Abstract
Binding of the internal ribosome entry site (IRES) of the hepatitis C virus (HCV) RNA to the eIF-free 40S ribosomal subunit is the first step of initiation of translation of the viral RNA. Hairpins IIId and IIIe comprising 253–302 nt of the IRES are known to be essential for binding to the 40S subunit. Here we have examined the molecular environment of the HCV IRES in its binary complex with the human 40S ribosomal subunit. For this purpose, two RNA derivatives were used that bore a photoactivatable perfluorophenyl azide cross-linker. In one derivative the cross-linker was at the nucleotide A296 in hairpin IIIe, and in the other at G87 in domain II. Site-specific introduction of the cross-linker was performed using alkylating derivatives of oligodeoxyribonucleotides complementary to the target RNA sequences. No cross-links with the rRNA were detected with either RNA derivative. The RNA with the photoactivatable group at A296 cross-linked to proteins identified as S5 and S16 (major) and p40 and S3a (minor), while no cross-links with proteins were detected with RNA modified at G87. The results obtained indicate that hairpin IIIe is located on the solvent side of the 40S subunit head on a site opposite the beak.
INTRODUCTION
It is now well established that initiation of protein synthesis can occur via two alternative mechanisms. The conventional cap-dependent mechanism is typically used by capped eukaryotic cellular mRNAs and is mediated by a number of initiation factors that taken together are responsible for the ultimate correct positioning of the start AUG codon on the 40S ribosomal P site (1,2). The alternative pathway called internal translation initiation is based on the cap-independent recruitment of the so-called internal ribosomal entry site (IRES), a specifically structured RNA segment in the 5′-untranslated region (5′-UTR) upstream the initiation AUG codon. This RNA segment is responsible for positioning of the start AUG codon in the P site mediated by a reduced number of initiation factors as compared to the conventional mechanism, or in several cases even without any initiation factors. A number of viral and some eukaryotic cellular mRNAs contain such IRES elements (3).
Hepatitis C virus (HCV) is one of the most dangerous human pathogens that can lead to cyrrosis, hepatocellular carcinoma and other damages of the liver. The genomic RNA of HCV contains an IRES element possessing a complex secondary (Figure 1a) and tertiary structure essential for effective translation of the viral RNA (3,4). This RNA is able to form a stable binary complex with the 40S ribosomal subunit providing the positioning of the initiation AUG codon at or close to the P site without any initiation factors (5). Interaction of the HCV IRES with the 40S subunit has been extensively studied using various approaches. By direct ultraviolet (UV)-induced cross-linking, it was reported that in the binary complex with the 40S subunit IRES cross-links with only one protein. This protein was initially identified as S9 (5), but another group then reported that it was actually S5 (6). At the same time, data obtained by cryo-electron microscopy (cryo-EM) (7) pointed to multiple contacts of the IRES with the 40S subunit. The footprinting (8) and the fingerprinting (9) results also indicated that the area of IRES interaction with the 40S ribosomal subunit is not restricted to one protein. HCV IRES with uridines randomly substituted by 4-thiouridines cross-linked to proteins S2, S3, S10, S15, S16, S18 and S27 in the binary complex with the 40S subunit (10). Moreover, it was observed that the ribosomal proteins S3, S5, S7, S18 and p40 can bind to the HCV IRES out of the ribosome in cell extracts (11). However, the relationships between these interactions and initiation of HCV RNA translation are as yet not clear.
Figure 1.
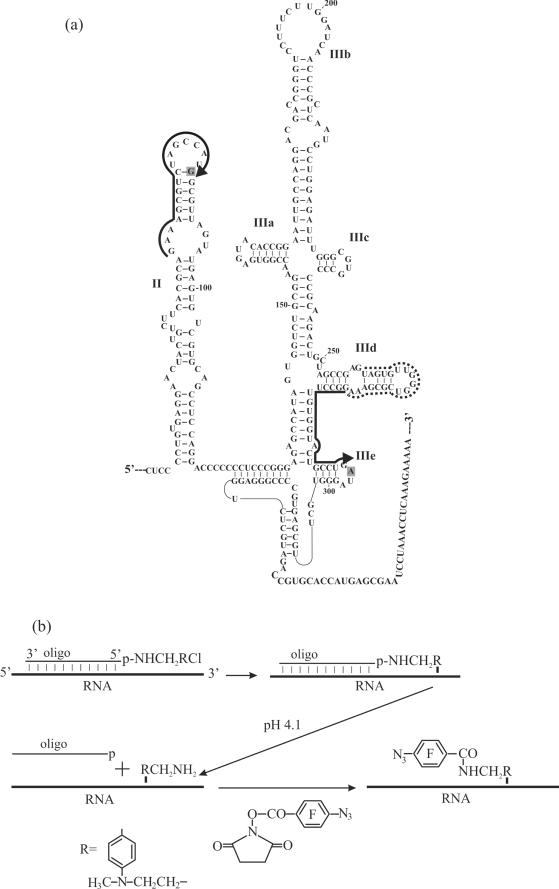
(a) Structural organization of the HCV IRES (3). RNA sequences complementary to deoxy oligomers used for the site-specific modification of the HCV IRES are marked with hard lines. A sequence complementary to the helper oligomer used together with the alkylating derivative in hairpin IIIe is marked with dotted line. Arrows indicate 5′-termini of the oligomers derivatized with the alkylating groups. Nucleotides of the HCV IRES cross-linked to the oligonucleotide derivatives are shaded. (b) Scheme of site-specific introduction of a photoactivatable group into specific RNA sites based on the complementary-addressed alkylation of the RNA with [4-(N-2-chloroethyl-N-methylamino)benzyl]-phosphoramides of oligodeoxyribonucleotides.
Data concerning the involvement of large structural domains of the HCV IRES in particular steps of initiation of viral RNA translation have been reported [e.g. see (12–14)]. Recently, a revised model of this process was suggested (15) according to which the HCV IRES first binds to the 40S subunit to form the binary complex, and only the basal portion of domain III (mainly hairpins IIId and IIIe) is required for binding. Thereafter, eIF3 and the ternary complex (eIF2•Met- •GTP) assemble to form a 48S preinitiation complex dependent on both the base of domain III and a proper initiation codon. In addition, at this step subdomain IIIb is involved in eIF3 binding. Subsequent formation of the 80S complex is dependent on GTP hydrolysis and 60S subunit joining, and is regulated by domains II and IIIb. It also remains unclear how particular structural elements of the HCV IRES interact with the 40S subunit and what ribosomal components are involved in the particular interactions at the various steps of translation initiation. The molecular environment of the HCV IRES domains on the ribosome may be studied with RNA derivatives bearing a cross-linker at specific locations. Such derivatives could be obtained by site-specific alkylation of an RNA with [4-(N-2-chloroethyl-N-methylamino)benzyl]phosphoramide derivatives of oligodeoxyribonucleotides complementary to a sequence adjacent to the target site. The phosphoramide bond in the covalent adducts formed would then be hydrolysed under mild acidic conditions, and aryl azide groups would be selectively introduced at the benzylamine groups liberated after hydrolysis. Initially, this approach was elaborated for the introduction of cross-linkers into specific sites of tRNAs (16); later this approach was shown to also be applicable to longer (up to 300 nt long) RNAs possessing complex secondary structure (17).
