Abstract
Chromatin structure plays a fundamental role in the regulation of nuclear processes such as DNA transcription, replication, recombination, and repair. Despite considerable efforts during three decades, the structure of the 30-nm chromatin fiber remains controversial. To define fiber dimensions accurately, we have produced very long and regularly folded 30-nm fibers from in vitro reconstituted nucleosome arrays containing the linker histone and with increasing nucleosome repeat lengths (10 to 70 bp of linker DNA). EM measurements show that the dimensions of these fully folded fibers do not increase linearly with increasing linker length, a finding that is inconsistent with two-start helix models. Instead, we find that there are two distinct classes of fiber structure, both with unexpectedly high nucleosome density: arrays with 10 to 40 bp of linker DNA all produce fibers with a diameter of 33 nm and 11 nucleosomes per 11 nm, whereas arrays with 50 to 70 bp of linker DNA all produce 44-nm-wide fibers with 15 nucleosomes per 11 nm. Using the physical constraints imposed by these measurements, we have built a model in which tight nucleosome packing is achieved through the interdigitation of nucleosomes from adjacent helical gyres. Importantly, the model closely matches raw image projections of folded chromatin arrays recorded in the solution state by using electron cryo-microscopy.
Keywords: chromatin structure, electron microscopy, linker histone, reconstitution
Eukaryotic chromosomes have a compact structure in which linear nucleosome arrays are first folded into a fiber of around 30-nm diameter (1, 2). The fundamental repeating unit of chromatin, the nucleosome core particle, organizes 147 bp of DNA in 1.7 left-handed superhelical turns around an octamer of the four core histones (H2A, H2B, H3, and H4) (3–5). Linker histone (H1/H5) binding organizes an additional 20 bp of DNA to complete the nucleosome containing 167 bp of DNA (6, 7). Such binding determines the geometry of the DNA entering and exiting the nucleosome core particle (8). In nucleosome arrays, adjacent nucleosomes are separated by linker DNA, varying in length between 0 and 80 bp in a tissue- and species-specific manner (9, 10). In vitro, linear nucleosome arrays fold into the “30-nm” fiber upon increasing ionic strength (11) in a process that depends on both the integrity of the core histone N-terminal tails (12, 13) and the presence of the linker histone (14, 15).
During the past three decades evidence from EM (14–23), x-ray and neutron scattering (24–27), electric and photochemical dichroism (28–31), sedimentation analysis (32–35), nuclease digestion (6, 9, 36), and x-ray crystallography (4, 5, 37, 38) has led to the proposal of a number of different models for the 30-nm fiber. These models fall into two main classes: the one-start helix or solenoid models, and the two-start helix models. The solenoid models are comprised of simple one-start helices in which successive nucleosomes are adjacent in the filament and connected by linker DNA that bends into the fiber interior to accommodate variable DNA lengths (14, 29, 34, 39). Importantly, such models predict that the dimensions of the fiber are determined by invariant nucleosome–nucleosome contacts and are therefore insensitive to variations in the nucleosome repeat length (29, 34). The two-start helix models are based on the zigzag arrangement of nucleosomes observed by EM in low-ionic-strength buffers (16, 18). A common feature of these models is the topology of the linker DNA, which, during compaction, remains extended and essentially straight (20, 22). The two-start helix models can be subdivided into the crossed-linker models and helical-ribbon models, in which the straight linker DNA is oriented essentially perpendicular (20, 22) and parallel (18) to the helix axis, respectively. In contrast to the solenoid models, the two-start helix models predict sensitivity to variations in the average linker DNA length, affecting either the diameter (crossed linker) or length (helical ribbon) of the resulting fiber. Recently, the 9-Å crystal structure of an array of four linked nucleosome cores lacking linker histone showed a nucleosomal arrangement that was interpreted as supporting the two-start helix class of chromatin models (38).
A great deal of controversy surrounds the question of whether the dimensions of the chromatin fiber are sensitive to variations in the average linker DNA length. A handful of studies have reported no significant difference in the diameter of chromatin fibers from species with average linker DNA lengths differing by >20 bp. In contrast, other studies have described variations in fiber diameter of 9 to 11 nm for chromatin differing in average linker length by 36 to 39 bp (19–22, 27). Such inconsistency is probably due to the inherent heterogeneity of the chromatin being analyzed, as well as variations in the experimental conditions used.
