Abstract
Recent studies on the contamination of groundwater with human enteric viruses have focused on public water systems, whereas little is known about the occurrence of viruses in private household wells. The objective of the present study was to estimate the incidence of viruses in Wisconsin household wells located near septage land application sites or in rural subdivisions served by septic systems. Fifty wells in seven hydrogeologic districts were sampled four times over a year, once each season. Reverse transcriptase PCR (RT-PCR), followed by Southern hybridization, was used to detect enteroviruses, rotavirus, hepatitis A virus (HAV), and Norwalk-like viruses (NLVs). In addition, cell culture was used to detect culturable enteroviruses. Companion water samples were collected for total coliforms, Escherichia coli, fecal enterococci, F-specific RNA coliphages, nitrate, and chloride analyses. Among the 50 wells, four (8%) were positive for viruses by RT-PCR. Three wells were positive for HAV, and the fourth well was positive for both rotavirus and NLV in one sample and an enterovirus in another sample. Contamination was transient, since none of the wells was virus positive for two sequential samples. Culturable enteroviruses were not detected in any of the wells. Water quality indicators were not statistically associated with virus occurrence, although some concordance was noted for chloride. The present study is the first in the United States to systematically monitor private household wells for virus contamination and, combined with data for public wells, provides further insight on the extent of groundwater contamination with human enteric viruses.
Groundwater is a common transmission route for waterborne infectious disease in the United States. Surveillance data since 1981 have shown that approximately half of all waterborne disease outbreaks were associated with contaminated groundwater (18, 39, 43, 46). For 1997 and 1998, the years for which the data have been most recently compiled, 80% (12 of 15) waterborne outbreaks linked to an infectious agent were attributed to drinking contaminated well water (9). Norwalk-like viruses (NLVs) (9, 10, 32, 41) and hepatitis A virus (HAV) (11, 12, 21) have been the most frequently reported viral etiologic agents of groundwater-related outbreaks. Oftentimes, an etiologic agent was not identified in a groundwater-related outbreak, and some of these outbreaks were presumably viral in origin (18). Public health officials suspect that groundwater is responsible for many cases of endemic enteric disease that are too sporadic to easily identify the infection source.
Enteric viruses are the most likely human pathogens to contaminate groundwater. Their extremely small size (25 to 100 nm) allows them to infiltrate soils, eventually reaching aquifers. Depending on factors such as rainfall, temperature, soil structure, organic carbon content, soil pore water pH, cation concentrations, ionic strength, and virus taxon-specific factors such as capsid diameter and isoelectric point, viruses can move considerable distances in the subsurface environment (22, 26, 27, 49, 55, 64). Penetration to depths as great as 67 m and horizontal migration as far as 408 m in glacial till and 1,600 m in fractured limestone have been reported (38, 50). Viruses can persist for several months in soils and groundwater when temperatures are low and soils are moist (35, 50, 58, 65). Enteric viruses are shed in enormous quantities in feces (109 to 1010/g) and have an infectious dose on the order of tens to hundreds of virions (23, 44), so that even an 8-log reduction in virus concentration during transport could still result in infectious virus present in potable groundwater.
Recent studies monitoring groundwater for enteric viruses have focused on public water systems (61). Private household wells, however, may be more vulnerable to viral contamination because they may be maintained less carefully and tested less frequently for water sanitary quality. Moreover, although most states regulate the minimum setback distance between a household well and the closest septic system or field with land-applied wastes, the total number of septic systems surrounding a household or the total volume of land-applied wastes may still result in substantial loading of human fecal wastes in proximity of a well. The primary objective of the present study was to estimate the incidence of human enteric viruses in household wells located near septage land application sites or in subdivisions served by septic systems. The rationale for this approach was that if viruses could not be found in household wells near identifiable fecal sources, contamination would be even less likely in other regions with lower fecal loading rates. Secondary objectives included comparing the occurrence of enteric viruses among wells in different hydrogeologic settings and assessing the predictive value of water quality indicators for virus contamination.
MATERIALS AND METHODS
Well selection and homeowner enrollment.
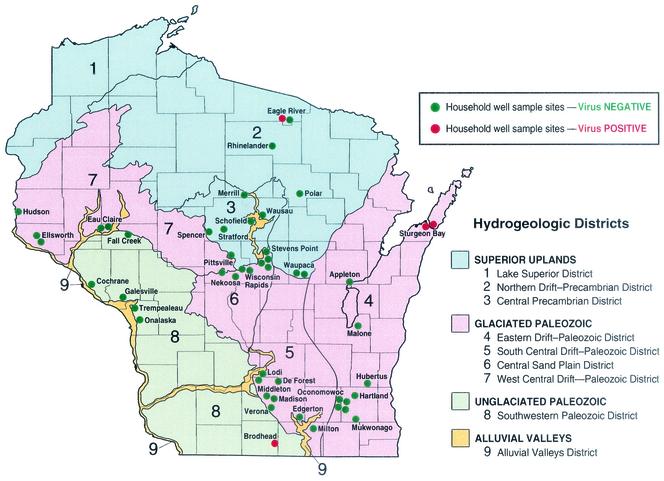
Well selection was stratified by major hydrogeologic districts so that representative wells could be sampled for viruses in seven of the nine major hydrogeologic districts in Wisconsin (66) (Fig. 1). These included district 2 (Northern Drift-Precambrian deposits, specifically the eastern portion with permeable sand and gravel deposits), district 3 (Central Precambrian, fractured Precambrian igneous rock overlain by thin, low permeability till), district 4 (Eastern Drift-Paleozoic sandstone and dolomite, the Silurian dolomite near Sturgeon Bay is fractured and soils are thin), district 5 (South Central Drift-Paleozoic, specifically the southern portion with permeable soils and high water production), district 6 (Central Sand Plain, sand-gravel aquifer that is highly productive and permeable), district 7 (West Central Drift-Paleozoic, sandstone aquifer overlain with Pleistocene deposits, specifically the high-yielding Western portion), and district 8 (Southwestern Paleozoic [Driftless region], sandstone or dolomite aquifers overlain by thin soil). District 1 (Lake Superior) and district 9 (alluvial valleys) were not included in the study.
FIG. 1.
Location of household well sampling sites and virus-positive wells. This illustration was modified from that of Zaporozec and Cotter (66) and is reprinted with permission from the Wisconsin Natural History and Geological Survey.
Wells located near sites with high volumes of land-applied septage or in subdivisions served by septic systems were eligible for virus sampling. Septage is defined as untreated wastewater from septic tanks and does not include sludge or animal manure. Land application sites were identified from Wisconsin Department of Natural Resources records, and sites were ranked in each hydrogeologic district by the total volume of septage applied. The maximum allowable application rate was 13,000 gal/acre/week (i.e., 0.5 in./week) (63). Except under specific conditions that limit human exposures, Wisconsin law requires septage to be limed before application (63). However, the effectiveness of lime in inactivating the full spectrum of virus types in septage has not been fully investigated, and we assumed these sites had high loading rates of infectious viruses. Sites were excluded if septage was not applied the year previous to well selection or if there was not at least one home within 500 m. Septic system sites were identified from Wisconsin Department of Natural Resources well construction reports, and sites were ranked in each hydrogeologic district by the number of septic systems per square mile. Households in areas with the highest septic system densities in a hydrogeologic district were favored for selection into the study. Homeowners were sent a letter describing the study and a few days later were telephoned to request their participation and confirm that their house was near a septage application site or in a subdivision with septic systems. Only one well was selected per septic system or septage land application site.
Sampling schedule.