Here, we present a study of the environment of subdomain IIIe of the HCV IRES on the human 40S ribosomal subunit in the binary complex that is formed as the first step of translation initiation of the HCV IRES. Hairpin IIIe belongs to the base of domain III, which is involved in binary complex formation (see above). For this purpose, the respective RNA transcript bearing a photoactivatable perfluorophenyl azide group at the nucleotide A296 in subdomain IIIe was used. In parallel, control experiments were carried out with a derivative of HCV RNA with the cross-linker at G87 in domain II which is involved only in the late stage of the initiation of HCV IRES translation and is not required for binding of the IRES to the 40S subunit (see above). The identification of cross-linked proteins was based on their electrophoretic analysis by one and two dimensional (1,2D) PAGE, a methodology widely used to identify ribosomal proteins cross-linked with various components of the translational machinery, e.g. with HCV IRES (10), and with mRNA analogues (18).
MATERIALS AND METHODS
Materials
Acrylamide, N,N′-methylene-bis-acrylamide and urea were purchased from AppliChem, ribonucleoside triphosphates and avidin agarose were from Sigma, biotin-16-uridine triphosphate from Roche, Sephadex G-75 from Pharmacia, AMV reverse transcriptase from Life Science and T7 RNA polymerase from NEB. T4 polynucleotide kinase was prepared in the Laboratory of Bioorganic Chemistry of enzymes at the Institute of Chemical Biology and Fundamental Medicine SB RAS; DNA oligomers complementary to definite sequences of the HCV IRES were synthesized in the Laboratory of Medical Chemistry, [α-32P]GTP (1 mCi/nmol) was synthesized in the Laboratory of Biotechnological Chemistry, 4-(N-2-chloroethyl-N-methylamino)benzylamine (ClRCH2NH2) and N-oxysuccinimide ester of 4-azidotetrafluorobenzoic acid were kind gifts from Dr T.M. Ivanova of this Institute.
Labelled HCV IRES RNA was obtained by in vitro transcription of plasmid pXL40-372.NS′ (19) linearized by digestion with BamH1 (‘Fermentas’, Lietuva) as described (14). The specific radioactivity of the RNA transcript at the time of isolation was typically about 25 000 c.p.m./pmol.
Slightly labelled biotinylayted HCV IRES was obtained as described above but adding 0.2 mM biotin-16-uridine triphosphate to the reaction mixture and using 5 µCi instead of 0.1 mCi [α-32P]GTP. The specific radioactivity of the RNA transcript at the time of isolation was typically about 1000 c.p.m./pmol.
4-(N-2-chloroethyl-N-methylamino)benzyl-5′-phosphoramides of oligodeoxyribonucleotides were synthesized and purified as described (14).
Site-specific modification of HCV IRES RNA was carried out as described (14) starting with 300 pmol of the RNA transcript.
Nucleotides of the IRES RNA cross-linked to the oligodeoxyribonucleotide derivatives were determined by reverse transcription as described (20).
To hydrolyse the phosphoramide bond in the deoxyribonucleotide derivatives attached to the RNA, the covalent RNA adducts were incubated in 300 mM NaOAc (pH 4.1) at 50°C for 6 h. The reaction was stopped by ethanol precipitation; the modified RNA was purified by 8% denaturing PAGE and eluted from the gel as described above. The mobility in the gel of the resulting modified RNA was the same as that of the unmodified IRES RNA and was evidence of the removal the oligodeoxyribonucleotide moiety and liberation of the RCH2NH2 group attached to the RNA.
Selective introduction of perfluorophenyl azide at the RCH2NH2 group attached to the RNA. RNA obtained after the hydrolysis of the phosphoramide bond was dissolved in 50 µl of 50 mM HEPES-KOH (pH 9.0) and mixed with an equal volume of 20 mM N-oxysuccinimide ester of 4-azidotetrafluorobenzoic acid in dimethyl sulfoxide (DMSO). The mixture was incubated in the dark at room temperature for 90 min, and low molecular mass organic compounds were then extracted with 3 volumes of ether. The modified RNA was precipitated from the water phase with ethanol, reprecipitated twice with ethanol from 50% DMSO in water, dissolved in water and stored in liquid nitrogen until used in cross-linking experiments.
Ribosomes, ribosomal complexes and cross-linking procedures
Ribosomes
40S ribosomal subunits with intact rRNA were isolated from unfrozen human placenta as indicated (21). Prior to use, the subunits were re-activated by incubation in binding buffer A [20 mM Tris–HCl (pH 7.5), 100 mM KCl, 2.5 mM MgCl2 and 0.25 mM spermidine] at 37°C for 10 min.
Ribosomal complexes
Binary complexes of HCV IRES and 40S subunits were obtained by incubating the subunits (1.5 × 10−6 M if not specified otherwise) with IRES (0.5 × 10−6 M if not specified otherwise) in buffer A as described (14). The extent of binding of the 5′-32P-labelled RNAs to 40S subunits was examined by nitrocellulose filtration assay using alkali pretreated filters as described (22).
Cross-linking
UV-irradiation of the complexes was carried out according to (18). The distribution of the cross-linked labelled IRES between the rRNA and proteins was examined by centrifugation in a 5–20% sucrose density gradient in the presence of SDS and EDTA as described (23).