To provide a model system for the analysis of the 30-nm chromatin fiber, we have developed an in vitro reconstitution system that generates regularly spaced nucleosome arrays from purified histones and tandem arrays of a strong nucleosome positioning sequence (11). Importantly, our nucleosome arrays contain a native-like composition of both the histone octamer and linker histone and consequently reach a level of compaction equivalent to native chromatin at physiological ionic strength (11). Here, we have extended our in vitro reconstitution method to produce much longer model 30-nm chromatin fibers that are less affected by destabilizing end effects and to which it is much easier to assign directionality. With the aim of distinguishing between the two main model classes, we have used these long model fibers to unambiguously establish the relationship between linker DNA length and the physical dimensions of the chromatin fiber. We have constructed arrays with up to 72 nucleosomes and with a range of different nucleosomal repeat lengths equivalent to linker DNA of between 10 and 70 bp. Our EM measurements establish that there is no linear relationship between linker length and fiber diameter. Instead, we find that two families of chromatin structure are produced with markedly different dimensions. Furthermore, length measurements reveal that in the fully compact state, chromatin fibers are almost twice as compact as generally assumed. From the strong constraints provided by these measurements, we have constructed a model for the 30-nm chromatin fiber. Importantly, different views of the model very closely match raw image projections of fully folded chromatin visualized by electron cryo-microscopy.
Results
Production of Long, Reconstituted 30-nm Chromatin Fibers.
To address the question of whether the dimensions of the 30-nm chromatin fiber are sensitive to variations in linker length, we have produced very long nucleosome arrays containing different nucleosome repeat lengths. The arrays are all based on the strong nucleosome positioning 601 DNA sequence (40) and share the same dyad position. The choice of the nucleosome repeat lengths (177, 187, 197, 207, 217, 227, and 237 bp) was based on two separate experimental findings: (i) DNA lengths found in nature appear to be quantized with a 10-bp repeat and phased such that the broad maxima satisfy the equation 147 bp + 10 n (10); (ii) our own experimental results show that an integer length linker favors folding into the 30-nm chromatin fiber (P.J.J.R. and D.R., unpublished work). The assembly of DNA arrays containing up to 72 tandem repeats of the 601 nucleosome positioning DNA was carried out in several steps to maximize the number of repeats within each array (see Materials and Methods). The inserts, each containing a different nucleosome repeat length DNA, were excised and purified, and their size was estimated from electrophoretic analysis against DNA markers of known size (see Fig. 5, which is published as supporting information on the PNAS web site). The DNA arrays are all very long: the 177-, 187-, 197-, 207-, 217-, 227-, and 237-bp arrays are estimated to contain 72, 52, 61, 47, 55, 66, and 56 nucleosomes, respectively. The different DNA arrays were reconstituted into nucleosome arrays by using histone octamer and linker histone H5 purified from chicken erythrocytes as described in ref. 11. Our reconstitution protocol produces nucleosome arrays with regularly spaced nucleosomes and containing one histone octamer and one linker histone H5 per 200 bp of 601 DNA repeat (11). Titrations with increasing concentrations of histone octamer and linker histone H5 show that in each case a clear point of saturation in binding is reached (Fig. 5). After reconstitution, all seven nucleosome arrays were folded in 1.6 mM MgCl2 (41).
Relationship Between Linker Length and the Diameter of the 30-nm Chromatin Fiber.
Fig. 1A shows a gallery of EM images of negatively stained 30-nm chromatin fibers produced by the seven different nucleosome repeat lengths arrays. Because of their length, ranging from 47 to 72 nucleosomes, the folded fibers lie flat on the EM grid, permitting an unambiguous definition of the path of the fiber axis, and hence the fiber diameter. All seven arrays form very compact structures with apparently uniform diameters. With the exception of the 177-bp nucleosome repeat length (Fig. 1, row 177), the folded arrays exist both as monomeric structures and longer fibers formed through the end-to-end self-association of individual fibers.
Fig. 1.
Relationship between linker length and chromatin fiber diameter. (A) Gallery of side views of negatively stained fibers of 72 × 177-bp (177), 52 × 187-bp (187), 61 × 197-bp (197), 47 × 207-bp (207), 55 × 217-bp (217), 66 × 227-bp (227), and 56 × 237-bp (237) 601 nucleosome arrays reconstituted with saturating concentrations of histone octamer and linker histone H5 and folded in 1.6 mM MgCl2. Images show both individual fibers and longer aggregates formed through the end-to-end stacking of folded arrays as described for native chromatin containing linker histone (48, 53). (B) Images of the 72 × 177-bp fibers captured in the frozen hydrated state by using electron cryo-microscopy. (Scale bar: 100 nm.)
To obtain an accurate estimate of the average fiber diameter, 150 to 300 measurements were taken for each nucleosome repeat length. To ensure consistency between the measurements, tobacco mosaic virus (TMV) particles were added to a subset of the chromatin preparations (177-, 207-, and 237-bp arrays) to act as an internal size standard (Fig. 2A). The average TMV diameter was found to be 18.6 (SD = 1.4), 19.2 (SD = 1.4), and 17.9 (SD = 1.0) nm from the three samples. These measurements are in good agreement with the well established value of 18.0 nm, determined by x-ray fiber diffraction (42), and therefore give confidence that the error associated with all aspects of the measurement process is insignificant (±0.65 nm).