Two sets of 25 wells (50 unique wells) were sampled; one set was sampled from January 1999 to November 1999, and a second set was sampled from November 1999 to June 2000. Wells were sampled four times, once per season: spring (April), summer (June, July, August), autumn (October, November), and winter (January, February). All samples during a given season were collected within a 3- to 6-week period.
Virus concentration, elution, and flocculation.
A laboratory technician collected all samples from a household tap, selecting taps, when possible, that bypassed water softening or home filtration units. Samples were collected aseptically: gloves were worn, taps were disinfected with 70% ethyl alcohol, and the entire virus filtration apparatus was sterilized with 0.1% chlorine for at least 30 min (24). Enteric viruses were concentrated by passing ca. 1,500-liter tap water through a 1 MDS cartridge filter (CUNO, Inc., Meriden, Conn.) according to established procedures (5). Filters were transported to the laboratory on ice on the day of sample collection and stored <2 days at 4°C. Viruses were eluted with 1 liter of sterile beef extract (1.5% [wt/vol]) with 0.05 M glycine (pH 9.5) passed through the filter twice. The eluate was flocculated by acidification and resuspended in 15 to 30 ml of sterile 0.15 M Na2HPO4 (final pH 7.0 to 7.5) (5). Final concentrated samples were stored at −80°C.
A virus recovery control was performed every 3 to 4 months during the 18-month study period for a total of five recovery controls. After the quality control procedure of the virus-monitoring protocol for the Information Collection Rule (24), 40 liters of dechlorinated tap water was seeded with 200 PFU of attenuated poliovirus type 1 (strain LSc). This test volume was filtered, eluted, and flocculated as described for the field samples. Viruses recovered in the concentrate were enumerated by using Buffalo green monkey kidney cells, and the most-probable-number total culturable virus assay. The recovery efficiency during the study period was 102% ± 43% (mean ± one standard deviation, n = 5).
Viral RNA extraction and purification.
Five hundred microliters of final concentrated sample was extracted with 500 μl of 4 M guanidine isothiocyanate, vortexed for 3 min, combined with 1 ml of buffered acidic phenol-chloroform (5:1), and then vortexed again. After centrifugation (12,000 × g for 20 min), the aqueous portion was combined with an equal volume of chloroform-isoamyl alcohol. Vortexing and centrifugation steps were repeated, and 750 μl of the aqueous layer was applied to a sterile column of 3 ml of DNA-grade Sephadex G-100 (Sigma Chemical Co., St. Louis, Mo.) in a 5-ml syringe barrel. The first 750 μl of column eluate (fraction 1) was discarded. Three successive 750-μl aliquots of Tris-EDTA buffer were applied, and these fractions (fractions 2, 3, and 4) were collected in separate microcentrifuge tubes with 50 μl of Chelex 100 resin (20% [wt/vol]) and stored at −80°C. Preliminary studies in our laboratory examined six column fractions and found that the majority of viral RNA was eluted in fraction 3, lesser amounts were found in fractions 2 and 4, and no RNA was detected in fractions 1, 5, or 6.
RT-PCR.
Reverse transcription-PCR (RT-PCR) was performed to detect five groups of enteric viruses: panenteroviruses (i.e., poliovirus, echoviruses, and coxsackieviruses), rotavirus, HAV, and NLV genogroups 1 and 2. We used a single-tube, large-volume RT-PCR format that has been previously described by Abbaszadegan et al. (2). Reactions were not multiplexed. In brief, 50 μl of chromatography column eluate, 50 μl of nuclease-free water, and 4 μl (2 μg) of random hexamers (Promega, Madison, Wis.) were mixed, heated for 4 min at 99°C, placed on ice, and then supplemented with 186 μl of RT reaction mixture. The mixture components and their final concentrations were as follows: 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 70 μM concentrations of each deoxynucleoside triphosphate (Applied Biosystems, Foster City, Calif.), 200 U of RNasin (Promega), and 500 U of SuperScript II reverse transcriptase (Life Technologies, Rockville, Md.). Reaction tubes were inserted into a thermal cycler (RoboCycler; Stratagene, La Jolla, Calif.), and the following thermal profile was run: 25°C for 15 min, 42°C for 60 min, and 99°C for 5 min and then 4°C until PCR amplification. After the RT reaction, an 8.6-μl PCR cocktail was added containing 10 U of Taq DNA polymerase (Applied Biosystems) and 0.4 μM concentrations of each primer (Integrated DNA Technologies, Coralville, Iowa). Primer pairs are listed in Table 1. Amplification conditions for enteroviruses, rotavirus, and HAV included an initial denaturation step for 4 min at 96°C, followed by 35 cycles of denaturation (94°C for 75 s), annealing (55°C for 75 s), and extension (72°C for 75 s). The amplification conditions for NLVs G1 and G2 were similar to the other virus groups, except that there were 40 cycles and the annealing and extension temperatures were 50 and 60°C, respectively. All amplifications ended with a final extension period of 72°C for 7 min. Reaction products were electrophoresed by using a 1.6% agarose gel containing ethidium bromide, and an amplicon of the size expected for the virus group tested (Table 1) was detected by UV light illumination (Gel-Doc System; Bio-Rad Laboratories, Hercules, Calif.).
TABLE 1.
Primers and probes for enteric virus detection by RT-PCR and Southern hybridization
| Virus group | Primer pairs | Product size (bp) | Primer citation | Internal oligoprobe | Probe citation |
|---|---|---|---|---|---|
| Enteroviruses | CCTCCGGCCCCTGAATG | 196 | DeLeon et al. (20) | CCCAAAGTAGTCGGTTCCGC | Abbaszadegan et al. (1) |
| ACCGGATGGCCAATCCAA | |||||
| Human rotavirus | TTGCCACCAAATTCAGAATAC | 211 | Gentsch et al. (25) | AGAGAGCACAAGTTAATGAAG | |
| ATTTCGGACCATTTATAACC | |||||
| HAV | CAGCACATCAGAAAGGTGAG | 192 | Jaykus et al. (36) | AATGTTTATCTTTCAGCAA | |
| CTCCAGAATCATCTCCAAC | |||||
| NLV G1 | TGTCACGATCTCATCATCACC | 123 | Ando et al. (6) | ACATCAGGAGAGTGCCCACT | Ando et al. (6) |
| GTGAACAGCATAAATCACTGG | ACATCAGGTGATAAGCCAGT | ||||
| GTGAACAGTATAAACCACTGG | ACATCGGGTGATAGGCCTGT | ||||
| GTGAACAGTATAAACCATTGG | |||||
| NLV G2 | TGTCACGATCTCATCATCACC | 123 | Ando et al. (6) | ATGTCAGGGGACAGGTTTGT | Ando et al. (6) |
| TGGAATTCCATCGCCCACTGG | ATGTCGGGGCCTAGTCCTGT | ||||
| Norwalk internal standard | CTTGTTGGTTTGAGGCCATAT | 347 | Schwab et al. (52) | ||
| ATAAAAGTTGGCATGAACA |
Note that a separate RT reaction with 50 μl of chromatography column eluate was run for each of the five virus groups tested. Given the range in final concentrated sample volumes obtained in the present study (11 to 36 ml), the extraction volume of 500 μl, and the column eluate volume of 750 μl, each RT-PCR assay analyzed 0.1 to 0.3% of the original sample volume.
RT-PCR controls included a negative control of the beef extract eluent, a negative control of the RT and PCR cocktails, and a positive control of each virus tested, seeded into beef extract and carried through the same RNA extraction and RT-PCR steps as the field samples. RT-PCRs were batched by using one positive control per batch to minimize the possibility of amplicon contamination.
RT-PCR inhibition control.
