and these sequences were assessed for termination efficiency. This tested the hypothesis as to whether signal abundance in the genome correlated with termination efficiency. In the E.coli test system, contexts that contained UAA or UGA stop codons showed excellent correlation between stop signal abundance and efficiency (Figure 6A and B) that was emphasized when the termination decoding mechanism signal was challenged by a competing cognate suppressor tRNA. Readthrough of UAG stop codons did not correlate with sequence abundance when challenged by the competing cognate suppressor tRNAs, SupD (Figure 6C) and SupE (data not shown), suggesting that there might be other selective pressures on UAG sequences in the genome. It was also observed that the E.coli K12 high CAI dataset contained only two genes that terminate with a TAG codon, both of which possess tandem termination codons (TAG TAG and TAG TAA).
The consequences of an inefficient signal in E.coli has been demonstrated as an effect on gene expression, particularly when mRNA levels are low and particularly for smaller genes where the initiation and termination signals are not so widely separated. The implication is that inefficient termination can cause ribosomal pausing and queuing of ribosomes in the coding region and affect initiation on the same mRNA (71).
To test whether the kinetics of recognition of the stop codon by the RF might be the key determinant for stop signal efficiency, rare and more abundant signals were subjected to an E.coli in vivo environment where the concentration of the cognate RF was increased by about 3- to 5-fold. Increased RF concentration improved termination efficiency at the rare signal dramatically. The termination efficiency of the more abundant signals were relatively less affected by the increased RF concentration, suggesting that the recognition of the signal by RF was less rate limiting. These data supported the model that RF binding to the ribosomal A site is influenced by the mRNA contexts 5′ and 3′ of the stop codon. It is proposed that the translation termination signal may influence RF binding either directly or indirectly by providing specific determinants or dictating subtle changes in the conformational state of the ribosomal A site. This might come in part from the P site tRNA (28). The fact that an inefficient termination signal arising from a particular ribosomal P site tRNA/codon combination can be corrected by a variant RF (67), implies that a P site tRNA might compromise the binding of the RF into the A site by steric inhibition. Indeed, we have shown that the P site tRNA can crosslink to the decoding RF by zero-length site-directed reagents indicating very close contact between the elbow and the anticodon region of the P site tRNA and the factor (E.S. Poole and W.P. Tate, unpublished data).
The recognition and binding of the decoding RFs for termination of protein synthesis may be the major, if not the sole determinant of the extended length of the stop signal in prokaryotes. In prokaryotes, transcription and translation are spatially and temporally linked, and efficient recognition of stop codons by the decoding factors is correlated with translation rate. In bacterial species that have undergone positive selection for factors that influence translation accuracy and efficiency, there has also been selection for translation termination signals that in vivo confer improved termination efficiency.
A wider role for eRF in the eukaryotic cell
Abundant or rare stop signal sequences from the eukaryotic genomes investigated were also assessed experimentally to determine their ability to direct termination efficiently. The results indicated that differences in the nucleotide sequences in both 3′ and 5′ contexts could affect signal efficiency but this did not correlate with bias and/or abundance. However, the efficiency of signals could be improved by increasing the concentration of the decoding RF as was the case for the prokaryotic sequences implying this is a rate-limiting step in the termination mechanism, and that modulation of the decoding factor concentration could influence gene expression for specific genes.
In eukaryotes, where transcription and translation are not spatially and temporally linked, the fact that the observed bias 5′ and 3′ of stop codons in the genomes are not directly correlated with termination efficiency implies termination of protein synthesis is not the only determinant that is important for the function of the translation termination signal. The answer to this enigma may lie in additional roles the translation termination sequence plays in the eukaryotic cell. One possible role for the sequence is in the formation of the proposed mRNA recycling loop that facilitates efficient ribosomal recycling. The binding of eRF1 and eRF3 to the 80S ribosomal complex is intrinsically involved in the formation of the recycling loop (72). Abundant termination signals might bind eRF1 and eRF3 more rapidly, thereby expediting the formation of this loop. Co-ordinated termination and formation of the recycling loop is important for maintaining mRNA stability as premature termination of translation triggers nonsense-mediated mRNA decay (73). Therefore, the existence of preferred extended termination signals may be important at the cellular level as a means for the translational machinery to distinguish between mature and premature stop codons (74). The formation of the recycling loop will decrease the rate of mRNA decay and will significantly increase the amount of protein expressed per transcript.