Fig. 2.
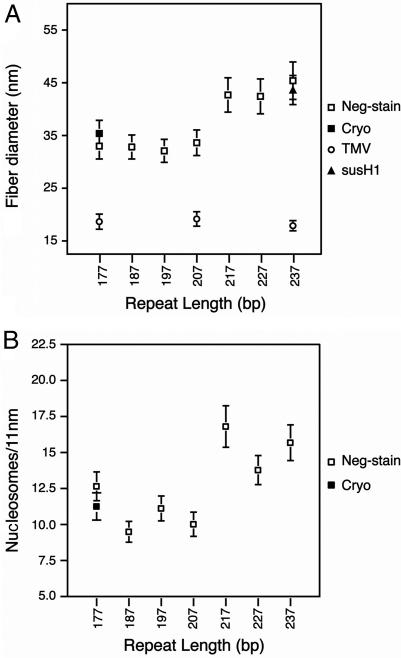
Plots of the diameter and length of the reconstituted and folded 601-nucleosome arrays shown in Fig. 1. (A) Relationship between fiber diameter and linker length. The average diameter dimensions shown are calculated from 150–300 measurements of negatively stained (open squares) and frozen hydrated (filled square) chromatin arrays folded in 1.6 mM MgCl2. Average diameters of TMV (open circles) and the 56 × 237-bp 601 array reconstituted with linker histone H1 purified from sea urchin sperm (filled triangle) are also included. (B) Relationship between the number of nucleosomes and chromatin fiber length. The average length dimensions shown are calculated from 70 to 100 measurements of negatively stained (open squares) and frozen hydrated (filled square) chromatin arrays folded in 1.6 mM MgCl2.
The chromatin diameter measurements plotted in Fig. 2A show the formation of two distinct structural classes. The first class is composed of the repeat lengths 177 to 207 bp, and the second is composed of the 217- to 237-bp repeats. The average fiber diameters of the first structural class are essentially constant: the 177-, 187-, 197-, and 207-bp nucleosome repeat length fibers have average diameters of 33.0 (SD = 2.5), 32.8 (SD = 2.3), 32.1 (SD = 2.2), and 33.6 (SD = 2.4) nm, respectively (Fig. 2A). The differences between these values are smaller than their associated standard deviations and hence are insignificant. Remarkably, an increase of 10 bp of DNA (3.4 nm in length) from 207- to 217-bp repeat length is accompanied by a dramatic increase of ≈10 nm to the average fiber diameter, creating a second class of chromatin fiber. As with the first class, the members of the second structural class have an essentially constant fiber diameter: the 217-, 227-, and 237-bp repeat fibers have average diameters of 42.7 (SD = 3.3), 42.4 (SD = 3.3), and 45.4 (SD = 3.6) nm, respectively (Fig. 2A).
Because the 237-bp nucleosome repeat length is typical of sea urchin sperm (43), we investigated whether reconstitution with the sea urchin specific linker histone H1, which is larger than the H5 used here, would affect the dimensions of the resulting folded 30-nm chromatin fiber. The resulting 237-bp repeat chromatin fibers have an average diameter of 43.6 (SD = 2.8) nm, which lies within the error range for measurements of the 237-bp chromatin fibers containing H5 (Fig. 2A). Therefore, under our experimental conditions, the diameter of the reconstituted and folded chromatin fiber is unaffected by the type of linker histone incorporated.
To evaluate the effects of heavy metal staining, the folded 177-bp nucleosome repeat length array containing 72 nucleosomes was also visualized as an unstained sample by electron cryo-microscopy (Fig. 1B). Because accurate measurements required that the fibers be orientated with their fiber axis perpendicular to the direction of the electron beam, the glutaraldehyde fixed arrays were adsorbed onto a thin carbon layer covering the surface of a standard holey carbon EM grid. From the resulting micrographs, 100 fibers were selected for diameter measurement. In the frozen hydrated state, these fibers have an average diameter of 35.4 (SD = 2.5) nm, which is 7% greater than that observed for the negatively stained 177-bp nucleosome repeat array sample (Fig. 2A). This result suggests that the chromatin fibers undergo a small degree of compaction or shrinkage during the uranyl acetate staining process. However, there is no reason to think that this effect would vary between the different chromatin samples analyzed.
The 30-nm Chromatin Fiber Is Unexpectedly Compact.