Isolation and analysis of ribosomal proteins cross-linked to biotinylated IRES RNAs
Irradiated 40S subunits were ethanol precipitated and dissolved in 20 µl of 20 mM Tris–HCl (pH 7.5) containing 1 mM EDTA. The mixture was supplied with 20 µl of an avidin agarose suspension, incubated at 37°C for 2 h under stirring, and the avidin agarose with bound biotinylated RNA was pelleted by centrifugation. To destroy all possible RNA–protein complexes, the pellet was incubated with 40 µl of 6 M urea containing 1% SDS at 50°C for 15 min, the avidin agarose was pelleted by centrifugation and the supernatant containing proteins not covalently bound to the IRES was discarded. The pellet was washed three times with 40 µl of the same solution and three times with 100 µl of water. To hydrolyse the RNA bound to avidin agarose, the pellet was mixed with 10 µl of 20 mM Tris–HCl (pH 7.5) containing 1 mM EDTA and 10 µg RNase A and incubated at 37°C for 3 h. The avidin agarose was then removed by centrifugation and to the supernatant were added 10 µl of buffer containing 120 mM of Tris–HCl (pH 7.5), 4% SDS, 40% glycerol, 0.1% bromphenol blue and 2% 2-mercaptoethanol for the subsequent analysis of the proteins by a standard one dimensional (1D) electrophoresis in a 150 × 180 × 0.75 mm polyacrylamide gel containing SDS (SDS–PAGE); the gel was silver stained and dried. Total human 40S protein isolated as indicated (21) was used for the marker lane.
Isolation and analysis of ribosomal proteins cross-linked to labelled IRES RNAs
Irradiated 40S subunits were precipitated with ethanol and dissolved in 30 µl of water, supplied with 1 µl of 2-mercaptoethanol, 90 µl of 8 M urea and 3.6 µl of 0.1 M EDTA-NaOH (pH 7.5) and incubated at 37°C for 5 min to dissociate the subunits into rRNA and proteins. The RNA was completely hydrolysed by addition of 3.2 µl of 0.7 M sodium citrate (pH 5.0), and 3 µl of RNAse A (10 mg/ml) followed by the incubation at 37°C for 1 h. The proteins were precipitated by cold acetone; after centrifugation the pellet was dissolved in 100 µl of 6 M guanidine hydrochloride containing 0.1% of 2-mercaptoethanol and incubated at 37°C for 30 min to completely denature the proteins. The proteins were separated from the labelled short oligoribonucleotides by gel filtration on a 2 ml Sephadex G-50 superfine column; elution was with 6 M guanidine hydrochloride, 25 mM Tris–HCl (pH 7.5), 0.1 mM EDTA, 25 mM NaCl and 5% ethanol. Fractions of 100 µl were collected and their radioactivity was measured by Cherenkoff counting. Typically, labelled proteins were eluted in fractions 3–6 corresponding to the void volume of the column. After 1–2 intermediate fractions, radioactivity strongly increased indicating elution of oligonucleotides. The proteins were precipitated from the respective fractions by addition of 9 vol of cold ethanol. Each pellet was washed with 200 µl of 90% ethanol, dried and dissolved in 7 µl of 8 M urea for subsequent analysis by 2D PAGE in an alkaline-SDS system (24). The separation in the first dimension was in an 8% gel containing 8 M urea at pH 8.6; in the second dimension, separation was in a 12.5% gel containing SDS at pH 6.7. To identify the cross-linked proteins, the radioautograms were superimposed on the respective stained gels and the positions of the radioactive bands with respect to those of unmodified proteins were determined.
RESULTS
Site-specific introduction of photoactivatable groups into the HCV IRES
The scheme for the introduction of a photoactivatable group in selected positions of RNA transcripts containing the complete 5′-UTR of HCV RNA is presented in Figure 1b. Site-specific alkylation of the RNA was performed using 4-(N-2-chloroethyl-N-methylamino)benzyl-5′-phosphoramides of oligodeoxyribonucleotides complementary to positions 277–295 and 71–87 of the HCV IRES. Sequence 277–295 is located in the region of the stem–loop IIIe (Figure 1a) which together with the hairpin IIId plays critical role in the formation of the binary complex of the HCV IRES with 40S subunits and is the object of the present study. In contrast, sequence 71–87 belongs to the apical loop of domain II (Figure 1a), which is not essential for binding to 40S subunits and the corresponding IRES derivative bearing the cross-linker in domain II was used here as control.
Alkylation of the IRES was carried out at 25°C for a period sufficiently long to almost completely convert the 2-chloroethylamine group to the ethylene immonium cation, which is an active intermediate (25). The ClRCH2NH- moiety coupled to the 5′-terminal nucleotide of the oligonucleotide is known to cross-link to the nucleotide base paired with it or to the neighboring unpaired nucleotide (17,26). Any RNA base at such positions may be cross-linked with the exception for uracil, which is unable to react with aromatic 2-chloroethyl amines under the conditions used (27).
To increase the yield of the covalent adduct with the derivative of the oligonucleotide complementary to positions 277–295, a helper oligomer complementary to positions 259–276 was used that forms a tandem with the oligomer bearing the alkylating group (Figure 1a) (14).
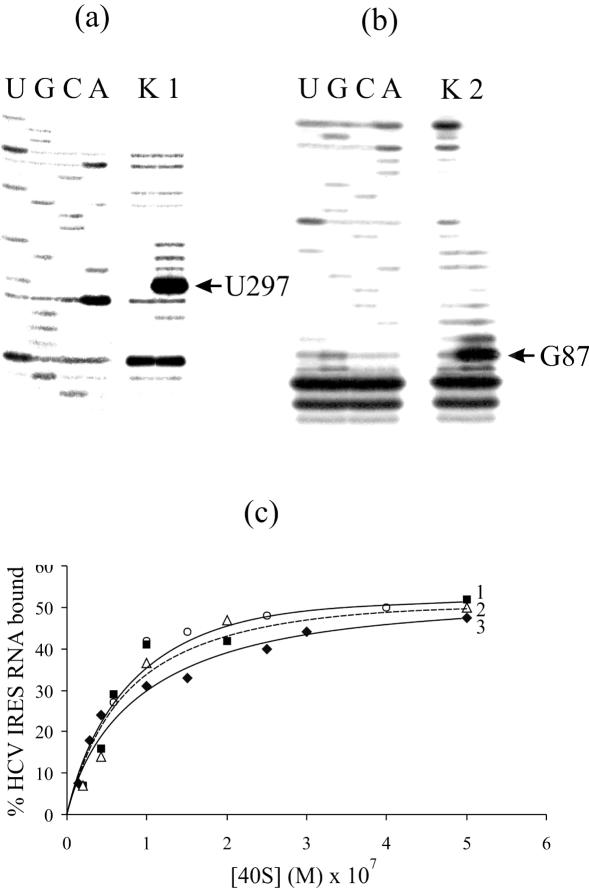
The covalent adducts resulting from IRES alkylation with the oligonucleotide derivatives were separated from unmodified RNA by denaturing PAGE on an 8% gel (the respective electrophoregrams are not shown). The extents of alkylation with the derivatives of oligomers complementary to positions 71–87 and 277–295 were 0.4 and 0.6 mol of oligomer residues per mol of RNA. In our previous study (14) we showed that the cross-linked oligomers actually form heteroduplexes with the respective complementary RNA sequences. Cross-linked nucleotides of the IRES were identified here by reverse transcription. Primer extension leads to a stop or pause at the cross-linked rRNA nucleotide. The cross-linking site is generally assumed to be the nucleotide 5′ of the primer extension stop site. Therefore, cross-linked nucleotides of the HCV IRES should be A296 (Figure 2a, lane 1) and U86 (Figure 2b, lane 2). However, taking into account that uracil could not be alkylated under the conditions used (see above), in the latter case the cross-linked nucleotide was G87 rather than U86. The reverse transcriptase most probably pauses at G87 (this is a pause rather than stop since the N7 of the guanine is not involved in Watson–Crick base pairing).