Eukaryotic cells exist within multicellular organisms and as such are subjected to greater regulation of gene expression than prokaryotic cells. The translation termination machinery appears to have taken on new roles as part of this regulation function in the eukaryotic kingdom, and this may explain some of the divergence between prokaryotes and eukaryotes with respect to their translational termination signals.
Acknowledgments
We would like to thank Dr Chris Brown and Dr Peter Stockwell for their expert help with the TransTerm database. A.G.C was supported by a Royal Society of New Zealand Marsden Fund postgraduate scholarship. L.L.M was supported by a University of Otago postgraduate scholarship. A grant from the Royal Society of New Zealand Marsden Fund supported W.P.T and E.S.P. Funding to pay the Open Access publication charges for this article was provided by University of Otago, and Royal Society of New Zealand Marsden Fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Poole E., Tate W. Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim. Biophys. Acta. 2000;1493:1–11. doi: 10.1016/s0167-4781(00)00162-7. [DOI] [PubMed] [Google Scholar]
- 2.Nakamura Y., Ito K. Making sense of mimic in translation termination. Trends Biochem. Sci. 2003;28:99–105. doi: 10.1016/S0968-0004(03)00006-9. [DOI] [PubMed] [Google Scholar]
- 3.Kisselev L., Ehrenberg M., Frolova L. Termination of translation: interplay of mRNA, rRNAs and release factors? EMBO J. 2003;22:175–182. doi: 10.1093/emboj/cdg017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Buckingham R.H., Grentzmann G., Kisselev L. Polypeptide chain release factors. Mol. Microbiol. 1997;24:449–456. doi: 10.1046/j.1365-2958.1997.3711734.x. [DOI] [PubMed] [Google Scholar]
- 5.Inge-Vechtomov S., Zhouravleva G., Philippe M. Eukaryotic release factors (eRFs) history. Biol. Cell. 2003;95:195–109. doi: 10.1016/s0248-4900(03)00035-2. [DOI] [PubMed] [Google Scholar]
- 6.Scolnick E., Tompkins R., Caskey T., Nirenberg M. Release factors differing in specificity for terminator codons. Proc Natl Acad. Sci. USA. 1968;61:768–774. doi: 10.1073/pnas.61.2.768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Frolova L., Le Goff X., Rasmussen H.H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A.-L., Celis J.E., Philippe M., et al. A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature. 1994;372:701–703. doi: 10.1038/372701a0. [DOI] [PubMed] [Google Scholar]
- 8.Dontsova M., Frolova L., Vassilieva J., Piendl W., Kisselev L., Garber M. Translation termination factor aRF1 from the archaeon Methanococcus jannaschii is active with eukaryotic ribosomes. FEBS Lett. 2000;472:213–216. doi: 10.1016/s0014-5793(00)01466-6. [DOI] [PubMed] [Google Scholar]
- 9.Kisselev L.L. Translation termination and yeast prions. Biochemistry. 1999;64:1337–1341. [PubMed] [Google Scholar]
- 10.Tate W.P., Poole E.S., Dalphin M.E., Major L.L., Crawford D.J., Mannering S.A. The translational stop signal: codon with a context, or extended factor recognition element? Biochimie. 1996;78:945–952. doi: 10.1016/s0300-9084(97)86716-8. [DOI] [PubMed] [Google Scholar]
- 11.Brown C.M., Stockwell P.A., Trotman C.N., Tate W.P. The signal for the termination of protein synthesis in procaryotes. Nucleic Acids Res. 1990;18:2079–2086. doi: 10.1093/nar/18.8.2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arkov A.L., Korolev S.V., Kisselev L.L. Termination of translation in bacteria may be modulated via specific interaction between peptide chain release factor 2 and the last peptidyl-tRNA(Ser/Phe) Nucleic Acids Res. 1993;21:2891–2897. doi: 10.1093/nar/21.12.2891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alff-Steinberger C., Epstein R. Codon preference in the terminal region of E.coli genes and evolution of stop codon usage. J. Theor. Biol. 1994;168:461–463. doi: 10.1006/jtbi.1994.1124. [DOI] [PubMed] [Google Scholar]