Length measurements were taken for ≈100 monomeric and negatively stained fibers from each of the seven different nucleosome arrays (Fig. 1A), as well as ≈70 frozen-hydrated 177-bp nucleosome repeat length fibers (Fig. 1B). By dividing these length measurements by the estimated number of 601 DNA repeats in each array, these data can be represented as an accurate estimate of the nucleosome packing ratio (nucleosomes per 11 nm) (Fig. 2B). The plot of the nucleosome packing ratios shows a similar pattern to that seen in the diameter measurements (Fig. 2A), with the same two structurally distinct classes observed. Again, the first class is composed of the nucleosome repeat lengths 177 to 207 bp with essentially constant values for nucleosome density: the 177-, 187-, 197- and 207-bp repeat length fibers have 12.7 (SD = 1.0) (neg-stain), 9.5 (SD = 0.7), 11.1 (SD = 0.9), and 10.0 (SD = 0.8) nucleosomes per 11 nm, respectively (Fig. 2B). The second class is composed of the 217- to 237-bp repeat length fibers, which have a strikingly different nucleosome density: the 217-, 227-, and 237-bp nucleosome repeat length fibers have 16.8 (SD = 1.4), 13.8 (SD = 1.0), and 15.7 (SD = 1.2) nucleosomes per 11 nm, respectively. Length measurements for this second structural class of fiber involved a more detailed statistical approach (see Materials and Methods).
Discussion
Chromatin Fibers form Two Discrete Structural Classes with High Nucleosome Density.
The production of very homogeneous and tightly folded 30-nm chromatin fibers from long nucleosome arrays with different nucleosome repeat lengths has permitted us to obtain accurate measurements of fiber dimensions and hence nucleosome packing ratios. Importantly, unlike previous chromatin reconstitution experiments (35, 38, 44), our nucleosome arrays contain a native-like complement of linker histone (11), which is critical for obtaining correct folding and maximal compaction (15, 32, 45). We find that over the range of nucleosome repeat lengths analyzed, there are two discrete classes of fiber structure, one 33 nm in diameter and with ≈11 nucleosomes per 11 nm, and the other ≈44 nm in diameter and with ≈15 nucleosomes per 11 nm. This finding resolves the controversy in the literature, showing that the fiber diameter does not increase linearly with linker length. The diameter of our reconstituted fibers containing 40 bp of linker DNA or less is consistent with previous measurements of chromatin isolated from mudpuppy erythrocytes (28-bp linker) (20–22, 27), rat liver (31-bp linker) (17) and chicken erythrocytes (42-bp linker) (17, 19, 23). Furthermore, the larger diameter of ≈44 nm observed for fibers with linker DNA between 50 and 70 bp is in good agreement with measurements from sea urchin sperm chromatin (67-bp linker) (17, 20–22, 27).
The long chromatin fibers of known nucleosome content have also enabled us to determine unambiguously the nucleosome density of fully folded chromatin containing linker histone. We find that the 33-nm-diameter fibers compact to 11 ± 1 nucleosomes per 11 nm, almost twice the value generally accepted. We believe that this value closely represents the nucleosome-packing ratio at physiological ionic strength. We have shown previously that our reconstituted nucleosome arrays have the ionic-strength-dependent compaction properties of native chromatin and, importantly, absolute S20,w values that closely match those from native nucleosome arrays of comparable size (11). Furthermore, folding in MgCl2 concentrations similar to those used in this study results in only a minor increase in compaction from that obtained at physiological ionic strength (≈120 mM).
Previously, Gerchman and Ramakrishnan (25) had reported that the nucleosome packing ratio within isolated chicken erythrocyte chromatin reaches a plateau of 6 to 7 nucleosomes per 11 nm at a salt concentration of 80 mM NaCl (25). However, this value is unlikely to represent fully compact chromatin because sedimentation velocity analysis of both native (32) and reconstituted nucleosome arrays (11) shows that chromatin continues to compact up to an ionic strength of 120 mM. Furthermore, our value of nucleosome density is in close agreement with measurements from selected regions of compact native chromatin using scanning tunneling EM (18) and also satisfies measurements of the local concentration of DNA in metaphase chromosomes (46).
A Compact, One-Start Helical Model for the 30-nm Chromatin Fiber.