Inhibition of the RT-PCR was checked for every well water sample by seeding 50 μl of the chromatography column eluate (fraction 3) with a synthetic RNA control constructed from an amplicon of the Norwalk virus polymerase gene. The control included a 123-bp deletion so that it could be distinguished from wild-type virus amplicon (52). Instead of running the control internally as described previously (52), it was amplified in separate reactions with Norwalk-specific primers NVp35 and NVp36 (Table 1). Master mix composition and thermal cycling conditions were the same as for NLVs G1 and G2. A sample was classified as inhibited if the expected RT-PCR product was not evident after gel electrophoresis.
Southern hybridization.
The amplicon was transferred from the gel to a nylon membrane (Amersham Pharmacia Biotech, Piscataway, N.J.) in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) by using a model 785 vacuum blotter (Bio-Rad) with vacuum applied (130 mm Hg) for 90 min. Gels were depurinated (0.4 M HCl for 15 min) and denatured (0.4 M NaOH for 15 min) before blotting. DNA was cross-linked to the membrane with 1.2 kJ of UV light m−2 (UV Stratalinker 2400; Stratagene). Membranes were prehybridized with hybridization buffer (ExpressHyb; Clontech, Palo Alto, Calif.) in a rotisserie hybridization incubator for 60 min at 42°C. The buffer was replaced with hybridization buffer containing 500 ng of oligonucleotide probe (Table 1) labeled with digoxigenin (DIG). DIG-labeling was performed with the DIG oligonucleotide 3′-end labeling kit (Boehringer Mannheim, Mannheim, Germany), according to the manufacturer's instructions and incorporating the manufacturer's controls. Hybridization was conducted overnight at 42°C. The membrane was washed twice at room temperature with 50 to 100 ml of 2× SSC-0.1% sodium dodecyl sulfate, and then washed an additional two times at 42°C with 50 to 100 ml of 0.1× SSC-0.1% sodium dodecyl sulfate. Hybridized probes were detected by using the DIG nucleic acid detection kit (Boehringer Mannheim), an enzyme-linked immunoassay using anti-DIG alkaline phosphatase conjugate and colorimetric substrates. The color reaction was allowed to proceed for 16 h.
Well water samples were classified as virus positive only when an oligoprobe was hybridized. If an amplicon of the correct molecular weight was observed on the gel, but the probe did not hybridize, the samples were classified as negative. Positive results were confirmed by reanalyzing the sample beginning with a new RNA extract from the final concentrated sample.
Indicator assays.
Samples for water quality indicators were taken at the same time as the virus samples. Microbial indicators included total coliforms, Escherichia coli, fecal enterococci, and F-specific RNA (FRNA) coliphages, and chemical indicators included chloride anion and nitrate. Two chromogenic substrate assays, Colisure (IDEXX Laboratories, Wesbrook, Maine) and Colilert (IDEXX), both of which test total coliforms and E. coli simultaneously, were performed in parallel, and if either assay was positive, a sample was classified as positive. Fecal enterococci were tested by using Enterolert (IDEXX), another chromogenic substrate assay, and scored as present or absent. The sample volume for bacterial indicators was 100 ml per assay.
FRNA coliphages were enumerated by a filtration-elution method and the double-agar-layer plaque assay, modified from methods described by Sinton et al. (53) and Sobsey et al. (54). In brief, 1 liter of well water, with the pH adjusted to neutrality if necessary, was filtered through a sterile 47-mm (pore-size) 1 MDS filter disk (CUNO, Inc.) at a rate <300 ml min−1. Coliphages were eluted from the filter with two 5-ml aliquots of 3% beef extract-1.5% Tween 80 (pH 9.0). The host bacterium E. coli Famp in log-phase growth was added to the top agar layer (50°C, 0.7% agar). This was combined with 2 ml of the beef extract eluate, and the mixture was poured over the bottom nutrient agar layer (1.1% agar). The entire eluate was assayed by pouring five plates per sample at 2 ml of eluate per plate. Plates were incubated overnight at 37°C, and the number of plaques was counted. The positive control was beef extract seeded with MS2 coliphage (ATCC 15597-B1) and the negative control was unseeded beef extract. Recovery efficiency of the filter-elution method was checked each time a batch of coliphage samples was analyzed. The mean recovery efficiency was 83.7% ± 46.7% (mean ± one standard deviation, n = 42).
Chloride was measured with a chloride ion selective combination electrode (Orion model 96-17B; Thermo Orion, Beverly, Mass.) and an ISE meter (Orion model 720A). Nitrate was measured with a nitrate ion selective combination electrode (Orion model 9707 Ionplus) and an Orion ISE meter. A chloride ionic strength adjustor and nitrate interference suppressor solution (both from Thermo Orion) were added to all standards and samples to minimize interferences.
Culturable enteroviruses.
Buffalo green monkey kidney, rhabdomyosarcoma, and Caco-2 cell lines were grown to confluent monolayers in tissue culture flasks with Eagle minimal essential medium with Earle's salts, HEPES buffer, penicillin-streptomycin-fungizone solution, and 10% fetal bovine serum. A 1.0- to 2.5-ml aliquot of final concentrated sample was filtered (Millex-GV, 0.22 μm pore size; Millipore, Bedford, Mass.) and inoculated into each of six 175-cm2 flasks containing cell monolayers and 2.0 to 2.5 ml of sterile saline. There were two flasks per cell line, and thus 6.0 to 15.0 ml of the final concentrated sample was analyzed per sample, which represented 41 to 71% of the original sample volume. Two flasks from each cell line served as controls. The negative control was inoculated with 0.15 M Na2HPO4, the diluent of the final concentrated sample, and the positive control was inoculated with 20 PFU of poliovirus type 1 ml−1. All flasks were rocked for 90 min at room temperature. Cell monolayers were washed with prewarmed saline with 2% fetal bovine serum, and then 50 ml of Eagle minimal essential medium with 2% fetal bovine serum was added to each flask. Flasks were incubated at 37°C for 14 days and examined on an inverted microscope (Nikon Eclipse TS100) for viral cytopathic effects. Positive cultures were frozen at −70°C when 75% of the monolayer showed cytopathic effects. Negative cultures were frozen at −70°C after removal of 40 ml of media and then thawed to release any unobserved virus present in the cells. One milliliter of freeze-thaw supernatant was passed into a 25-cm2 flask containing the same cell line. Flasks were incubated at 37°C and observed for another 14 days to confirm the first passage results.
Statistical analysis.
For each water quality indicator its true-positive rate, true-negative rate, positive predictive value, and negative predictive value were calculated relative to the virus occurrence results (56). These measures of concordance were calculated on a per sample basis, comparing indicator and virus tests for the same sample, and on a per-well basis, comparing indicator and virus tests for the same well, with the well classified as indicator or virus positive if any of its samples were positive. The true-positive rate is defined as the percentage of virus-positive samples (or wells) that were also found to be positive by the indicator, whereas the true-negative rate is the percentage of virus-negative samples (or wells) that were also found to be negative by the indicator. The positive predictive value is the percentage of indicator-positive samples (or wells) that were positive for virus, whereas the negative predictive value is the percentage of indicator-negative samples (or wells) that were negative for virus. In addition, Kappa statistics (17) were calculated, together with 95% confidence limits, as a measure of agreement between the water quality indicators and the virus detection results on a per-sample basis (without adjustment for multiple samples per well) and on a per-well basis.
RESULTS
Well site and construction characteristics.