Figure 2.
Identification of sites of cross-linking of oligonucleotide derivatives to the HCV IRES by reverse transcription and binding properties of the HCV IRES derivatives. Extension of 5′-32P-labelled primers complementary to the HCV IRES sequences 331–350 (a) and 103–120 (b). Lanes 1, 2 and K, primer extension on RNA I, RNA II and RNA K, respectively. Lanes U, G, C, A, sequencing of HCV IRES. Positions of the reverse transcription stops caused by the cross-links are indicated by arrows. (c) Isotherms of binding of the HCV IRES and its derivatives (1.0 × 10−7 M) to 40S subunits in a buffer containing 100 mM K+, 2.5 mM Mg2+, 0.25 mM spermidine at 30°C. Solid lines 1 (filled squares) and 3 (filled diamonds), RNA II and RNA I, respectively; dotted line 2 (open triangles), RNA K; open circles without line correspond to untreated HCV IRES. Relative error was in average about 10%.
To introduce photoactivatable moieties at the modified RNA nucleotides, the phosphoramide bond in the covalent adducts was hydrolysed under mild acidic conditions (pH 4.1) that resulted in the release of the aliphatic amino group -RCH2NH2 linked to the RNA. The modified IRES RNAs were purified by denaturing PAGE in an 8% gel and the -RCH2NH2 moieties were selectively derivatized by treatment with the N-oxysuccinimide ester of 4-azidotetrafluorobenzoic acid. Thus, HCV IRES bearing photoactivatable groups at nucleotides A296 (designated RNA I) and G87 (designated RNA II) were obtained. These RNAs were used in the cross-linking experiments with human 40S ribosomal subunits. IRES RNAs (designated RNA K) treated under identical conditions but without addition of alkylating derivatives of oligodeoxyribonucleotides served as controls.
Binding and cross-linking of photoactivatable HCV IRES derivatives to 40S subunits
Filter retention assays revealed that affinities of RNA I, RNA II and RNA K for human 40S subunits are similar to each other and do not significantly differ from that of the untreated IRES (Figure 2c). For the cross-linking experiments, saturating concentration of 40S subunits was used. To obtain the cross-links, the binary complexes of RNA I and RNA II with 40S subunits were irradiated with mild UV light (wavelength >280 nm). The subsequent analysis of the distribution of the radioactive label between the rRNA and proteins by centrifugation in sucrose density gradients containing SDS and EDTA did not reveal radioactivity in the fractions containing 18S rRNA (the respective sedimentation profiles are not presented). Radioactivity was found only in the top fractions containing proteins and the IRES not cross-linked to ribosomal components. This indicated that both RNA I and RNA II did not cross-link to the rRNA.
Identification of ribosomal components cross-linked to photoactivatable HCV IRES derivatives
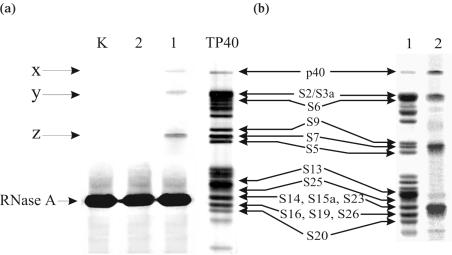
To identify the ribosomal proteins cross-linked to the derivatives of the IRES, two different approaches were used. The first approach was based on the application of the slightly labelled biotinylated RNA transcripts. After irradiation of the binary complexes of RNA I, RNA II and RNA K with 40S subunits, the RNA–protein cross-links were isolated by affinity chromatography on avidin agarose. The RNA cross-linked to the proteins was hydrolysed by RNase A and the proteins released from the adsorbent were resolved by 1D SDS–PAGE followed by silver staining. The results are presented in Figure 3a. In the experiments with RNA I, three bands x, y and z are visible that correspond to the proteins cross-linked to this RNA (lane 1). This cross-linking is specific since no protein bands are seen with RNA II (lane 2) or with RNA K (lane K). To identify the proteins cross-linked to RNA I, one should take into account the fact that the trinucleotide fragment of the RNA remains attached to these proteins after complete hydrolysis with RNase A, as may be easily inferred from the sequence of the HCV IRES around nucleotide A296 carrying the cross-linker (Figure 1a). Such a fragment does not noticeably affect the electrophoretic mobility of ribosomal proteins with molecular masses of 25 kDa and higher (18). The positions of the bands ‘x’ and ‘y’ in lane 1 correspond exactly to those of proteins p40 and S2/S3a, respectively, in lane TP40 (Figure 3a). Thus, bands ‘x’ and ‘y’ may be unambiguously assigned to proteins p40 and S2/S3a cross-linked to RNA I. As for ribosomal proteins with lower molecular masses, the cross-linked nucleotide residues slightly but noticeably retard the electrophoretic mobilities of proteins as compared to those of the unmodified proteins (18). Therefore, band ‘z’ in lane 1 whose position exactly coincides with that of protein S7 in lane TP40 (Figure 3a) should be assigned to the cross-linked S5 rather than the S7 protein (neighboring S9 is not a candidate for the cross-linked protein since product ‘z’ migrates even faster than unmodified S9 in lane TP40). Thus, 1D SDS–PAGE made it possible to identify proteins p40, S2/S3a and S5 as cross-linked to RNA I. At the same time, this set may be incomplete since the strong band corresponding to RNase A could mask bands of other cross-linked proteins, such as S16, S19, S26 and less probably, S20, S14, S15a and S23 (Figure 3a). To examine this possibility, we performed the 1D SDS–PAGE analysis of the proteins cross-linked to a highly labelled RNA I. This analysis (Figure 3b) revealed four main radioactive bands; the positions of the upper three bands are compatible with those of the above mentioned cross-linked proteins p40, S2/S3a and S5. The lower and most intense band is in the position that was occupied by RNase A in Figure 3a. These four intense bands are seen in the background of several weak bands that were not revealed in the experiment presented in Figure 3a. Their origin is not clear and hence their assignment was not done in this work.
Figure 3.