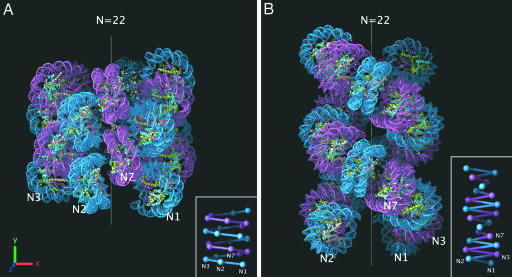
Our measurements of fiber dimensions and tight nucleosome packing provide strong constraints for modeling the 30-nm chromatin fiber. Initially, we attempted to construct a model using both one- and two-start helical arrangements. However, we found that the constraints could only be satisfied by a compact one-start helical structure (Fig. 3A). The model is based on a fiber with a 33-nm diameter because in nature the repeat lengths encompassing the 178-, 188-, and 197-bp maxima are by far the most abundant (10). The model describes a left-handed one-start helix with interdigitation of nucleosomes from successive gyres to achieve the high nucleosome packing ratio we have measured (Fig. 2B). Such interdigitation leads to tight nucleosome packing with regular nucleosome face-to-face contacts that closely relate to the packing interactions between Xenopus laevis nucleosome core particles observed in crystallographic studies (47). The linker DNA has not been modeled (discussed below). The helical parameters of outer radius (r) and subunit axial translation (h) were defined as 16.5 and 1 nm, respectively, being simple derivations from our measurements of diameter and nucleosome packing ratio (Fig. 2 A and B). For the azimuthal rotation angle between subunits (φ), a range of 64° to 83° was calculated based on (i) a lower limit originating from steric clashes, and (ii) an upper limit based on the length of available linker DNA in a 177-bp nucleosome repeat length array (see Fig. 6, which is published as supporting information on the PNAS web site). The model shown in Fig. 3A was constructed with φ equal to 66.4° and consequently has a helical pitch of ≈5.4 nm. To avoid steric clashes the model also incorporates a nucleosome tilt of 17° in relation to the fiber axis. This value corresponds well to the range (13° to 38°) measured for chromatin isolated from a number of different species by using both electric (28, 29) and photochemical dichroism (30, 31). The proposed nucleosome packing arrangement can easily be extended to the 44-nm fiber because the concomitant increase in volume and mass per unit length would result in an essentially constant nucleosome density.
Fig. 3.
Modeling the 30-nm chromatin fiber. (A) Interdigitated one-start helix model built by using the constraints imposed by our measurements of diameter and nucleosome packing ratio (Fig. 2). The helix contains 22 nucleosomes and has a diameter of 33 nm and height of ≈33 nm. Alternate helical gyres are colored marine and magenta. (B) Two-start helical crossed linker model adapted from the idealized model reported by Schalch et al. (38). The model maintains the same parameters for rise per nucleosome and azimuthal rotation angle while extrapolating to a 177-bp nucleosome repeat length by increasing the radius by 3.4 nm (roughly approximating the addition of 10 bp of extra linker DNA lying orthogonal to the fiber axis). The resulting helix contains 22 nucleosomes and has a 28.4-nm diameter and ≈47-nm height. Alternate nucleosome pairs are colored marine and magenta. The positions of the first, second, third, and seventh nucleosomes in the linear DNA sequence are marked on both models with N1, N2, N3, and N7. (Insets) Schematic representations of both atomic models showing the proposed DNA connectivity.
The arrangement of nucleosomes in our model accounts remarkably well for the x-ray diffraction pattern from oriented native chromatin fibers (24). The 55-Å pitch and 110-Å helical repeat of our model clearly explain the 1/55 Å−1 and 1/110 Å−1 meridional reflections in the observed diffraction pattern. The model also provides a convincing explanation for the observed jump in sedimentation coefficient (33) and the sudden increase in gel-migration (48) of arrays between 5 and 6 nucleosomes in length. Whereas five nucleosomes are not sufficient to complete one turn of the fiber, a sixth nucleosome will make intimate face–face contacts with the first, presumably stabilizing the split-ring structure produced and thereby reducing the frictional coefficient of the entire array.
Crucially, the interdigitated model we propose closely matches different views of tightly folded short chromatin arrays captured in an unperturbed solution state by using electron cryo-microscopy (compare Fig. 4A with B). When freely rotated, the atomic model containing 22 nucleosome core particles gives quasi-projections that closely resemble true EM projections of a folded chromatin array containing 22 nucleosomes (177-bp repeat) (Fig. 4B). The raw image projections are most consistent with a highly compact structure of ≈33-nm diameter, showing very limited internal detail. Our model clearly satisfies each of these criteria (Fig. 4B). Our model is most closely related to the family of interdigitated models proposed by Daban and Bermudez (39).
Fig. 4.
Comparison of models to raw images of folded chromatin. (A) Electron cryo-microscopy images of a folded chromatin array containing 22 nucleosomes with a repeat length of 177 bp. The particles are visualized unperturbed in the frozen hydrated state and oriented randomly in ice containing 1.0 mM Mg2+. The particles in each row represent similar views. (Scale bar: 100 nm.) (B) Views of the interdigitated one-start helix model (Fig. 3A) that closely match the EM images from the corresponding row in A. (C) Views of the adapted two-start crossed-linker model (Fig. 3B) that are equivalent to those of the interdigitated model from the corresponding row in B. The varying orientation of the coordinate axes describes the three-dimensional orientation of the models in the corresponding rows.
