Abstract
Lactobacillus salivarius subsp. salivarius strain UCC118 is a bacteriocin-producing strain with probiotic characteristics. The 2.13-Mb genome was shown by sequencing to comprise a 1.83 Mb chromosome, a 242-kb megaplasmid (pMP118), and two smaller plasmids. Megaplasmids previously have not been characterized in lactic acid bacteria or intestinal lactobacilli. Annotation of the genome sequence indicated an intermediate level of auxotrophy compared with other sequenced lactobacilli. No single-copy essential genes were located on the megaplasmid. However, contingency amino acid metabolism genes and carbohydrate utilization genes, including two genes for completion of the pentose phosphate pathway, were megaplasmid encoded. The megaplasmid also harbored genes for the Abp118 bacteriocin, a bile salt hydrolase, a presumptive conjugation locus, and other genes potentially relevant for probiotic properties. Two subspecies of L. salivarius are recognized, salivarius and salicinius, and we detected megaplasmids in both subspecies by pulsed-field gel electrophoresis of sizes ranging from 100 kb to 380 kb. The discovery of megaplasmids of widely varying size in L. salivarius suggests a possible mechanism for genome expansion or contraction to adapt to different environments.
Keywords: megaplasmid, probiotic, heterofermentation
Lactobacilli are widely used for fermenting foods products or as adjuncts to foodstuffs. Many Lactobacillus species have been shown to confer probiotic properties, meaning that, upon ingestion, they confer a range of benefits on the host (1). Lactobacilli are part of the normal human gastrointestinal (GI) microbiota, and they may also be found in the GI tracts of other mammalian species (2–4). Bacteria belonging to the species investigated in this study, Lactobacillus salivarius, have been isolated from the intestinal mucosa of 9% of human subjects examined (5), the tongue and rectum of 12% of healthy adults sampled (6), and feces of infants (7). L. salivarius subsp. salivarius strain UCC118 was isolated from the terminal ileum of a (otherwise healthy) patient undergoing urinary tract reconstructive surgery (8). This strain has been extensively studied for its probiotic properties in human trials and animal models (8–11). In addition, L. salivarius UCC118 produces a two-component bacteriocin Abp118 (12), which has broad spectrum activity against Gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (8).
The application of genomic technologies recently has led to major advances in our understanding of lactobacilli (13, 14), through genome sequence determination for Lactobacillus plantarum, Lactobacillus johnsonii, Lactobacillus acidophilus, and Lactobacillus sakei (15–18). L. plantarum has the largest Lactobacillus genome sequenced to date (3.3 Mb) and has a commensurate biochemical complexity. For example, L. plantarum possesses enzymes for the biosynthesis of all amino acids except leucine, isoleucine, and valine, whereas L. johnsonii is predicted to be incapable of synthesizing any amino acids (19). L. sakei, although displaying the highest levels of orthology with L. plantarum, is unable to synthesize 18 amino acids (18). L. johnsonii and L. acidophilus are more related to each other than to L. plantarum (see also below) and have smaller genomes, reflected in nutritional fastidiousness. L. acidophilus appears to be auxotropic for 14 amino acids (17). L. johnsonii lacks the ability for de novo synthesis of purines and cofactors (16, 19). In common with Streptococcus thermophilus (20), which has been passed for centuries in the nutrient-rich medium of milk, L. johnsonii and L. acidophilus appear to have undergone genome reduction in adapting to a lifestyle of close host association.
The genus Lactobacillus is very diverse, which is evident from the lack of long-range synteny observed between the four available complete Lactobacillus genomes and the assembled genome from Lactobacillus gasseri (refs. 13, 14, and 19 and unpublished analyses). The Lactobacillus 16S rRNA gene phylogeny (available as Fig. 4, which is published as supporting information on the PNAS web site) shows that L. salivarius is part of a distinct clade at the periphery of the genus that is not represented by completed or in-progress genome sequence projects. The genome of L. salivarius UCC118 described here reveals the presence of a 242-kb megaplasmid, which, although apparently dispensable for viability based on gene content, confers on the strain a large number of contingency metabolic capabilities and traits directly related to GI tract survival or competitiveness.
Results and Discussion
General Genome Features.