Analysis of 40S ribosomal proteins cross-linked to the derivatives of HCV IRES by 1D PAGE in the presence of SDS. (a) Silver stained gel obtained in experiments with biotinylated IRES derivatives. Lanes 1, 2 and K correspond to the proteins isolated from the irradiated complexes of 40S subunits with RNA I, RNA II and RNA K, respectively. Lane TP40 corresponds to the electrophoregram of total 40S protein; positions of the 40S ribosomal proteins are indicated (35,36). (b) Analysis of proteins cross-linked to labelled RNA I in the binary complex with the 40S subunit. Lane 1, Coomassie stained gel; lane 2, autoradiogram of the gel.
To unambiguously identify cross-linked protein(s) migrating as RNase A and that corresponding to band ‘y’ (S2/S3a) in Figure 3a, we resolved the proteins cross-linked to the highly labelled RNA I by 2D PAGE under alkaline conditions in the first dimension, and in a slightly acidic gel containing SDS (‘basic-SDS’ system) in the second dimension. This system does not allow detection of proteins p40 and S5 that do not enter the gel in the first dimension, but these proteins were already unambiguously identified by SDS–PAGE (see the text above and Figure 3a). On the other hand, the basic-SDS system is the best to resolve candidate proteins S14, S15a, S16, S19, S20, S23 and S26 (see above) and to make a choice between S2 and S3a. To avoid possible effects caused by unspecific binding of proteins with short labelled oligoribonucleotides resulting from hydrolysis of RNA with RNase A, the proteins were separated from the oligonucleotides by gel filtration under strongly denaturing conditions (6 M guanidine chloride) prior to the electrophoretic analysis. The 2D PAGE separation revealed one major and several minor radioactive spots (Figure 4a). It is known that in the basic-SDS system short cross-linked oligonucleotides detectably retard protein mobility almost exclusively in the first dimension (18). Consequently, the radioactive spot of the cross-linked protein should be somewhat shifted to the left of the respective stained spot of the unmodified protein. The only stained spot that is adjacent to, and to the right of the major radioactive spot, is S13/S16; S25 is also to the right but below the radioactive spot (Figure 4b). Therefore, the cross-linked protein is S13/S16 rather than S25. Moreover, cross-linking to protein S25 may be excluded as well since it does not belong to the list of candidate proteins whose bands could be masked by the RNase A band in the SDS–polyacrylamide gel (see above). For the same reason, protein S13 should not be regarded as cross-linked. Therefore, the major radioactive band in the 2D polyacylamide gel corresponds to the cross-linked protein S16. A minor radioactive spot in the upper part of the electrophoregram clearly corresponds to the cross-linked protein S3/S3a (Figure 4b). Thus, combining the sets of proteins identified by SDS–PAGE (p40, S2/S3a and S5) and by 2D PAGE (S3/S3a and S16), we conclude that the HCV IRES with the perfluorophenyl azide modified nucleotide A296 cross-links to proteins p40, S3a, S5 and S16 in the binary complex with the 40S subunit. Of these proteins, S5 and S16 seem to be the preferred targets for cross-linking. Proteins p40, S5 and S16 are homologous to prokaryotic S2, S7 and S9, respectively (28), and S3a has no prokaryotic counterpart (28).
Figure 4.
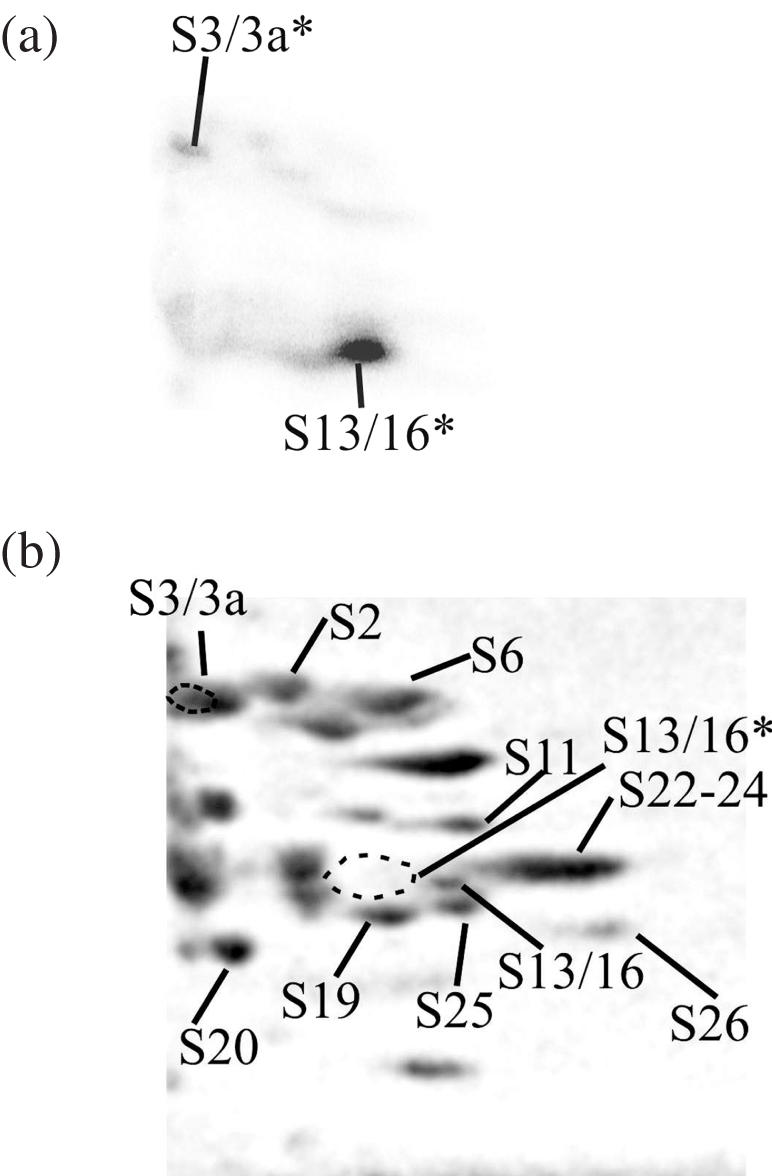
Analysis of proteins cross-linked to RNA I in the binary complex with the 40S subunit by 2D PAGE. (a), autoradiogram; (b), Coomassie stained gel, the positions of the proteins are indicated according to (24). The locations of the radioactive spots corresponding to the cross-linked proteins are indicated on the stained gel by dotted lines. The cross-linked proteins are marked with an asterisk (*).