To achieve the target enrollment of 50 wells, 82 households with private wells had to be contacted for participation in the study (i.e., participation rate = 61%). Five to eleven wells were located in each of seven hydrogeologic districts of Wisconsin (Fig. 1). Twenty-four wells were near septage land application sites (median application rate, 909,000 gal year−1; range, 0.2 to 3.8 million gal year−1), and 26 wells were located in subdivisions served by septic systems (median septic system density, 38 systems/mile2; range, 16 to 186 systems/mile2). Forty-seven wells were constructed by drilling, two were driven point, and one was hydrojetted. The depth, casing depth, and age of the wells varied widely (Table 2). One well was located 100 m from a lake. All other wells were not within sight of a river or lake, and the groundwater was assumed free of surface water influence.
TABLE 2.
Well construction and virus sample summary statisticsa
| Statistic | Well depth (m) (n = 44) | Casing depth (m) (n = 43) | Well age (yr) (n = 42) | Sample volume (liter) (n = 194) | Sample pH (n = 193) | Sample temp (°C) (n = 192) |
|---|---|---|---|---|---|---|
| Mean | 36.6 | 25.3 | 7 | 1,234 | 7.36 | 11.7 |
| Median | 33.2 | 17.1 | 6 | 1,325 | 7.41 | 11.6 |
| Minimum | 8.8 | 7.9 | 2 | 568 | 6.05 | 7.7 |
| Maximum | 93.0 | 73.2 | 22 | 1,605 | 8.54 | 18.6 |
Well construction reports were available for 44 of the 50 wells, and 194 samples were collected. n < 44 or n < 194 is due to missing data.
Sample characteristics.
Forty-six wells were sampled four times, once each season as originally planned, two wells were sampled three times, and two wells were sampled twice for a total of 194 samples. Six samples were missed due to homeowners withdrawing from the study or leaving their residence for the winter. Sample volume, pH, and temperature data are summarized in Table 2. Only two samples had an initial pH of >8.0, requiring addition of HCl during sample collection to facilitate virus adsorption. In homes with water softeners or plumbed filtration units, all samples except one were taken prior to softening, and all samples except seven were taken prior to filtration. Filter types included six cellulose heavy sediment filters and one activated charcoal filter. None of these eight samples were identified later as virus positive.
RT-PCR inhibition.
Inhibition of RT-PCR, as evidenced by failure of the internal standard, was observed in 8% (16 of 194) of the samples. The 16 inhibitory samples were from 14 different wells; two wells had two inhibitory samples. Inhibition appeared to be seasonal since one-half (8 of 16) of the inhibitory samples were collected during the winter, one-quarter (4 of 16) were collected in the fall, and 3 of 16 and 1 of 16 were collected in the summer and spring, respectively. RT-PCR was eventually performed successfully on all 16 samples by diluting the target fraction of the chromatography column 1:10 (13 samples), analyzing the chromatography fraction previous to the target fraction (2 samples), or by both using the earlier fraction and diluting it 1:10 (1 sample). One of the inhibited samples was later determined to be virus positive.
Virus incidence as determined by RT-PCR.
Of the 194 samples tested by RT-PCR for enteric viruses, five samples (3%) were virus positive (Table 3), as evidenced by a positive Southern hybridization blot. Four samples were positive for one virus, and one sample was positive for two viruses, rotavirus, and NLV genogroup 2. HAV was the most commonly identified virus, and each of the other virus groups tested was detected at least once, except for NLV genogroup 1. The amplicon from the enterovirus-positive sample was sequenced and found to have 98% identity with poliovirus type 3 (GenBank BLAST search, e-score = 2 × 10−96).
TABLE 3.
Incidence of enteric viruses in private household wells
| Virus tested | No. of positive samples (n = 194) | No. of positive wells (n = 50) |
|---|---|---|
| Enteroviruses | 1 | 1 |
| Rotavirus | 1 | 1 |
| HAV | 3 | 3 |
| NLV G1 | 0 | 0 |
| NLV G2 | 1 | 1 |
| Any virus | 5a | 4b |
One sample was positive for two viruses.
One well was positive for three viruses.
When virus incidence is expressed on a per-well basis, 4 of the 50 wells sampled (8%) were virus positive (Table 3). Virus-positive wells were found in three of the seven hydrogeologic districts included in the present study (Fig. 1). Two wells were located in the Door County Peninsula near Sturgeon Bay, one well was located in the northern part of the state near Eagle River, and the other well was located in the south near Brodhead.
The construction and site characteristics of the four virus-positive wells were similar (Table 4). All were drilled, had a casing depth in compliance with state code, and were located in subdivisions served by septic systems. The well construction reports for three of the wells described the surface geology. All three were drilled through a coarse textured surface, namely, sand and gravel.
TABLE 4.
Well construction and site characteristics of virus-positive wells
| Well site | Site type | Well type | Age at sampling (yr) | Well depth (m) | Casing depth (m) | Surface geology |
|---|---|---|---|---|---|---|
| Eagle River | SSa | Drilled | 5 | 13.1 | 12.2 | Sand, gravel, clay |
| Sturgeon Bay 1 | SS | Drilled | 22 | 71.3 | 51.8 | No report |
| Sturgeon Bay 2 | SS | Drilled | 3 | 73.2 | 51.8 | Clay-gravel |
| Brodhead | SS | Drilled | 9 | 16.8 | 15.8 | Sand |
SS, septic system site.
Virus occurrence in wells was intermittent. Three of the four positive wells had only one positive sample out of four collected. The other well was positive for enteroviruses in the summer, negative in the autumn, and then positive for rotavirus and Norwalk-like G2 virus in the winter. Viruses were detected in wells only during the summer and winter sampling periods.
Incidence of culturable viruses.
Of 194 samples tested by cell culture, all were negative for culturable viruses. Cytopathic effects were not observed in any of the three cell lines.
Water quality indicators.
The proportions of positive water sanitary quality indicators are reported on both a per-sample and a per-well basis (Table 5). Among microbial indicators, total coliform bacteria were the most common, whereas E. coli was detected in only one sample. FRNA coliphages were detected in two wells. Four wells exceeded the U.S. Environmental Protection Agency maximum contaminant level for NO3 (10 mg liter−1), and 20 wells had a chloride level of >28 mg liter−1. Alhajjar and colleagues (3) measured Cl concentrations in wells down gradient from septic systems at 17 sites in Wisconsin and found the median concentration ranged from 12 to 35 mg liter−1, depending on the distance from the septic system. Based on these numbers, and prior to the statistical analysis, we arbitrarily selected Cl concentrations of >28 mg liter−1 in household wells to indicate fecal contamination.
TABLE 5.
Microbial and chemical indicators of water sanitary quality
| Indicator | No. positive/total no. (%)
|
|
|---|---|---|
| Samples | Wells | |
| Total coliforms | 14/194 (7) | 14/50 (28) |
| E. coli | 1/193 (<1) | 1/50 (2) |
| Fecal enterococci | 5/188 (3) | 5/50 (10) |
| FRNA coliphages | 2/193 (1) | 2/50 (4) |
| NO3 > 10 mg liter−1 | 9/192 (5) | 4/50 (8) |
| Cl > 28 mg liter−1 | 60/192 (31) | 20/50 (40) |
The predictive accuracy of the water quality indicators was generally poor (Table 6). The true-positive rate was 0% on a per-well basis for three indicators: E. coli, fecal enterococci, and FRNA coliphages. In other words, these three indicators were never detected in wells that had a virus present sometime during the year. The highest true-positive rate was 75% for chloride on a per-well basis. However, given the small number of virus-positive results, estimates of true-positive rates were imprecise (e.g., the estimate of 75% has 95% confidence limits that ranges from 19.4 to 99.4%). On a per-well basis, the indicator true-negative rates ranged from 63% for chloride to 97.8% for FRNA coliphages, whereas the highest positive predictive value was only 15% for the chloride indicator. The maximum Kappa statistic was 0.135 for chloride, suggesting slight agreement between this indicator and virus detection results (40), but all Kappa statistics were nonsignificant.