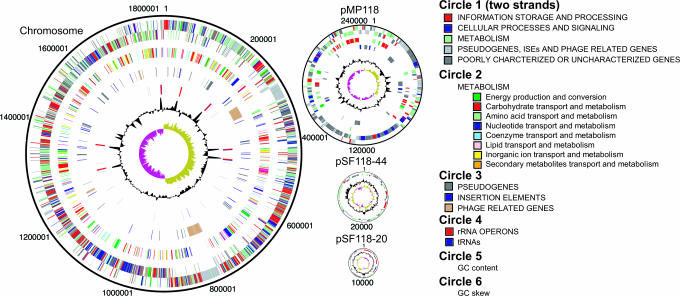
The genome sequence of L. salivarius subsp. salivarius strain UCC118 consists of 2,133,977 nucleotides with an average GC content of 33.04%. The genome comprises four replicons (Fig. 1), a circular chromosome of 1,827,111 nucleotides, a previously undescribed megaplasmid of 242,436 nucleotides designated pMP118, and two previously described plasmids of 20.4 kb (pSF118–20) and 44 kb (pSF118–44) (21). The two smaller plasmids have elevated GC contents (>39%), whereas the GC content of pMP118 is very close to that of the chromosome. The coding densities of the chromosome and pMP118 are 84% and 77%, respectively. Functions could be predicted for 71% of the chromosomal genes, whereas half of the genes on the megaplasmid had no predicted functions, and many of these genes were clustered in two distinct regions (Fig. 1). Seven rRNA operons and 78 tRNAs, representing all 20 amino acids, were detected in the genome and arranged in 25 clusters.
Fig. 1.
Genome atlas of L. salivarius UCC118. The color coding of the genomic features in circle 1 and 2 represents different Clusters of Orthologous Groups categories.
Multireplicon genome architecture increasingly has become recognized in bacteria (reviewed in refs. 22 and 23) and may include minichromosomes, which are difficult to formally distinguish from megaplasmids (24). We designated pMP118 as a megaplasmid for the following reasons: It contains neither tRNA nor rRNA genes; it has plasmid-related replication and partition proteins; and it does not contain the only copy in the genome of any known essential gene. Plasmid-encoded Rep proteins bind to specific sequences in the plasmid replication origin, initiating replication and facilitating binding of host proteins (25). The repA gene of pMP118 (LSL_1739) coincides with the position of a switch in GC skew that is characteristic of replication origins (23, 26). The repA gene product has the highest identity (33%) with the RepA/RepE protein of the Enterococcus faecalis theta-replicating plasmid pS86 (27). The pMP118 ori region includes a second Rep protein, LSL_1740, that is weakly related to staphylococcal plasmid replication proteins. The ori region extends to a gene LSL_1741, whose product contains the Pfam motif associated with presumptive chromosome partitioning ATPases, ParA. LSL_1741 is 41% identical to a copy number control protein of the Listeria innocua plasmid pLI100. The origin is disrupted by three pseudogenes between LSL_1740 and LSL_1741. RepA proteins usually bind to repetitive motifs called RepA boxes or iterons (25). We could not identify corresponding sequences in pMP118. The megaplasmid also has an asymmetric GC skew pattern (Fig. 1), which may be indicative of recent DNA acquisition or loss (28). Smaller plasmids like pSF118-20 and pSF118-44 are relatively common in lactobacilli (29).
Four extensive regions of bacteriophage-related DNA were detected in the genome (Fig. 1), two of which appeared to be intact prophage and two of which were remnants. The bacteriophage content of L. salivarius UCC118 will be described in detail elsewhere (M. Ventura, C.C., V. Bernini, E. Altermann, R. Barrangou, S. McGrath, M.J.C., Y.L., S.L., C. D. Walker, R. Zink, E. Neviani, J. Steele, J. Broadbent, T. R. Klaenhammer, G.F.F., P.W.O., and D.v.S., unpublished data). The other extended region of extensive GC content deviation was related to exopolysaccharide (EPS) production (EPS cluster 2; LSL_1547–LSL_1574; see below). Alignment of the concatenated L. salivarius UCC118 genome against those of L. plantarum, L. johnsonii, L. acidophilus, and L. sakei revealed no long-range synteny (Fig. 5, which is published as supporting information on the PNAS web site). The greatest overall number of orthologs was shared with L. plantarum (61.1% of L. salivarius proteins), followed by E. faecalis (50.4%), L. sakei (49.8%), L. acidophilus (46.3%), and L. johnsonii (45.1%).