DISCUSSION
Choice of sequences within the HCV IRES for the site-specific introduction of perfluorophenyl azide cross-linkers
Initially, derivatives of oligodeoxyribonucleotides complementary to specific sequences of the HCV IRES and bearing an alkylating group at the terminal phosphate were applied to obtain covalent adducts of the IRES with the deoxy oligomers. The purified adducts have been then used to study the role of the respective IRES sequences in the binding to 40S subunits (14). Affinity of 40S subunits for the HCV IRES covalent adducts bearing deoxy oligomers complementary to sequences in hairpin IIId and IIIe was drastically lower than that for the unmodified IRES. The lowest affinity was observed for the adduct with the deoxy oligomer complementary to positions 277–295. In contrast, attachment of a deoxy oligomer complementary to the sequence 71–87 in the apical loop of domain II barely affected the affinity of the IRES for the 40S subunits (14). These results were in excellent agreement with other data indicating that HCV IRES subdomains IIId and IIIe are the main determinants for binding to 40S subunits and that domain II is not required for the binding (12,13,15). Based on these results, we chose alkylating derivatives of deoxy oligomers complementary to the HCV IRES sequences 277–295 in hairpin IIIe and 71–87 in domain II to specifically introduce cross-linkers into hairpin IIIe and domain II (Figure 1a). The former oligomer was chosen since it had the strongest inhibitory effect on binding to 40S subunits, and the latter was used in parallel as control. The perfluorophenyl azide employed here as cross-linker makes it possible to reveal ribosomal components located within 14 Å of the specific HCV IRES nucleotide bearing the cross-linker. It can modify both rRNA and proteins as shown in studies with mRNA analogues bearing such cross-linkers (18). The cross-linker introduced at either A296 or G87 did not interfere with binding of the respective HCV IRES derivatives to the 40S subunit (Figure 2c). Thus, as opposed to previous studies, we now present data in which the cross-linked ribosomal components are attributed to specific sites within the IRES.
Ribosomal components neighboring the HCV IRES on the 40S subunit
Our finding that proteins p40, S3a, S5 and S16 lie close to hairpin IIIe of the IRES provides new details, in addition to the earlier cross-linking data (6,10), on the position of the IRES on the 40S subunit. As reported earlier (10), we did not find any contacts of the HCV IRES with the 18S rRNA. Two major proteins S5 and S16, had already been identified as interacting with the HCV IRES (6,10). Protein S5 was a target for UV (254 nm)-induced cross-linking with the HCV IRES (6), and protein S16 was among the proteins cross-linked to the IRES randomly substituted with 4-thiouridines (10). These proteins could not be attributed earlier to the neighbors of the specific structural elements of the IRES since the nucleotides involved in cross-linking remained unknown. Cross-linking of the HCV IRES to proteins p40 and S3a was detected here for the first time. We cannot exclude that these new proteins were cross-linked to the IRES derivative due to the relatively long and flexible perfluoropohenyl azide used here but that these proteins could not be reached by zero-length cross-linkers applied previously (6,10). However, these two reports clearly indicate that even zero-length cross-linking may result in quite different patterns depending on whether a modified (thioU-substituted) or non-modified RNA is used in the cross-linking studies. For instance, no protein S5 was cross-linked to the thio(U)- substituted HCV IRES (10) and no ribosomal proteins other than S5 were identified when using unmodified HCV IRES and UV cross-linking at 254 nm.
The cross-linking approach using dozens of reactive mRNA analogues has been extensively applied for the study of the human 80S ribosome mRNA binding channel [e.g. see (18,23,29,30)].Thus it is worth comparing sets of proteins cross-linked to mRNA analogues and to the HCV IRES derivative; proteins p40 and S16 have never been found to cross-link with mRNA analogues and minor cross-linking to S5/S7 was detected when the cross-linker was in the first (from the 5′ end) position of the E site codon (30). Protein S3a cross-linked to several mRNA analogues when a cross-linker was in the same position or 3 nt upstream (29). We suggest that the environment of hairpin IIIe of the HCV IRES on the 40S subunit overlaps to some extent with the mRNA binding channel in the region of the E site. This overlap seems not to be large and is probably limited to the only S3a protein since protein S26, the main target for cross-linking to perfluorophenyl azide modified nucleotides of the E site codon and upstream (18), was not found in this study.
Correlation of the cross-linking data with cryo-EM models
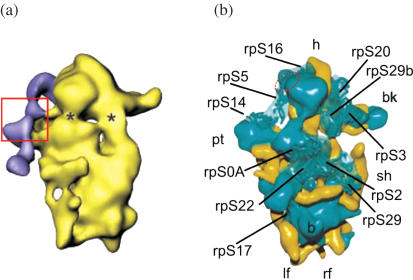
Models of eukaryotic ribosomes at atomic resolution based on X-ray crystallographic data are not yet available, so the cross-linking data may be applied to models of eukaryotic ribosomes created on the basis of cryo-EM data (7,31–33). The most detailed studies on the structural organization of HCV IRES-ribosome complexes were performed by cryo-EM of the binary complexes 40S•HCV IRES (7) and 80S•HCV IRES (33). Figure 5a shows one of views representing the arrangement of HCV IRES on the 40S subunit from rabbit reticulocytes according to (7), however, the model does not show positions of ribosomal proteins on the subunit surface. Proteins that have prokaryotic counterparts have been docked on the model of yeast 40S subunits (31) (Figure 5b); in particular, this model presents the locations of proteins S0A (a homologue of human p40 in yeast), S5 and S16. One can see that all of them are presumably located on the upper part of the solvent side of the subunit. This is basically consistent with the cryo-EM model of the HCV IRES location on the 40S subunit (7,33). Proteins S5 and S16 come very close to each other on the head of the subunit on the site opposite the beak near the exit tunnel for mRNA (7,32). On the other hand, S0A (p40) is located between the platform and the shoulder of the 40S subunit relatively far from S5 and S16 (Figure 5b), at a distance of about 50 Å, meaning several times more than the maximum length of the cross-linker (14 Å). The latter seems to be inconsistent with cross-linking of these proteins with a photoactivatable group at the same nucleotide of the IRES bound to the 40S subunit. Possible reasons for this discrepancy are not fully understood but several explanations for this phenomenon can be put forward. First and the most probable reason is that p40 is significantly larger than its part homologous to prokaryotic counterpart S2 that has been placed in the cryo-EM map. Therefore, the distance between p40 and proteins S5 and S16 might be significantly smaller than 50 Å. Alternatively, a part of p40 may come close to these proteins due to conformational alterations in the 40S subunit caused by binding of the IRES (7) since the location of the proteins mentioned has been proposed for the free 40S subunit (31). Finally, the discrepancies in the distances between the ribosomal components estimated from ‘frozen’ static models and those obtained by means of biochemical methods in solution under near physiological conditions (in particular, cross-linking) may reflect flexibility of the ribosomes, e.g. dynamic movements of the ribosomal parts with respect to each other (34). Although we cannot discriminate between these possibilities, based on the cross-linking results, the most probable site for the location of hairpin IIIe of the HCV IRES on the static model of the eukaryotic 40S subunit can be suggested and is presented in Figure 5b.