TABLE 6.
Predictive accuracy of water quality indicators for virus occurrence in the same sample or in the same well
| Basis | Indicator | True-positive rate (%)a | True-negative rate (%)b | Positive predictive value (%)c | Negative predictive value (%)d |
|---|---|---|---|---|---|
| Per sample | Total coliforms | 20.0 | 93.1 | 7.1 | 97.8 |
| E. coli | 0 | 99.5 | 0 | 97.9 | |
| Fecal enterococci | 0 | 97.3 | 0 | 97.8 | |
| FRNA coliphages | 0 | 98.9 | 0 | 97.4 | |
| NO3 > 10 mg liter−1 | 0 | 95.2 | 0 | 97.3 | |
| Cl > 28 mg liter−1 | 60.0 | 69.5 | 5.0 | 98.5 | |
| Per well | Total coliforms | 25.0 | 71.7 | 7.1 | 91.7 |
| E. coli | 0 | 97.8 | 0 | 91.8 | |
| Fecal enterococci | 0 | 89.1 | 0 | 91.1 | |
| FRNA coliphages | 0 | 95.7 | 0 | 91.7 | |
| NO3 > 10 mg liter−1 | 0 | 91.3 | 0 | 91.3 | |
| Cl > 28 mg liter−1 | 75.0 | 63.0 | 15.0 | 96.7 |
Percentage of virus-positive samples (or wells) that were also found to be positive by the indicator.
Percentage of virus-negative samples (or wells) that were also found to be negative by the indicator.
Percentage of indicator-positive samples (or wells) that were positive for virus.
Percentage of indicator-negative samples (or wells) that were negative for virus.
DISCUSSION
Virus incidence in household wells.
We believe this is the first study in the United States to have systematically sampled private household wells for human enteric viruses. Of the 50 wells sampled throughout the state of Wisconsin, 4 (8%) were positive for viruses, including HAV, rotavirus, poliovirus, or NLV (genogroup 2). If we express virus incidence on a per-sample basis, 5 of 194 samples (3%) were positive. By comparison, in a study of 30 municipal wells in 17 states and two U.S. territories, seven wells (23%) were positive for enteroviruses by cell culture (61). Using RT-PCR virus detection methods, Abbaszadegan et al. (2) analyzed 150 samples from municipal wells in 35 states and found 30.1, 13.8, and 8.6% to be positive for enteroviruses, rotavirus, and HAV, respectively. Although private household wells may be located closer to fecal sources and maintained less effectively than municipal wells, the household wells in the present study exhibited a lower virus contamination rate. One possible explanation is the lower volume of groundwater drawn from household wells, which would generate a smaller capture zone less likely to overlap with a fecal source and pull in viruses. Because the household wells were purposely selected to be near human fecal sources, 8% may represent the upper limit of contamination, and the actual statewide virus contamination rate in Wisconsin may be lower. On the other hand, the selected wells appear to have been representative of the level of groundwater sanitary quality generally found throughout the state. The Wisconsin State Laboratory of Hygiene tested ca. 15,000 household well water samples in calendar year 2000 and found 20% to be positive for total coliforms and 2% to be positive for E. coli (J. Standridge, unpublished data), similar to the 28 and 2% incidence rates, respectively, reported in the present study. Likewise, the proportion of wells exceeding the U.S. Environmental Protection Agency maximum contaminant level for NO3 (10 mg liter−1) was 8%, similar to the 6.6% statewide rate estimated from a 1994 survey of 534 wells in Wisconsin (15).
It is also possible that low levels of virus contamination were missed due to the small fraction of the water sample analyzed. If we assume a final concentrated sample volume of 30 ml, and after the concentrate is divided into aliquots for RNA extraction and the extract is aliquoted for the RT-PCR, the effective sample volume analyzed would be 1/900 of the sample volume collected. For a 1,500-liter sample, the fraction analyzed was equivalent to 1.7 liters.
Of the four virus-positive wells, three were positive for HAV. This virus has been responsible for a number of groundwater-related outbreaks (11, 12, 21). Compared to other members of the Picornaviridae, which includes enteroviruses, HAV is stable to high temperature and low pH (14). HAV incubated for 8 weeks at 5°C in groundwater was negligibly inactivated, whereas 8 weeks at 25°C were necessary for 99% inactivation, a much longer survival time than that of poliovirus and echovirus measured in the same study (55). The ability to withstand inactivation may explain the frequent occurrence of HAV in groundwater.
Incidence of culturable viruses.
The RT-PCR method for detecting RNA viruses in environmental samples is sensitive and specific but is limited in that it cannot distinguish between infectious and noninfectious viral particles. This limitation is moot if the virus is nonculturable, as are NLVs, and RT-PCR is the only method of detection available. Most enteroviruses, however, are easily cultured, and we used a standard procedure to detect infectious virions, intending to complement the RT-PCR data. Of the 194 samples from 50 wells, none were positive for enteroviruses by cell culture. The one sample positive for poliovirus by RT-PCR was negative by cell culture, suggesting that at the time of sampling the viruses detected were noninfectious. This discrepancy in results between methods highlights the difficulty in interpreting the public health significance of finding viral RNA in drinking water by RT-PCR. An epidemiologic study design, using gastrointestinal infections as the outcome measure and the RT-PCR method to assess virus exposure, would help to clarify whether the method is useful as a measure of public health risk.
Virus occurrence by hydrogeologic district.
Well susceptibility to viral contamination was not restricted to a specific major hydrogeologic district of the state, since three of seven districts had contaminated wells. On a smaller hydrogeological scale, the two contaminated wells near Sturgeon Bay in the Door County peninsula were consistent with previous studies documenting the vulnerability of the groundwater in this region. The peninsula has shallow topsoil underlain with extensively fractured dolomite, allowing contaminants to easily reach groundwater and travel far distances (48). Both wells were modern and conventionally drilled with 52-m deep casings. If we assume the casings were intact and the well covers were properly sealed, the viruses had to be transported to a 52-m depth before they could enter the household water supplies. The wells in the Door County peninsula are probably among the most vulnerable in the state for virus contamination, based upon the hydrogeologic features of this region.
Characteristics of virus-positive wells.
The present study was designed to estimate the incidence of viruses in household wells. The design was not directed at identifying the types of wells, soil factors, or fecal sources associated with virus occurrence. Nonetheless, several observations are noteworthy. First, all four virus-positive wells were relatively new (3 to 22 years old) and complied with Wisconsin state code (e.g., minimum casing depth of 40 ft), suggesting that current well construction practices do not prevent virus contamination. Noncompliant wells may have a greater frequency of virus contamination. Second, consistent with previous reports that virus transport is greatest in sand and gravel (64), three of the four virus-positive wells in the present study were located in coarse textured soils. Third, even though half of the 50 wells enrolled in the study were located near septage land application sites, all of the virus-positive wells were in subdivisions served by septic systems, suggesting that septic systems were more likely to be a contamination source than land application sites. Enteric viruses can pass through a septic drain field and reach groundwater (51, 62). In one study conducted in south-central Wisconsin, after dosing a properly functioning conventional septic system with polioviruses, the virus could be detected over 3 months in a monitoring well located 6 m from the drain field edge (4). The findings of the present study, however, do not preclude septage land application sites as a potential source of viral contamination. Groundwater beneath wastewater irrigation sites has become contaminated with enteric viruses (30, 38). Additional research is needed to identify the sources of virus contamination and the conditions under which household wells are most vulnerable.