- 14.Berezovsky I.N., Kilosanidze G.T., Tumanyan V.G., Kisselev L. COOH-terminal decamers in proteins are non-random. FEBS Lett. 1997;404:140–142. doi: 10.1016/s0014-5793(97)00112-9. [DOI] [PubMed] [Google Scholar]
- 15.Arkov A.L., Korolev S.V., Kisselev L.L. 5′ contexts of Escherichia coli and human termination codons are similar. Nucleic Acids Res. 1995;23:4712–4716. doi: 10.1093/nar/23.22.4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gutman G.A., Hatfield G.W. Nonrandom utilization of codon pairs in Escherichia coli. Proc. Natl Acad. Sci. USA. 1989;86:3699–3703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berezovsky I.N., Kilosanidze G.T., Tumanyan V.G., Kisselev L.L. Amino acid composition of protein termini are biased in different manners. Protein Eng. 1999;12:23–30. doi: 10.1093/protein/12.1.23. [DOI] [PubMed] [Google Scholar]
- 18.Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J. Mol. Biol. 1983;164:73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- 19.Edelmann P., Martin R., Gallant J. Nonsense suppression context effects in Escherichia coli bacteriophage T4. Mol. Gen. Genet. 1987;207:517–518. doi: 10.1007/BF00331625. [DOI] [PubMed] [Google Scholar]
- 20.Miller J.H., Albertini A.M. Effects of surrounding sequence on the suppression of nonsense codons. J. Mol. Biol. 1983;164:59–71. doi: 10.1016/0022-2836(83)90087-6. [DOI] [PubMed] [Google Scholar]
- 21.Stormo G.D., Schneider T.D., Gold L. Quantitative analysis of the relationship between nucleotide sequence and functional activity. Nucleic Acids Res. 1986;14:6661–6679. doi: 10.1093/nar/14.16.6661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin R., Weiner M., Gallant J. Effects of release factor context at UAA codons in Escherichia coli. J. Bacteriol. 1988;170:4714–4717. doi: 10.1128/jb.170.10.4714-4717.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Poole E.S., Brimacombe R., Tate W.P. Decoding the translational termination signal: the polypeptide chain release factor in Escherichia coli crosslinks to the base following the stop codon. RNA. 1997;3:974–982. [PMC free article] [PubMed] [Google Scholar]
- 24.Poole E.S., Major L.L., Mannering S.A., Tate W.P. Translational termination in Escherichia coli: three bases following the stop codon crosslink to release factor 2 and affect the decoding efficiency of UGA-containing signals. Nucleic Acids Res. 1998;26:954–960. doi: 10.1093/nar/26.4.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bjornsson A., Isaksson L.A. UGA codon context which spans three codons. Reversal by ms2i6A37 in tRNA, mutation in rpsD(S4) or streptomycin. J. Mol. Biol. 1993;232:1017–1029. doi: 10.1006/jmbi.1993.1457. [DOI] [PubMed] [Google Scholar]
- 26.Mottagui-Tabar S., Bjornsson A., Isaksson L.A. The second to last amino acid in the nascent peptide as a codon context determinant. EMBO J. 1994;13:249–257. doi: 10.1002/j.1460-2075.1994.tb06255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mottagui-Tabar S., Isaksson L.A. Only the last amino acids in the nascent peptide influence translation termination in Escherichia coli genes. FEBS Lett. 1997;414:165–170. doi: 10.1016/s0014-5793(97)00978-2. [DOI] [PubMed] [Google Scholar]