Chromatin Fiber Dimensions Are Inconsistent with the Two-Start Helical Models.
Recently, Richmond and colleagues (49) proposed a two-start crossed-linker model for the 30-nm chromatin fiber with a diameter of ≈25 nm and 5 to 6 nucleosomes per 11 nm. Our measurements suggest that a two-start crossed-linker model cannot represent the structure of the 30-nm chromatin fiber for a number of reasons. (i) A fundamental prediction of these models is that the fiber diameter will increase almost linearly with increasing linker length (20, 22). Our results clearly show that this is not the case, demonstrating instead the existence of two structural classes each with invariant fiber dimensions. (ii) These models cannot account for the compaction of an array containing 10 bp of linker DNA into a chromatin fiber measuring 33 nm in diameter. The 3.4-nm linker DNA length in this array is too short to span the ≈11-nm distance between the dyads of successive nucleosomes predicted by these models. For example, extrapolating Richmond’s idealized two-start helix model to a 177-bp nucleosome repeat produces a fiber with a maximum possible diameter of 28.4 nm (Fig. 3B), clearly much narrower than the 177-bp fiber measurements described here (Fig. 2A). Indeed, we could not match projection views of the Richmond model with any of our experimental EM data (compare Fig. 4 A with C). (iii) The density of nucleosomes in these models is significantly lower than the value we have determined by direct measurement (Fig. 2B).
Theoretically, it is also possible to construct a 33-nm-wide fiber from nucleosomes with a DNA repeat of 177 bp organized in a zigzag with straight linker DNA. This could be achieved if a zigzag of nucleosomes were coiled into a helical-ribbon structure, with straight linker DNA running almost parallel to the fiber axis (18). However, this type of model is also inconsistent with our data because we fail to observe the significant increase in fiber length that would be expected from this structure upon increasing linker length.
The Path of the Linker DNA.
The constancy of fiber dimensions over the linker length range 10 to 40 bp suggests both that the structure is dominated by conserved nucleosome–nucleosome contacts and that linkers of different lengths must bend to be accommodated in the interior of the fiber. Therefore, it is difficult to reconcile our results with models in which the linker DNA follows a straight path across the centre of the fiber (38). However, consideration of the significant force required to distort the DNA helix suggest that the straight linker models would be more energetically favorable. A solution to this apparent contradiction is likely to lie with the role of the linker histone, which has been shown to be critical in organizing (15) and stabilizing the maximal compaction (11, 32) of nucleosomes within the 30-nm fiber. We suggest that the linker histone is responsible for dictating the geometry of chromatin folding by altering the trajectory of the linker DNA (50) and consequently directing the relative positioning of successive nucleosomes and the pattern of nucleosome–nucleosome contacts. Because a reduction in linker histone content leads to chromatin structural rearrangement and decondensation (51), it is likely that the resulting structures are distinct from the 30-nm fiber investigated here. Full resolution of this issue awaits direct and more detailed structural elucidation of the 30-nm chromatin fiber by using high-resolution approaches.
Materials and Methods
Construction of 601 DNA Arrays.
The array containing 22 repeats of a 177-bp 601 DNA sequence was constructed in the pUC18 vector as described in ref. 11. The long arrays containing from 47 to 72 × 601 DNA tandem repeats were produced in several cloning steps by using the low-copy-number vector pETcoco-1 (Novagen).
Reconstitution and Folding of Nucleosome Arrays.
The histone octamer and linker histone H5 were purified from chicken erythrocyte nuclei as described in ref. 11. The short and long 601 DNA arrays were reconstituted at 125 and 25 μg/ml DNA, respectively, and the molar input ratios of the histone proteins were empirically determined as described in ref. 11. Competitor DNA (147 bp) was added in all reconstitutions at a competitor:601 DNA array weight ratio of 1:1. Folding of the arrays was carried out by dialysis into 1.0 to 1.6 mM MgCl2. Analyses of reconstitution and folding were by electrophoresis in native agarose gels of varying percentage (0.7% to 1.4%). In all cases, electrophoresis was carried out in 17 × 19-cm flat-bed gels, 0.2× TB (18 mM Tris/borate, pH 8.3) electrophoresis buffer at 20 V/cm.
EM.
For negative-stain microscopy, fibers were fixed and imaged as described in ref. 11. For electron cryo-microscopy, fibers produced from arrays containing 22 repeats of a 177-bp 601 DNA sequence were vitrified and imaged within the holes of a holey carbon film by using standard methods (52). Fibers containing 72 × 177-bp repeats were captured and imaged in a vitrified state on a thin carbon film covering the surface of a thicker holey carbon support.
Fiber Measurements.