Intergenic shuffling that modulates restriction-modification (R/M) activity is a recognized phenomenon in lactic acid bacteria and other groups (30, 31). We identified a type I R/M system (LSL_0915–LSL_0920) in the L. salivarius genome that appeared to be subject to phase variation controlled by DNA inversion events at intragenic inverted repeat sites (Fig. 6, which is published as supporting information on the PNAS web site). The 9,360-bp shufflon contains genes for two extra complete HsdS subunits, downstream and outside of the R/M operon, that allow for nine combinations of the active HsdS subunit. Interestingly, sequence reads confirmed the presence of all nine HsdS combinations in the clonal growth used for DNA preparation. Based on sequence reads, the combination A″B″ was most abundant at 38%, followed by AB and A′B′ (15% each), AB′ and A′B (6% each), and almost equal representations of other combinations. This sequence read distribution indicates that a culture of L. salivarius UCC118 contains a heterogenous collection of different restriction enzymes and differently methylated chromosomes, thus providing a highly effective barrier against bacteriophage infection.
Pseudogenes and Transposable Elements.
The genome includes 73 pseudogenes (Table 1, which is published as supporting information on the PNAS web site), defined as genes that have frameshifts, stop codons, large deletions, insertion sequence (IS) insertions, or truncations of at least 10% of the sequence. A disproportionately high fraction (27%) are located on pMP118. Among pseudogenes, genes with unknown function are the most common, followed by transposase-encoding genes, genes predicted to encode surface proteins, restriction/modification systems, and ABC transporters. Frame shifting is the most common cause of gene inactivation. For example, the megaplasmid harbors a full-length prtP gene homolog (putatively encoding the major caseinolytic cell-surface protease PrtP; LSL_1774b) that contains a single frameshift.
Transposable elements and, in particular, IS element are common features in bacterial genomes, and transposons have been shown to be means of gene transfer. Scattered across the two larger replicons, we identified 16 different IS elements, present in a total of 43 copies (11 of which are nonfunctional because of frameshifts, truncations, and in-frame stop codons) representing 10 IS families (Table 2, which is published as supporting information on the PNAS web site). The largest number of IS elements belonged to the ISLasa3 homology group, with 21 copies of this IS element uniformly distributed over the chromosome and one additional copy on the megaplasmid. Remaining IS elements were found in either single or double copies. Two of these tandem pairs were present as almost identical arrangements, together with flanking DNA, in both the chromosome and the megaplasmid (LSL_732–LSL_733 corresponding with LSL_1779–LSL_1780, and LSL_0049–LSL_0049b corresponding with LSL_1957–LSL_1958). The megaplasmid contains almost 25% of the IS element content of the genome. It is possible that IS elements contributed to formation of pMP118 by a cointegration mechanism, which was suggested for the formation of an archaeal megaplasmid (24). Overall, L. salivarius possesses both the highest number and diversity of IS elements in Lactobacillus genomes sequenced to date.
Biosynthetic Capabilities.
Major pathways are provided as Figs. 7–14, which are published as supporting information on the PNAS web site. L. salivarius has an alanine dehydrogenase gene (EC1.4.1.1; LSL_1768) located on the megaplasmid, which is unique among published Lactobacillus genomes but present in Lactobacillus casei and Lactobacillus delbrueckii draft genomes (www.jgi.doe.gov). This enzyme catalyzes the NAD+-dependent reversible reductive amination of pyruvate into alanine. It is most frequently found in the genus Bacillus and has been exploited for engineering of L. lactis to produce l-alanine (32). By virtue of the pMP118-encoded enzyme, l-alanine can be synthesized from pyruvate. Linked to this alanine dehydrogenase gene is a gene encoding a putative alanine permease (LSL_1767); both genes show elevated GC content (40.37% and 41.5% for LSL_1767 and LSL_1768, respectively), indicating lateral transfer. The megaplasmid also encodes a paralog (LSL_1927) for one of the two enzymes required for conversion of pyruvate to l-aspartate.