- 28.Zhang S., Ryden-Aulin M., Isaksson L.A. Functional interaction between release factor one and P-site peptidyl-tRNA on the ribosome. J. Mol. Biol. 1996;261:98–107. doi: 10.1006/jmbi.1996.0444. [DOI] [PubMed] [Google Scholar]
- 29.Bjornsson A., Mottagui-Tabar S., Isaksson L.A. Structure of the C-terminal end of the nascent peptide influences translation termination. EMBO J. 1996;15:1696–1704. [PMC free article] [PubMed] [Google Scholar]
- 30.Brown C.M., Stockwell P.A., Trotman C.N., Tate W.P. Sequence analysis suggests that tetra-nucleotides signal the termination of protein synthesis in eukaryotes. Nucleic Acids Res. 1990;18:6339–6345. doi: 10.1093/nar/18.21.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kohli J., Grosjean H. Usage of the three termination codons: compilation and analysis of the known eukaryotic and prokaryotic translation termination sequences. Mol. Gen. Genet. 1981;182:430–439. doi: 10.1007/BF00293932. [DOI] [PubMed] [Google Scholar]
- 32.Cavener D.R., Ray S.C. Eukaryotic start and stop translation sites. Nucleic Acids Res. 1991;19:3185–3192. doi: 10.1093/nar/19.12.3185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sharp P.M., Burgess C.J., Cowe E., Lloyd A.T., Mitchell K.J. Selective use of termination codons and variations in codon choice. In: Hatfield D.L., Lee B.J., Pirtle R.M., editors. Transfer RNA in protein synthesis. Boca Raton: CRC Press; 1992. pp. 397–425. [Google Scholar]
- 34.Bonetti B., Fu L.W., Moon J., Bedwell D.M. The efficiency of translation termination is determined by a synergistic interplay between upstream and downstream sequences in Saccharomyces cerevisiae. J. Mol. Biol. 1995;251:334–345. doi: 10.1006/jmbi.1995.0438. [DOI] [PubMed] [Google Scholar]
- 35.Angenon G., Van M.M., Depicker A. Analysis of the stop codon context in plant nuclear genes. FEBS Lett. 1990;271:144–146. doi: 10.1016/0014-5793(90)80392-v. [DOI] [PubMed] [Google Scholar]
- 36.McCaughan K.K., Brown C.M., Dalphin M.E., Berry M.J., Tate W.P. Translational termination efficiency in mammals is influenced by the base following the stop codon. Proc. Natl Acad. Sci. USA. 1995;92:5431–5435. doi: 10.1073/pnas.92.12.5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Konecki D.S., Aune K.C., Tate W., Caskey C.T. Characterization of reticulocyte release factor. J. Biol. Chem. 1977;252:4514–4520. [PubMed] [Google Scholar]
- 38.Chavatte L., Frolova L., Kisselev L., Favre A. The polypeptide chain release factor eRF1 specifically contacts the s4UGA stop codon located in the A site of eukaryotic ribosomes. Eur. J. Biochem. 2001;268:2896–2904. doi: 10.1046/j.1432-1327.2001.02177.x. [DOI] [PubMed] [Google Scholar]
- 39.Mottagui-Tabar S., Tuite M.F., Isaksson L.A. The influence of 5′ codon context on translation termination in Saccharomyces cerevisiae. Eur. J. Biochem. 1998;257:249–254. doi: 10.1046/j.1432-1327.1998.2570249.x. [DOI] [PubMed] [Google Scholar]
- 40.Namy O., Hatin I., Rousset J.P. Impact of the six nucleotides downstream of the stop codon on translation termination. EMBO Rep. 2001;2:787–793. doi: 10.1093/embo-reports/kve176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cassan M., Rousset J.P. UAG readthrough in mammalian cells: effect of upstream and downstream stop codon contexts reveal different signals. BMC Mol. Biol. 2001;2:3. doi: 10.1186/1471-2199-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tork S., Hatin I., Rousset J.P., Fabret C. The major 5′ determinant in stop codon read-through involves two adjacent adenines. Nucleic Acids Res. 2004;32:415–421. doi: 10.1093/nar/gkh201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown C.M., Jacobs G., Stockwell P., Schreiber M. Detection of signals in mRNAs that influence translation. Appl. Bioinformatics. 2003;2:S47–51. [PubMed] [Google Scholar]