Intermittent virus contamination of groundwater.
Only the samples collected during summer and winter months were virus positive. However, whether virus occurrence in Wisconsin household wells is truly seasonal could not be inferred from the small number of viral detects. What can be concluded is that virus occurrence in a particular well is intermittent, and the vulnerability of a well to viral contamination cannot be characterized from a single sample. A priori, sampling on a seasonal basis would appear justified given that hydrogeological parameters that affect virus transport and survival vary seasonally (e.g., recharge rate and groundwater temperature).
RT-PCR inhibition.
Natural waters contain dissolved compounds, such as humic and fulvic acids, that may inhibit the PCR (34, 59). In the present study of well water the presence of inhibitory compounds was sample specific. Oftentimes when one sample was inhibited to RT-PCR, the other three samples from the same well were uninhibited. The majority of inhibited samples were collected during the winter months, the period in Wisconsin with the lowest rate of groundwater recharge (57). The concentration of inhibitory compounds in well water may peak at this time due to the absence of recharge dilution or perhaps due to an influx of dissolved organic matter leached from deciduous leaves dropped several months previously. These findings underscore the importance of checking each well water sample for RT-PCR inhibition.
Inhibition may also depend on the specific set of primers and nucleic acid target of the PCR so, ideally, inhibition should be checked for each virus type tested. The Norwalk virus inhibition control used in the present study included a 123-bp deletion, so if there had been amplicon contamination, it could be distinguished from wild-type virus. Similarly modified controls were not available for the other viruses. Instead, we chose to rely on this single control to assess inhibition and avoid seeding all 194 samples with wild-type strains from each virus group, which would have increased the risk of false positives. The tradeoff was that virus-specific inhibition of some samples may have gone undetected, and the study results then may have included some false negatives.
Association of indicators with virus occurrence.
Ideally, virus presence should be accurately predicted from a microbial or chemical indicator, thereby avoiding the expense, time, and technical expertise involved with virus testing. In previous studies, standard bacterial indicators, such as total coliform bacteria, fecal coliforms, and E. coli, have not been associated with virus occurrence (28, 31, 37), although total coliforms have been associated with waterborne disease outbreaks caused by viruses (19). Similarly, fecal enterococci appear to be a good predictor of gastrointestinal disease risk (8, 13, 45) (M. A. Borchardt et al., unpublished data), but thus far an association between fecal enterococci and the presence of specific viruses in water has not been demonstrated (37). Recently, the focus has been on FRNA and somatic coliphages as potential indicators because they are shed in human feces and, since the size and morphology of some coliphages are similar to the enteric pathogenic virus groups, they presumably also possess similar survival and transport characteristics (7, 42). When the present study was designed, we opted to omit somatic coliphages from the panel of indicator tests because they can infect nonfecal coliform bacteria in the environment, resulting in low fecal specificity (42). FRNA coliphages, which require bacterial F pili expressed only under the elevated temperature found in warm-blooded animals, are presumably more specific to fecal contamination. However, FRNA coliphages are shed by only 3% of humans (42) and may occur infrequently in groundwater (16).
We assessed the predictive accuracy of four microbial indicators and two chemical indicators for virus contamination. The true-positive rate and positive predictive values of all six indicators were low or zero, although the small number of virus-positive samples and wells limited this assessment. Likewise, although the true-negative rates and negative predictive values were generally >90%, these parameters may be biased upward by virtue of the large number of negative results for both viruses and indicators. Taking a more biological instead of statistical perspective, it is plausible for a fecal indicator to be present and pathogenic viruses absent simply due to the lack of infections and virus shedding (i.e., indicator has low positive predictive value). In contrast, if an enteric virus were present, recently shed in feces, one would expect the fecal indicator to be present as well (i.e., indicator has a high true-positive rate). That was not the case in the present study. Of the virus-positive samples or wells, none were positive for E. coli, fecal enterococci, or FRNA coliphages (i.e., true-positive rate = 0%), and the fourth microbial indicator, total coliform, had a true-positive rate of only 25%. While the low predictive accuracy of the bacterial indicators is consistent with previous studies (cited above), the findings for FRNA coliphages may be due to the small number of positive samples. Havelaar et al. (33) reported a strong correlation between the density of FRNA coliphages and the densities of enteroviruses and reoviruses in lake and river water, even though in some samples enteroviruses were present whereas FRNA coliphages were absent. Morinigo et al. (47) also found enteroviruses in the absence of FRNA coliphages in environmental waters.
The comparatively high true-positive rate of the chloride indicator (i.e., when a virus was present, the chloride concentration was elevated) suggests that the virus-positive wells were in a fecal plume. Chloride is excreted in elevated concentrations in human feces and, as a conservative anion when released to groundwater, it is attenuated only by dilution, which makes it an excellent marker for locating the subsurface fecal plume emanating from a septic system (3). Its usefulness as a virus indicator, however, was diminished by its low positive predictive value, only 15%. It may be difficult to find a fecal indicator with a high positive predictive value for the reason given earlier, pathogenic viruses are not present in all feces and also because there may be nonfecal confounding sources of the indicator. In the case of chloride, other anthropogenic sources include deicing salt and fertilizers. The best hope in the face of these difficulties may be an indicator that has some acceptable level of positive predictive value and yet is highly concordant with virus presence (i.e., high true-positive and true-negative rates) and is always absent when viruses are absent (i.e., high negative predictive value).
Epidemiologic implications.
In the United States there are an estimated 267 million episodes of acute diarrhea each year (29). The majority of diarrheal illnesses are endemic (i.e., nonoutbreak). How many of these are attributable to drinking water is unknown, let alone the fraction attributable to drinking from contaminated household wells. Borchardt et al. (unpublished) provided an initial estimate, reporting that in a defined population of children in central Wisconsin, 11% of acute diarrhea of unidentifiable etiology was attributable to drinking from household wells that were positive for fecal enterococci. Alternatively, the relative importance of household wells as a disease transmission route can be gauged from the potential number of people exposed. Fifteen million households in the United States use a private well as their primary drinking water source (60). If we assume the 8% virus contamination rate determined in the present study can be generalized to the nation, then 1.2 million households may be exposed to enteric viruses through their private wells. The RT-PCR assay does not indicate infectivity, and not all exposures result in infection so the actual number of infected households is likely lower. The generalizability of the 8% rate is uncertain. It may be underestimated given that the sanitary quality of Wisconsin groundwater is relatively high compared to that of other Midwest states (15). It may be overestimated because the wells in the present study were deliberately selected to be located near fecal sources. What is certain is that some household wells are contaminated with human enteric viruses, presenting a risk for disease transmission that should be investigated further.
Acknowledgments
This study was funded by grants from the Wisconsin Department of Natural Resources and Marshfield Clinic.
The Norwalk virus internal standard was kindly provided by Mary Estes at the Baylor College of Medicine. Ken Bradbury assisted with site selection, Brad Argue collected samples, Carla Finck provided data management services, and Richard Berg conducted the statistical analyses. Alice Stargardt assisted with manuscript preparation. We sincerely thank Kellogg Schwab for providing technical advice and reviewing an earlier version of the manuscript.
REFERENCES
- 1.Abbaszadegan, M., M. S. Huber, C. P. Gerba, and I. L. Pepper. 1993. Detection of enteroviruses in groundwater with the polymerase chain reaction. Appl. Environ. Microbiol. 59:1318-1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbaszadegan, M., P. Stewart, and M. LeChevallier. 1999. A strategy for detection of viruses in groundwater by PCR. Appl. Environ. Microbiol. 65:444-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alhajjar, B. J., G. Chesters, and J. M. Harkin. 1990. Indicators of chemical pollution from septic systems. Ground Water 28:559-568. [Google Scholar]
- 4.Alhajjar, B. J., S. L. Stramer, D. O. Cliver, and J. M. Harkin. 1988. Transport modeling of biological tracers from septic systems. Water Res. 22:907-915. [Google Scholar]
- 5.American Public Health Association. 1995. Standard methods for the examination of water and wastewater, 19th ed. American Public Health Association, Washington, D.C.