Dimensions were measured from digitized micrographs by using the web graphics user interface of the spider image-processing package. Pixel measurements were converted into nanometers by calculation of the pixel resolution as follows: pixel resolution = scanner sampling (nm)/magnification. For the 177- to 207-bp nucleosome repeat length arrays the length and width dimensions of the resulting chromatin fibers were unambiguous. However, for fibers containing the 217- to 237-bp repeat lengths, in which the length and width dimensions are similar, a statistical approach was used to distinguish between the average values for both dimensions. For each fiber, perpendicular measurements were recorded along the width/length axes, and each measurement was plotted independently in a histogram. For the three fibers with repeats between 217- and 237-bp DNA, each histogram showed two clear peaks, one of which corresponded closely to the diameter average determined accurately from measurements of long end-to-end stacked fibers, and the other corresponding to the average fiber length (data not shown).
Additional details on array construction and imaging are provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Supplementary Material
Acknowledgments
We thank A. Klug, T. Crowther, and V. Ramakrishnan for critically reading the manuscript and for helpful discussion, and J. Widom and J. Thomas for providing the 601 DNA sequence and sea urchin sperm H1, respectively. This work was supported by the Medical Research Council.
Footnotes
Conflict of interest statement: No conflicts declared.
References
- 1.Davies H. G., Small J. V. Nature. 1968;217:1122–1125. doi: 10.1038/2171122a0. [DOI] [PubMed] [Google Scholar]
- 2.Ris H., Kubai D. F. Annu. Rev. Genet. 1970;4:263–294. doi: 10.1146/annurev.ge.04.120170.001403. [DOI] [PubMed] [Google Scholar]
- 3.Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- 4.Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Nature. 1984;311:532–537. doi: 10.1038/311532a0. [DOI] [PubMed] [Google Scholar]
- 5.Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 6.Noll M., Kornberg R. D. J. Mol. Biol. 1977;109:393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- 7.Allan J., Hartman P. G., Crane-Robinson C., Aviles F. X. Nature. 1980;288:675–679. doi: 10.1038/288675a0. [DOI] [PubMed] [Google Scholar]
- 8.Hamiche A., Schultz P., Ramakrishnan V., Oudet P., Prunell A. J. Mol. Biol. 1996;257:30–42. doi: 10.1006/jmbi.1996.0144. [DOI] [PubMed] [Google Scholar]
- 9.Kornberg R. D. Annu. Rev. Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- 10.Widom J. Proc. Natl. Acad. Sci. USA. 1992;89:1095–1099. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huynh V. A., Robinson P. J., Rhodes D. J. Mol. Biol. 2005;345:957–968. doi: 10.1016/j.jmb.2004.10.075. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Ramirez M., Dong F., Ausio J. J. Biol. Chem. 1992;267:19587–19595. [PubMed] [Google Scholar]
- 13.Moore S. C., Ausio J. Biochem. Biophys. Res. Commun. 1997;230:136–139. doi: 10.1006/bbrc.1996.5903. [DOI] [PubMed] [Google Scholar]
- 14.Finch J. T., Klug A. Proc. Natl. Acad. Sci. USA. 1976;73:1897–1901. doi: 10.1073/pnas.73.6.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thoma F., Koller T., Klug A. J. Cell Biol. 1979;83:403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Worcel A., Strogatz S., Riley D. Proc. Natl. Acad. Sci. USA. 1981;78:1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zentgraf H., Franke W. W. J. Cell Biol. 1984;99:272–286. doi: 10.1083/jcb.99.1.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woodcock C. L., Frado L. L., Rattner J. B. J. Cell Biol. 1984;99:42–52. doi: 10.1083/jcb.99.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Widom J., Finch J. T., Thomas J. O. EMBO J. 1985;4:3189–3194. doi: 10.1002/j.1460-2075.1985.tb04064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Williams S. P., Athey B. D., Muglia L. J., Schappe R. S., Gough A. H., Langmore J. P. Biophys. J. 1986;49:233–248. doi: 10.1016/S0006-3495(86)83637-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Athey B. D., Smith M. F., Rankert D. A., Williams S. P., Langmore J. P. J. Cell Biol. 1990;111:795–806. doi: 10.1083/jcb.111.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith M. F., Athey B. D., Williams S. P., Langmore J. P. J. Cell Biol. 1990;110:245–254. doi: 10.1083/jcb.110.2.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Woodcock C. L. J. Cell Biol. 1994;125:11–19. doi: 10.1083/jcb.125.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widom J., Klug A. Cell. 1985;43:207–213. doi: 10.1016/0092-8674(85)90025-x. [DOI] [PubMed] [Google Scholar]