The megaplasmid also harbors two genes (LSL_1931 and LSL_1932) predicted to encode the alpha and beta subunits of l-serine dehydratase (EC 4.3.1.17), which catalyzes pyruvate-serine interconversion. Serine formed from pyruvate can subsequently be converted to glycine by a chromosomally encoded enzyme. Serine may be thiolated to cysteine by CysK (EC 2.5.1.47; LSL_0026 and LSL_1718), and cysteine can be converted to methionine by using four chromosomally encoded enzymes. Genes whose products are predicted to synthesize aspartate from pyruvate, and lysine and threonine from aspartate, were also annotated. Unlike L. plantarum, L. salivarius appears to lack the genes required for synthesis of tryptophan and related amino acids. In summary, L. salivarius UCC118 can synthesize de novo or by interconversion nine amino acids, can convert glutamine to three more, and is theoretically auxotrophic for eight amino acids. This level of auxotrophy makes it clearly more dependent on uptake of extracellular amino acids than L. plantarum, less dependent than L. acidophilus (auxotrophic for 14), and far less dependent than L. johnsonii or L. sakei (auxotrophic for 18). However, L. sakei has numerous other adaptations for living in the particular nutrient-rich meat environment (18) where extracellular compounds, including amino acids, are freely available.
In L. salivarius UCC118, the pentose phosphate pathway (see below) may be used for phosphoribosyl pyrophosphate (PRPP) generation. The L. salivarius UCC118 chromosome encodes a complete pathway for the biosynthesis of UMP that can be further converted to UTP and CTP via a pathway apparently requiring the megaplasmid gene LSL_1936 (nucleoside-diphosphate kinase; Dnk). However, as discussed in ref. 33, other enzymes, including pyruvate kinase, can catalyze this reaction. L. salivarius UCC118 contains a complete purine biosynthetic pathway for the creation of IMP and the pathway branches AMP and GMP. L. acidophilus and L. johnsonii are auxotrophic for pyrimidines and purines, respectively (16, 17), whereas L. plantarum can synthesize pyrimidines and purines de novo (19) from PRPP, as can L. sakei (18). Thus, although other “small-genome” intestinal lactobacilli are auxotrophic for one or other nucleotide class, L. salivarius UCC118 appears to be independent of a host provision of both.
Peptidoglycan and Exopolysaccharide Biosynthesis.
All of the genes involved in peptidoglycan biosynthesis are located on the L. salivarius chromosome. Exopolysaccharide (EPS) genes in the L. salivarius UCC118 genome are mainly clustered in two regions, at positions 0.99–1.101 Mb and 1.62–1.65 Mb in the chromosome. This arrangement is in contrast to species such as L. johnsonii (16), L. acidophilus (17), and Lactobacillus rhamnosus (34), in which most of the EPS genes are clustered in a single operon. Cluster 1 (LSL_0977–LSL_0997) contains 21 genes that include two putative chain length determinators, an oligosaccharide translocase or flippase, and 12 glycosyltransferases (Fig. 9). A homolog encoding a priming glucose phosphotransferase appears to be absent from this cluster. Cluster 1 shows low level synteny with other bacterial EPS clusters (e.g., Oceanobacillus iheyensis), which is restricted to a minority of the glycosyltransferase genes and biosynthesis genes at either end of the cluster (data not shown). Cluster 2 (LSL_1547-LSL_1574) has lower GC content (29%) compared with the rest of the genome and comprises 21 EPS-related genes, interrupted by 7 genes of unknown or unrelated function; 14 of the genes in this cluster show significant homology to genes in the major EPS cluster of L. plantarum, but there is no extended synteny. This region of the L. plantarum genome also shows aberrant base composition and has recently been subdivided into three clusters, the second of which is up-regulated in a quorum-sensing mutant (35). L. salivarius EPS cluster 2 includes genes for a LytR-type transcriptional regulator, a flippase, and seven glycosyltransferases, including a priming glucose phosphotransferase (EpsE homolog; LSL_1550), and, thus, represents a more complete unit than Cluster 1, relative to characterized EPS operons.
Carbohydrate Metabolism and Transport.