Figure 5.
(a) Surface representation of the 40S ribosomal subunit from rabbit reticulocytes in complex with the HCV IRES [adapted from ref. (7)]. The cryo-EM map is shown in yellow. The difference map corresponding to the HCV IRES is superimposed and given in purple. The view from the solvent side. The suggested location of hairpins IIId/e/f is boxed. Asterisk show the proposed entry (right) and exit (left) sites of the mRNA. (b) Structural model of yeast 40S ribosomal subunit obtained from cryo-EM data (solvent side view) [adapted from ref. (31)] in which locations of several ribosomal proteins that have prokaryotic counterparts are shown. Abbreviations used: rp, ribosomal protein; h, head; bk, beak; sh, shoulder; pt, platform; b, body; lf, left foot; rf, right foot. Red dotted oval shows the suggested location of hairpin IIIe.
Protein S3a found here to be a minor target for cross-linking to the IRES derivative has never been docked on cryo-EM models of the eukaryotic 40S subunit since it has no prokaryotic counterparts (28). Data on cross-linking of the HCV IRES derivatives and mRNA analogues to this protein [This report and ref. (29)] now provide a reasonable idea on the location of protein S3a on the ribosome. Most probably, S3a is on the solvent side of the 40S subunit, on its upper part aside from the beak close to the E site and mRNA exit tunnel.
The lack of cross-linking to 40S subunits of the HCV IRES with a photoactivatable group in G87 in domain II is in agreement with the data discussed above that this domain does not contribute to the affinity of the IRES for the 40S subunit. On the other hand, two cryo-EM reconstructions from independent labs (7,33) show a very strong connection between domain II and the head of the 40S at the position of S5, which is powerful evidence for a direct interaction. But in the earlier cross-linking report (10) in which HCV IRES with zero-length cross-linker was applied, no cross-linking to protein S5 was detected. There may be different explanations for these discrepancies. For example, binding of the IRES could block the access of the cross-linking reagent to protein S5, or the protein lacks suitable targets for cross-linking. In any case, the absence of the cross-link cannot indicate lack of interaction, so our results as well as data of (10) do not exclude possible interaction of protein S5 with domain II of HCV IRES. At the same time, our cross-linking results indicate that protein S5 is located also near hairpin IIIe. Thus, the results obtained provide new details concerning the structural organization of the HCV IRES•40S complex and give an idea of the ribosomal area responsible for the initial binding of HCV IRES (Figure 5b).
Our results confirm the data (10) indicating that the 18S rRNA is not determinant for HCV IRES binding to the 40S subunit and are consistent with the suggestion made there that important determinants for IRES binding are most likely formed by amino acid sequences specific of mammalian 40S ribosomal proteins. The latter suggestion was based on the inability of yeast 40S subunits to bind HCV IRES and on large differences in amino acid sequences of the proteins interacting with the IRES and their yeast counterparts. All these data imply that the ability of the small ribosomal subunits to bind HCV IRES appeared at late stages in evolution, whereas ribosomal sites operating during ‘classical’ translation, which are organized mainly by the rRNA (rRNA is much more conserved as compared to ribosomal proteins) were formed at early stages.
Acknowledgments
The authors gratefully thank Anne-Lise Haenni for critical reading of this manuscript. This work was supported by the Russian Foundation for Basic Research (grant #05-04-48357-a to G.K.). The Open Access publication charges for this article were waived by Oxford University Press.
Conflict of interest statement. None declared.
REFERENCES
- 1.Merrick W.C. Mechanism and regulation of eukaryotic protein synthesis. Microbiol. Rev. 1992;56:291–315. doi: 10.1128/mr.56.2.291-315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gingras A.C., Raught B., Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Annu. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 3.Hellen C.U.T., Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 4.Tsukiyama-Kohara K., Iizuka N., Kohara M., Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J. Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pestova T.V., Shatsky I.N., Fletcher S.P., Jackson R.J., Hellen C.U.T. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukushi S., Okada M., Stahl J., Kageyama T., Hoshino F.B., Katayama K. Ribosomal protein S5 interacts with the internal ribosomal entry site of hepatitis C virus. J. Biol. Chem. 2001;276:20824–20826. doi: 10.1074/jbc.C100206200. [DOI] [PubMed] [Google Scholar]
- 7.Spahn C.M.T., Kieft J.S., Grassucci R.A., Penczek P.A., Zhou K., Doudna J.A., Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40S ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 8.Kolupaeva V.G., Pestova T.V., Hellen C.U. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lytle J.R., Wu L., Robertson H.D. Domains on the hepatitis C virus internal ribosome entry site for 40S subunit binding. RNA. 2002;8:1045–1055. doi: 10.1017/s1355838202029965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto G.A., Lukavsky P.J., Lancaster A.M., Sarnow P., Puglisi J.D. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu H., Li W., Noble W.S., Payan D., Anderson D.C. Riboproteomics of the hepatitis C virus internal ribosomal entry site. J. Proteome Res. 2004;3:949–957. doi: 10.1021/pr0499592. [DOI] [PubMed] [Google Scholar]
- 12.Kieft J.S., Zhou K., Jubin R., Doudna J.A. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tallet-Lopez B., Aldaz-Carroll L., Chabas S., Dausse E., Staedel C., Toulme J.J. Antisense oligonucleotides targeted to the domain IIId of the hepatitis c virus IRES compete with 40S ribosomal subunit binding and prevent in vitro translation. Nucleic Acids Res. 2003;31:734–742. doi: 10.1093/nar/gkg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malygin A.A., Graifer D.M., Laletina E.S., Shatsky I.N., Karpova G.G. An approach to identify the functionally important RNA sites by complementary addressed modification. Mol. Biol. 2003;37:873–879. (Translated from Molekulyarnaya Biologiya, 37, 1027–1034) [PubMed] [Google Scholar]