- 6.Ando, T., S. S. Monroe, J. R. Gentsch, Q. Jin, D. C. Lewis, and R. I. Glass. 1995. Detection and differentiation of antigenically distinct small round-structured viruses (Norwalk-like viruses) by reverse transcription-PCR and southern hybridization. J. Clin. Microbiol. 33:64-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Armon, R., and Y. Kott. 1996. Bacteriophages as indicators of pollution. Crit. Rev. Environ. Sci. Technol. 26:299-335. [Google Scholar]
- 8.Barrell, R. A., P. R. Hunter, and G. Nichols. 2000. Microbiological standards for water and their relationship to health risk. Commun. Dis. Public Health 3:8-13. [PubMed] [Google Scholar]
- 9.Barwick, R. S., D. A. Levy, G. F. Craun, M. J. Beach, and R. L. Calderon. 2000. Surveillance for waterborne-disease outbreaks—United States, 1997-1998. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 49:1-21. [PubMed] [Google Scholar]
- 10.Beller, M., A. Ellis, S. H. Lee, M. A. Drebot, S. A. Jenkerson, E. Funk, M. D. Sobsey, O. D. Simmons III, S. S. Monroe, T. Ando, J. Noel, M. Petric, J. P. Middaugh, and J. S. Spika. 1997. Outbreak of viral gastroenteritis due to a contaminated well: international consequences. JAMA 278:563-568. [PubMed] [Google Scholar]
- 11.Bloch, A. B., S. L. Stramer, J. D. Smith, H. S. Margolis, H. A. Fields, T. W. McKinley, C. P. Gerba, J. E. Maynard, and R. K. Sikes. 1990. Recovery of hepatitis A virus from a water supply responsible for a common source outbreak of hepatitis A. Am. J. Public Health 80:428-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowen, G. S., and M. A. McCarthy. 1983. Hepatitis A associated with a hardware store water fountain and a contaminated well in Lancaster County, Pennsylvania, 1980. Am. J. Epidemiol. 117:695-705. [DOI] [PubMed] [Google Scholar]
- 13.Cabelli, V. J., A. P. Dufour, L. J. McCabe, and M. A. Levin. 1982. Swimming-associated gastroenteritis and water quality. Am. J. Epidemiol. 115:606-616. [DOI] [PubMed] [Google Scholar]
- 14.Cederna, J. B., and J. T. Stapleton. 1995. Hepatitis A virus, p. 1025-1032. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 6th ed. American Society for Microbiology, Washington, D.C.
- 15.Centers for Disease Control and Prevention. 1998. A survey of the quality of water drawn from domestic wells in nine midwestern states. [Online.] http://www.cdc.gov/nceh/emergency/wellwater/midwestwell.htm.
- 16.Cliver, D. O., and M. A. Woody. 1995. Survey of Wisconsin groundwater for F-specific RNA coliphages. Technical report WIS WRC 95-02. Water Resources Center, University of Wisconsin, Madison.
- 17.Cohen, J. 1960. A coefficient of agreement for nominal scales. Educ. Psychol. Measure. 20:37-46. [Google Scholar]
- 18.Craun, G. F. 1992. Waterborne disease outbreaks in the United States of America: causes and prevention. World Health Stat. Q. 45:192-199. [PubMed] [Google Scholar]
- 19.Craun, G. F., P. S. Berger, and R. L. Calderon. 1997. Coliform bacteria and waterborne disease outbreaks. J. Am. Water Works Assoc. 89:96-104. [Google Scholar]
- 20.De Leon, R., C. Shieh, R. S. Baric, and M. D. Sobsey. 1990. Detection of enteroviruses and hepatitis A virus in environmental samples by gene probes and polymerase chain reaction, p. 833-853. Proceedings of the American Water Works Association Water Quality and Technology Conference, Denver, Co.
- 21.De Serres, G., T. L. Cromeans, B. Levesque, N. Brassard, C. Barthe, M. Dionne, H. Prud'homme, D. Paradis, C. N. Shapiro, O. V. Nainan, and H. S. Margolis. 1999. Molecular confirmation of hepatitis A virus from well water: epidemiology and public health implications. J. Infect. Dis. 179:37-43. [DOI] [PubMed] [Google Scholar]
- 22.Dowd, S. E., S. D. Pillai, S. Wang, and M. Y. Corapcioglu. 1998. Delineating the specific influence of virus isoelectric point and size on virus adsorption and transport through sandy soils. Appl. Environ. Microbiol. 64:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fields, B. N., D. M. Knipe, P. M. Howley, R. M. Chanock, J. L. Melnick, T. P. Monath, B. Roizman, and S. E. Straus (ed.). 1996. Fields virology, vol. 1, 3rd ed. Lippincott/Williams & Wilkins, Philadelphia, Pa.
- 24.Fout, G. S., F. W. Schaefer III, J. W. Messer, D. R. Dahling, and R. E. Stetler. 1996. ICR microbial laboratory manual. National Exposure Research Laboratory, Office of Research and Development, U.S. Environmental Protection Agency, Cincinnati, Ohio.
- 25.Gentsch, J. R., R. I. Glass, P. Woods, V. Gouvea, M. Gorziglia, J. Flores, B. K. Das, and M. K. Bhan. 1992. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J. Clin. Microbiol. 30:1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerba, C. P., and G. Bitton. 1984. Microbial pollutants: their survival and transport pattern to groundwater, p. 65-68. In G. Bitton and C. P. Gerba (ed.), Groundwater pollution microbiology. John Wiley & Sons, Inc., New York, N.Y.
- 27.Gerba, C. P., and J. B. Rose. 1990. Viruses in source and drinking water, p. 380-396. In G. A. McFeters (ed.), Drinking water microbiology: progress and recent developments. Springer-Verlag, New York, N.Y.