- 44.Sharp P.M., Li W.-H. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Major L.L., Edgar T.D., Yip P.Y., Isaksson L.A., Tate W.P. Tandem termination signals: myth or reality? FEBS Lett. 2002;514:84–89. doi: 10.1016/s0014-5793(02)02301-3. [DOI] [PubMed] [Google Scholar]
- 46.Neidhardt F.C., Bloch P.L., Smith D.F. Culture medium for enterobacteria. J. Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coulondre C., Miller J.H. Genetic studies of the lac repressor. III. Additional correlation of mutational sites with specific amino acid residues. J. Mol. Biol. 1977;117:525–567. doi: 10.1016/0022-2836(77)90056-0. [DOI] [PubMed] [Google Scholar]
- 48.Grentzmann G., Ingram J.A., Kelly P.J., Gesteland R.F., Atkins J.F. A dual-luciferase reporter system for studying recoding signals. RNA. 1998;4:479–486. [PMC free article] [PubMed] [Google Scholar]
- 49.Bjornsson A., Mottagui-Tabar S., Isaksson L.A. The analysis of translational activity using a reporter gene constructed from repeats of an antibody-binding domain from protein A. Methods Mol. Biol. 1998;77:75–91. doi: 10.1385/0-89603-397-X:75. [DOI] [PubMed] [Google Scholar]
- 50.Frolova L.Y., Merkulova T.I., Kisselev L.L. Translation termination in eukaryotes: polypeptide release factor eRF1 is composed of functionally and structurally distinct domains. RNA. 2000;6:381–390. doi: 10.1017/s135583820099143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gluzman Y. SV40-transformed simian cells support the replication of early SV40 mutants. Cell. 1981;23:175–182. doi: 10.1016/0092-8674(81)90282-8. [DOI] [PubMed] [Google Scholar]
- 52.Matthews J.C., Hori K., Cormier M.J. Purification and properties of Renilla reniformis luciferase. Biochemistry. 1977;16:85–91. doi: 10.1021/bi00620a014. [DOI] [PubMed] [Google Scholar]
- 53.Srikantha T., Klapach A., Lorenz W.W., Tsai L.K., Laughlin L.A., Gorman J.A., Söll D.R. The sea pansy Renilla reniformis luciferase serves as a sensitive bioluminescent reporter for differential gene expression in Candida albicans. J. Bacteriol. 1996;178:121–129. doi: 10.1128/jb.178.1.121-129.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tanguay R.L., Gallie D.R. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 1996;16:146–156. doi: 10.1128/mcb.16.1.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ikemura T. Codon usage and tRNA content in unicellular and multicellular organisms. Mol. Biol. Evol. 1985;2:13–34. doi: 10.1093/oxfordjournals.molbev.a040335. [DOI] [PubMed] [Google Scholar]
- 56.Percudani R., Pavesi A., Ottonello S. Transfer RNA gene redundancy and translational selection in Saccharomyces cerevisiae. J. Mol. Biol. 1997;268:322–330. doi: 10.1006/jmbi.1997.0942. [DOI] [PubMed] [Google Scholar]
- 57.Wilson D.N., Guevremont D., Tate W.P. The ribosomal binding and peptidyl-tRNA hydrolysis functions of Escherichia coli release factor 2 are linked through residue 246. RNA. 2000;6:1704–1713. doi: 10.1017/s135583820000131x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dincbas-Renqvist V., Engstrom A., Mora L., Heurgue-Hamard V., Buckingham R., Ehrenberg M. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 2000;19:6900–6907. doi: 10.1093/emboj/19.24.6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sharp P.M., Bailes E., Grocock R.J., Peden J.F., Sockett R.E. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–1153. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Urbonavicius J., Qian O., Durand J.M.B., Hagervall T.G., Bjork G.R. Improvement of reading frame maintenance is a common function for several tRNA modifications. EMBO J. 2001;20:4863–4873. doi: 10.1093/emboj/20.17.4863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fahlman R.P., Dale T., Uhlenbeck O.C. Uniform binding of aminoacylated transfer RNAs to the ribosomal A and P sites. Mol. Cell. 2004;16:799–805. doi: 10.1016/j.molcel.2004.10.030. [DOI] [PubMed] [Google Scholar]