- 25.Gerchman S. E., Ramakrishnan V. Proc. Natl. Acad. Sci. USA. 1987;84:7802–7806. doi: 10.1073/pnas.84.22.7802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Graziano V., Gerchman S. E., Schneider D. K., Ramakrishnan V. Nature. 1994;368:351–354. doi: 10.1038/368351a0. [DOI] [PubMed] [Google Scholar]
- 27.Williams S. P., Langmore J. P. Biophys. J. 1991;59:606–618. doi: 10.1016/S0006-3495(91)82276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGhee J. D., Rau D. C., Charney E., Felsenfeld G. Cell. 1980;22:87–96. doi: 10.1016/0092-8674(80)90157-9. [DOI] [PubMed] [Google Scholar]
- 29.McGhee J. D., Nickol J. M., Felsenfeld G., Rau D. C. Cell. 1983;33:831–841. doi: 10.1016/0092-8674(83)90025-9. [DOI] [PubMed] [Google Scholar]
- 30.Mitra S., Sen D., Crothers D. M. Nature. 1984;308:247–250. doi: 10.1038/308247a0. [DOI] [PubMed] [Google Scholar]
- 31.Sen D., Mitra S., Crothers D. M. Biochemistry. 1986;25:3441–3447. doi: 10.1021/bi00359a052. [DOI] [PubMed] [Google Scholar]
- 32.Butler P. J., Thomas J. O. J. Mol. Biol. 1980;140:505–529. doi: 10.1016/0022-2836(80)90268-5. [DOI] [PubMed] [Google Scholar]
- 33.Thomas J. O., Butler P. J. J. Mol. Biol. 1980;144:89–93. doi: 10.1016/0022-2836(80)90215-6. [DOI] [PubMed] [Google Scholar]
- 34.Butler P. J. EMBO J. 1984;3:2599–2604. doi: 10.1002/j.1460-2075.1984.tb02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tse C., Hansen J. C. Biochemistry. 1997;36:11381–11388. doi: 10.1021/bi970801n. [DOI] [PubMed] [Google Scholar]
- 36.Staynov D. Z. Nucleic Acids Res. 2000;28:3092–3099. doi: 10.1093/nar/28.16.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramakrishnan V., Finch J. T., Graziano V., Lee P. L., Sweet R. M. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 38.Schalch T., Duda S., Sargent D. F., Richmond T. J. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 39.Daban J. R., Bermudez A. Biochemistry. 1998;37:4299–4304. doi: 10.1021/bi973117h. [DOI] [PubMed] [Google Scholar]
- 40.Lowary P. T., Widom J. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 41.Bartolome S., Bermudez A., Daban J. R. J. Cell Sci. 1994;107:2983–2992. doi: 10.1242/jcs.107.11.2983. [DOI] [PubMed] [Google Scholar]
- 42.Namba K., Stubbs G. Science. 1986;231:1401–1406. doi: 10.1126/science.3952490. [DOI] [PubMed] [Google Scholar]
- 43.Spadafora C., Bellard M., Compton J. L., Chambon P. FEBS Lett. 1976;69:281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- 44.Dorigo B., Schalch T., Bystricky K., Richmond T. J. J. Mol. Biol. 2003;327:85–96. doi: 10.1016/s0022-2836(03)00025-1. [DOI] [PubMed] [Google Scholar]
- 45.Carruthers L. M., Hansen J. C. J. Biol. Chem. 2000;275:37285–37290. doi: 10.1074/jbc.M006801200. [DOI] [PubMed] [Google Scholar]
- 46.Daban J. R. Biochemistry. 2000;39:3861–3866. doi: 10.1021/bi992628w. [DOI] [PubMed] [Google Scholar]
- 47.White C. L., Suto R. K., Luger K. EMBO J. 2001;20:5207–5218. doi: 10.1093/emboj/20.18.5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bartolome S., Bermudez A., Daban J. R. J. Biol. Chem. 1995;270:22514–22521. doi: 10.1074/jbc.270.38.22514. [DOI] [PubMed] [Google Scholar]
- 49.Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. Science. 2004;306:1571–1573. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 50.Sivolob A., Prunell A. J. Mol. Biol. 2003;331:1025–1040. doi: 10.1016/s0022-2836(03)00831-3. [DOI] [PubMed] [Google Scholar]
- 51.Fan Y., Nikitina T., Zhao J., Fleury T. J., Bhattacharyya R., Bouhassira E. E., Stein A., Woodcock C. L., Skoultchi A. I. Cell. 2005;123:1199–1212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 52.Dubochet J., Adrian M., Chang J. J., Homo J. C., Lepault J., McDowall A. W., Schultz P. Q. Rev. Biophys. 1988;21:129–228. doi: 10.1017/s0033583500004297. [DOI] [PubMed] [Google Scholar]
- 53.Thomas J. O., Rees C., Pearson E. C. Eur. J. Biochem. 1985;147:143–151. doi: 10.1111/j.1432-1033.1985.tb08730.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.