- 28.Gerba, C. P., S. M. Goyal, R. L. LaBelle, I. Cech, and G. F. Bodgan. 1979. Failure of indicator bacteria to reflect the occurrence of enteroviruses in marine waters. Am. J. Public Health 69:1116-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glass, R. I., J. Bresee, B. Jiang, J. Gentsch, T. Ando, R. Fankhauser, J. Noel, U. Parashar, B. Rosen, and S. S. Monroe. 2001. Gastroenteritis viruses: an overview, p. 5-25. In D. Chadwick and J. A. Goode (ed.), Gastroenteritis viruses. John Wiley & Sons, Ltd., Chichester, England. [DOI] [PubMed]
- 30.Goyal, S. M., B. H. Keswick, and C. P. Gerba. 1984. Viruses in groundwater beneath sewage irrigated cropland. Water Res. 18:299-302. [Google Scholar]
- 31.Griffin, D. W., C. J. Gibson III, E. K. Lipp, K. Riley, J. H. Paul III, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Häfliger, D., P. Hübner, and J. Lüthy. 2000. Outbreak of viral gastroenteritis due to sewage-contaminated drinking water. Int. J. Food Microbiol. 54:123-126. [DOI] [PubMed] [Google Scholar]
- 33.Havelaar, A. H., M. van Olphen, and Y. C. Drost. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl. Environ. Microbiol. 59:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ijzerman, M. M., D. R. Dahling, and G. S. Fout. 1997. A method to remove environmental inhibitors prior to the detection of waterborne enteric viruses by reverse transcription-polymerase chain reaction. J. Virol. Methods 63:145-153. [DOI] [PubMed] [Google Scholar]
- 35.Jansons, J., L. W. Edmonds, B. Speight, and M. R. Bucens. 1989. Survival of viruses in groundwater. Water Res. 23:301-306. [Google Scholar]
- 36.Jaykus, L. A., R. De Leon, and M. D. Sobsey. 1996. A virion concentration method for detection of human enteric viruses in oysters by PCR and oligoprobe hybridization. Appl. Environ. Microbiol. 62:2074-2080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Keswick, B. H., and C. P. Gerba. 1980. Viruses in groundwater. Environ. Sci. Technol. 14:1290-1297. [Google Scholar]
- 39.Kramer, M. H., B. L. Herwaldt, G. F. Craun, R. L. Calderon, and D. D. Juranek. 1996. Surveillance for waterborne-disease outbreaks—United States, 1993-1994. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 45:1-33. [PubMed] [Google Scholar]
- 40.Landis, J. R., and G. G. Koch. 1977. The measurement of observer agreement for categorical data. Biometrics 33:159-174. [PubMed] [Google Scholar]
- 41.Lawson, H. W., M. M. Braun, R. I. Glass, S. E. Stine, S. S. Monroe, H. K. Atrash, L. E. Lee, and S. J. Englender. 1991. Waterborne outbreak of Norwalk viral gastroenteritis at a southwest US resort: role of geological formations in contamination of well water. Lancet 337:1200-1204. [DOI] [PubMed] [Google Scholar]
- 42.Leclerc, H., S. Edberg, V. Pierzo, and J. M. Delattre. 2000. Bacteriophages as indicators of enteric viruses and public health risk in groundwaters. J. Appl. Microbiol. 88:5-21. [DOI] [PubMed] [Google Scholar]
- 43.Levy, D. A., M. S. Bens, G. F. Craun, R. L. Calderon, and B. L. Herwaldt. 1998. Surveillance for waterborne-disease outbreaks—United States, 1995-1996. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 47:1-34. [PubMed] [Google Scholar]
- 44.Melnick, J. L., and C. P. Gerba. 1980. Viruses in water and soil. Public Health Rev. 9:185-213. [PubMed] [Google Scholar]
- 45.Moe, C. L., M. D. Sobsey, G. P. Samsa, and V. Mesolo. 1991. Bacterial indicators of risk of diarrhoeal disease from drinking-water in the Philippines. Bull. W. H. O. 69:305-317. [PMC free article] [PubMed] [Google Scholar]
- 46.Moore, A. C., B. L. Herwaldt, G. F. Craun, R. L. Calderon, A. K. Highsmith, and D. D. Juranek. 1993. Surveillance for waterborne disease outbreaks—United States, 1991-1992. Morb. Mortal. Wkly. Rep. CDC Surveill. Summ. 42:1-22. [PubMed] [Google Scholar]
- 47.Morinigo, M. A., D. Wheeler, C. Berry, C. Jones, M. A. Munoz, R. Cornax, and J. J. Borrego. 1992. Evaluation of different bacteriophage groups as faecal indicators in contaminated natural waters in Southern England. Water Res. 26:267-271. [Google Scholar]
- 48.Rayne, T. W., K. R. Bradbury, and M. A. Muldoon. 2001. Delineation of capture zones for municipal wells in fractured dolomite, Sturgeon Bay, Wisconsin, USA. Hydrogeol. J. 9:432-450. [Google Scholar]
- 49.Redman, J. A., S. B. Grant, T. M. Olson, J. M. Adkins, J. L. Jackson, M. S. Castillo, and W. A. Yanko. 1999. Physicochemical mechanisms responsible for the filtration and mobilization of a filamentous bacteriophage in quartz sand. Water Res. 33:43-52. [Google Scholar]
- 50.Robertson, J. B., and S. C. Edberg. 1997. Natural protection of spring and well drinking water against surface microbial contamination. I. Hydrogeological parameters. Crit. Rev. Microbiol. 23:143-178. [DOI] [PubMed] [Google Scholar]
- 51.Scandura, J. E., and M. D. Sobsey. 1997. Viral and bacterial contamination of groundwater from on-site sewage treatment systems. Water Sci. Tech. 35:141-146. [Google Scholar]
- 52.Schwab, K. J., M. K. Estes, F. H. Neill, and R. L. Atmar. 1997. Use of heat release and an internal RNA standard control in reverse transcription-PCR detection of Norwalk virus from stool samples. J. Clin. Microbiol. 35:511-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinton, L. W., R. K. Finlay, and A. J. Reid. 1996. A simple membrane filtration-elution method for the enumeration of F-RNA, F-DNA and somatic coliphages in 100-ml water samples. J. Microbiol. Methods 25:257-269. [Google Scholar]
- 54.Sobsey, M. D., K. J. Schwab, and T. R. Handzel. 1990. A simple membrane filter method to concentrate and enumerate male-specific RNA coliphages. J. Am. Water Works Assoc. 82:52-59. [Google Scholar]
- 55.Sobsey, M. D., P. A. Shields, F. H. Hauchman, R. L. Hazard, and L. W. Caton III 1986. Survival and transport of hepatitis A virus in soils, groundwater and wastewater. Water Sci. Tech. 18:97-106. [Google Scholar]
- 56.Sox, H. C., Jr., M. A. Blatt, M. C. Higgins, and K. I. Marton (ed.). 1998. Medical decision making. Butterworth-Heinemann, Boston, Mass.
- 57.Steuer, J. J., and R. J. Hunt. 2001. Use of a watershed-modeling approach to assess hydrologic effects of urbanization, North Fork Pheasant Branch Basin near Middleton, Wisconsin. USGS Water-Resources Investigations Report 01-4113. U.S. Geological Survey, Washington, D.C.
- 58.Straub, T. M., I. L. Pepper, and C. P. Gerba. 1993. Virus survival in sewage sludge amended desert soil. Water Sci. Tech. 27:421-424. [Google Scholar]
- 59.Tsai, Y. L., and B. H. Olson. 1992. Rapid method for separation of bacterial DNA from humic substances in sediments for polymerase chain reaction. Appl. Environ. Microbiol. 58:2292-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.U.S. Bureau of the Census. 1993. 1990 Census of population. Characteristics of the population, vol. 1. U.S. Government Printing Office, Washington, D.C.
- 61.U.S. Environmental Protection Agency. 2000. Proposed rules. Fed. Regist. 65:30193-30274. [PubMed] [Google Scholar]
- 62.Vaughn, J. M., E. F. Landry, and M. Z. Thomas. 1983. Entrainment of viruses from septic tank leach fields through a shallow, sandy soil aquifer. Appl. Environ. Microbiol. 45:1474-1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wisconsin Administrative Code. 1996. Chapter NR 113. Servicing septic or holding tanks, pumping chambers, grease interceptors, seepage beds, seepage pits, seepage trenches, privies, or portable restrooms. Register, September, no. 489. Wisconsin Department of Natural Resources, Madison, Wis.
- 64.Yates, M. V., and S. R. Yates. 1988. Modeling microbial fate in the subsurface environment. Crit. Rev. Environ. Control 17:307-344. [Google Scholar]
- 65.Yates, M. V., C. P. Gerba, and L. M. Kelley. 1985. Virus persistence in groundwater. Appl. Environ. Microbiol. 49:778-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zaporozec, A., and R. D. Cotter. 1985. Major ground-water units of Wisconsin. University of Wisconsin-Extension, Geological and Natural History Survey, Madison, Wis.