- 62.Murphy F.V., Ramakrishnan V., Malkiewicz A., Agris P.F. The role of modifications in codon discrimination by tRNALysUUU. Nature Struct. Mol. Biol. 2004;11:1186–1191. doi: 10.1038/nsmb861. [DOI] [PubMed] [Google Scholar]
- 63.Vestergaard B., Van L.B., Andersen G.R., Nyborg J., Buckingham R.H., Kjeldgaard M. Bacterial polypeptide release factor RF2 is structurally distinct from eukaryotic eRF1. Mol. Cell. 2001;8:1375–1382. doi: 10.1016/s1097-2765(01)00415-4. [DOI] [PubMed] [Google Scholar]
- 64.Rawat U.B.S., Zavialov A.V., Sengupta J., Valle M., Grassucci R.A., Linde J., Vestergaard B., Ehrenberg M., Frank J. A cryo-electron microscopic study of ribosome-bound termination factor RF2. Nature. 2003;421:87–90. doi: 10.1038/nature01224. [DOI] [PubMed] [Google Scholar]
- 65.Klaholz B.P., Pape T., Zavialov A.V., Myasnikov A.G., Orlova E.V., Vestergaard B., Ehrenberg M., Van Heel M. Structure of the Escherichia coli ribosomal termination complex with release factor 2. Nature. 2003;421:90–94. doi: 10.1038/nature01225. [DOI] [PubMed] [Google Scholar]
- 66.Song H.W., Mugnier P., Das A.K., Webb H.M., Evans D.R., Tuite M.F., Hemmings B.A., Barford D. The crystal structure of human eukaryotic release factor eRF1 - Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell. 2000;100:311–321. doi: 10.1016/s0092-8674(00)80667-4. [DOI] [PubMed] [Google Scholar]
- 67.Zhang S., Ryden-Aulin M., Isaksson L.A. Functional interaction between tRNA2Gly at the ribosomal P-site and RF1 during termination at UAG. J. Mol. Biol. 1998;284:1243–1246. doi: 10.1006/jmbi.1998.2319. [DOI] [PubMed] [Google Scholar]
- 68.Petry S., Broderson D.E., Murphy F.V., Dunham C.M., Selmer M., Tarry M.J., Kelley A.C., Ramakrishnan V. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell. 2005;123:1–12. doi: 10.1016/j.cell.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 69.Ozawa Y., Saito R., Washio T., Tomita M. Comparative study of translation termination sites and release factors (RF1 and RF2) in procaryotes. J. Mol. Evol. 2003;56:665–672. doi: 10.1007/s00239-002-2435-9. [DOI] [PubMed] [Google Scholar]
- 70.Liu Q. Comparative analysis of biases around stop codons in six eukaryotes. BioSystems. 2005;81:281–289. doi: 10.1016/j.biosystems.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 71.Jin H.N., Bjornsson A., Isaksson L.A. Cis control of gene expression in E.coli by ribosome queuing at an inefficient translational stop signal. EMBO J. 2002;21:4357–4367. doi: 10.1093/emboj/cdf424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sonenberg N., Dever T.E. Eukaryotic translation initiation factors and regulators. Curr. Opin. Struct. Biol. 2003;13:56–63. doi: 10.1016/s0959-440x(03)00009-5. [DOI] [PubMed] [Google Scholar]
- 73.Jacobson A., Peltz S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 74.Amrani N., Ganesan R., Kervestin S., Mangus D.A., Ghosh S., Jacobson A. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 75.Sprinzl M., Vassilenko K.S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 2005;33:D139–140. doi: 10.1093/nar/gki012. [DOI] [PMC free article] [PubMed] [Google Scholar]