- 15.Otto G.A., Puglisi J.D. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 16.Venkstern T.V., Graifer D.M., Karpova G.G., Morozov I.A. Studying interaction of a derivative of tRNAPhe bearing an aryl azide group at the G-24 with Escherichia coli ribosomes and with tRNA-(adenine-1)methyltransferase from Thermus thermophilus. Biopolymery i kletka (Kiev) 1990;6:59–65. [Google Scholar]
- 17.Zenkova M., Ehresmann C., Caillet J., Springer M., Karpova G., Ehresmann B., Romby P. A novel approach to introduce site-directed specific cross-links within RNA–protein complexes. Application to the Escherichia coli threonyl-tRNA synthetase/translational operator complex. Eur. J. Biochem. 1995;231:726–735. doi: 10.1111/j.1432-1033.1995.0726d.x. [DOI] [PubMed] [Google Scholar]
- 18.Graifer D., Molotkov M., Styazhkina V., Demeshkina N., Bulygin K., Eremina A., Ivanov A., Laletina E., Ven'yaminova A., Karpova G. Variable and conserved elements of human ribosomes surrounding the mRNA at the decoding and upstream sites. Nucleic Acids Res. 2004;32:3282–3293. doi: 10.1093/nar/gkh657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reinolds J.E., Kaminsky A., Kettinen H.J., Grace K., Clarke B.E., Carroll A.R., Rowlands D.J., Jackson R.J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wollenzien P. Isolation and identification of RNA cross-links. Meth. Enzymol. 1988;164:319–329. doi: 10.1016/s0076-6879(88)64052-3. [DOI] [PubMed] [Google Scholar]
- 21.Matasova N.B., Myltseva S.V., Zenkova M.A., Graifer D.M., Vladimirov S.N., Karpova G.G. Isolation of ribosomal subunits containing intact rRNA from human placenta. Estimation of functional activity of 80S ribosomes. Anal. Biochem. 1991;198:219–223. doi: 10.1016/0003-2697(91)90416-q. [DOI] [PubMed] [Google Scholar]
- 22.Graifer D.M., Malygin A.A., Matasova N.B., Mundus D.A., Zenkova M.A., Karpova G.G. Studying functional significance of the sequence 980–1061 in the central domain of human 18S rRNA using complementary DNA probes. Biochim. Biophys. Acta. 1997;1350:335–344. doi: 10.1016/s0167-4781(96)00176-5. [DOI] [PubMed] [Google Scholar]
- 23.Malygin A.A., Graifer D.M., Bulygin K.N., Zenkova M.A., Yamkovoy V.I., Stahl J., Karpova G.G. Arrangement of mRNA at the decoding site of human ribosomes. 18S rRNA nucleotides and ribosomal proteins cross-linked to oligouridylate derivatives with alkylating groups at either the 3′ or the 5′-termini. Eur. J. Biochem. 1994;226:715–723. doi: 10.1111/j.1432-1033.1994.tb20100.x. [DOI] [PubMed] [Google Scholar]
- 24.Madjar J.-J., Arpin M., Buisson M., Reboud J.-P. Spot position of rat liver ribosomal proteins by four different two-dimensional electrophoresis in polyacrylamide gel. Mol. Gen. Genet. 1979;171:121–134. doi: 10.1007/BF00269998. [DOI] [PubMed] [Google Scholar]
- 25.Grineva N.I., Lomakina T.S., Tigeeva N.T., Chimitova T.A. Kinetics of ionization of the C-Cl bond in 4-(N-2-chloroethyl-N-methylamino)benzyl-5′-phosphamides of nucleosides and oligonucleotides. Bioorganicheskaya Khimiya (Moscow) 1977;3:210–214. [Google Scholar]
- 26.Bulygin K.N., Malygin A.A., Karpova G.G., Westermann P. Site specific modification of 4.5S RNA apical domain by complementary oligodeoxynucleotides carrying an alkylating group. Eur. J. Biochem. 1998;251:175–180. doi: 10.1046/j.1432-1327.1998.2510175.x. [DOI] [PubMed] [Google Scholar]
- 27.Karpova G.G. Chemical aspects of complementary-addressed modification of nucleic acids. Izv. Sib. Otd. Akad. Nauk (Novosibirsk) 1987;12:82–95. [Google Scholar]
- 28.Wool I.G., Chan Y.-L., Glueck A. Mammalian ribosomes: the structure and the evolution of the proteins. In: Hershey J.W.B., Matthews M.B., Sonenberg N., editors. Translational control. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1996. pp. 685–732. [Google Scholar]
- 29.Bulygin K.N., Matasova N.B., Graifer D.M., Veniyaminova A.G., Yamkovoy V.I., Stahl J., Karpova G.G. Protein environment of mRNA at the decoding site of 80S ribosomes from human placenta as revealed from affinity labeling with mRNA analogs—derivatives of oligoribonucleotides. Biochim. Biophys. Acta. 1997;1351:325–332. doi: 10.1016/s0167-4781(96)00224-2. [DOI] [PubMed] [Google Scholar]
- 30.Demeshkina N.A., Laletina E.S., Meschaninova M.I., Repkova M.N., Ven'yaminova A.G., Graifer D.M., Karpova G.G. The mRNA codon environment at the E and P sites of human ribosomes deduced from photocrosslinking with pUUUGUU derivatives. Mol. Biol. 2003;37:132–139. (Translated from Molekulyarnaya Biologiya, 37, 147–155) [PubMed] [Google Scholar]
- 31.Spahn C.M.T., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccaromyces serevisiae—tRNA-ribosome and subunit-subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- 32.Morgan D.G., Menetret J.-.F, Neuhof A., Rapoport T.A., Akey C.W. Structure of the mammalian ribosome-channel complex at 17 A resolution. J. Mol. Biol. 2002;324:871–886. doi: 10.1016/s0022-2836(02)01111-7. [DOI] [PubMed] [Google Scholar]
- 33.Boehringer D., Thermann R., Ostareck-Lederer A., Lewis J.D., Stark H. Structure of the hepatitis C virus IRES bound to the human 80S ribosome: remodeling of the HCV IRES. Structure. 2005;13:1695–1706. doi: 10.1016/j.str.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 34.Ivanov V., Mears J. Using cross-links to study ribosomal dynamics. J. Biomol. Struct. Dyn. 2004;21:691–698. doi: 10.1080/07391102.2004.10506962. [DOI] [PubMed] [Google Scholar]
- 35.Collatz E., Ulbrich N., Tsurugi K., Lightfoot H.N., MacKinlay W., Lin A., Wool I. Isolation of eukaryotic ribosomal proteins. J. Biol. Chem. 1977;252:9071–9080. [PubMed] [Google Scholar]
- 36.Malygin A.A., Shaulo D.D., Karpova G.G. Proteins S7, S10, S16 and S19 of the human 40S ribosomal subunit are most resistant to dissociation by salt. Biochim. Biophys. Acta. 2000;1494:213–216. doi: 10.1016/s0167-4781(00)00252-9. [DOI] [PubMed] [Google Scholar]