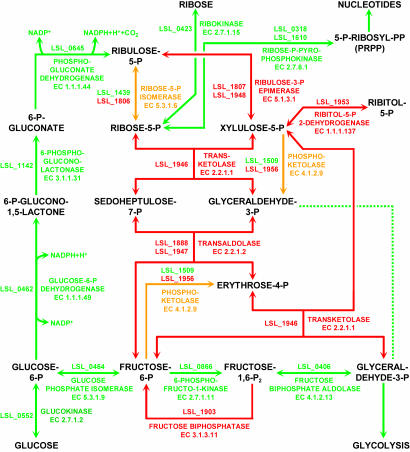
Genes in the Clusters of Orthologous Groups top-level category of metabolism were enriched in the half of the L. salivarius chromosome closest to the origin (Fig. 1), as previously reported for L. plantarum (15), although carbon metabolism and transport genes are more clustered at positions 400 kb, 500 kb, and 1.35 Mb. L. salivarius is currently regarded as homofermentative (36), meaning that sugars can be fermented only via the Embden-Meyerhof-Parnas pathway, and genes for the complete glycolysis pathway are present in the chromosome. Interestingly, genes for the pentose phosphate pathway also were found in the L. salivarius UCC118 genome, suggesting that it should be grouped among the facultatively heterofermentative lactobacilli. The genome sequence suggested that L. salivarius UCC118 would be able to assimilate ribose (Fig. 2). This suggestion was experimentally confirmed by growth on ribose as a sole carbon source, and, furthermore, we detected lactate, acetate, and ethanol by HPLC in the culture medium, confirming the heterofermentative status of this strain. The two key enzymes of the pentose phosphate pathway, transketolase (LSL_1946) and transaldolase (LSL_1888, LSL_1947), are encoded by the megaplasmid (Fig. 2). The gene products of pMP118 are not essential for nucleotide biosynthesis when L. salivarius UCC118 is grown on glucose, but the presence of an additional copy of a gene for ribose-5-phosphate isomerase on pMP118 (LSL_1806) may increase the flexibility and flux of this pathway. The presence of pMP118 would be essential for the growth of UCC118 if pentose were used as the sole carbon source. Growth on ribose, aided by genes resident on pMP118, may confer a competitive advantage on the strain when living in the human GI tract, because ribose might be an abundant carbon source in the GI tract because of RNA degradation.
Fig. 2.
Genes for completing the pentose phosphate pathway are on pMP118. Numbers and arrows in red or green represent genes and pathways encoded by pMP118 or the chromosome, respectively. Features in amber have genes coded by both replicons.
A gene on pMP118 encoding fructose biphosphatase (LSL_1903) and a chromosomally encoded phosphoenolpyruvate carboxykinase (LSL_0395) were identified. The first enzyme catalyzes the formation of fructose-6-phosphate, which is required for the synthesis of glucosamine-6-phosphate and its derivatives involved in peptidoglycan formation. The second enzyme catalyzes the formation of phosphoenolpyruvate by decarboxylation of oxaloacetate, while hydrolyzing ATP, a rate-limiting step in gluconeogenesis. Although L. sakei 23k has fructose biphosphatase and L. plantarum WCFS1 has phosphoenolpyruvate carboxykinase, L. salivarius UCC118 and L. casei ATCC334 (www.jgi.doe.gov) seem to be the only sequenced Lactobacilli that have both enzymes (with the assistance of pMP118 in L. salivarius), suggesting the presence of a functional gluconeogenesis pathway (Figs. 11 and 12). When glucose is exhausted, as can be encountered in certain regions of the GI tract, the gluconeogenesis pathway might be activated. The presence of a complete pentose phosphate pathway to generate glyceraldehyde-3-phosphate, coupled with gluconeogenesis from pyruvate, could be an adaptation to pentose-based growth.
In addition to glucose and ribose, data obtained from API 50 sugar fermentation tests showed that L. salivarius UCC118 is able to ferment a number of other monosaccharides (adonitol, galactose, fructose, mannose, mannitol, sorbitol, and N-acetylglucosamine) and disaccharides (maltose, lactose, sucrose, and trehalose). Good growth of L. salivarius UCC118 (i.e., at least 109 colony-forming units/ml) was achieved in MRS broth supplemented with each of the listed carbohydrates (data not shown). Most of these carbohydrates are likely found in, or can be generated from, human dietary components, reflecting the adaptation of L. salivarius to the human GI tract. Putative phosphotransferase transporters or ABC families of transporters for using these sugars were identified in the L. salivarius UCC118 genome. In addition, genes encoding enzymes involved in rhamnose and N-acetylneuraminic acid (sialic acid) catabolism as well as sorbitol utilization are present on pMP118. Collectively, the presence of pMP118 appears likely to increase the metabolic flexibility and, thus, competitiveness of L. salivarius UCC118.
Secreted Proteins and Transporters.
The predicted secreted protein complement of L. salivarius UCC118 contains 119 proteins (J.P.v.P., C.C., K. A. Ryan, Y.L., M.J.C., B. Sheil, L. Steidler, L. O’Mahony, G.F.F., D.v.S., and P.W.O., unpublished data). Among these 119 proteins, only 10 are likely sortase-dependent proteins. The megaplasmid is predicted to encode eight of the secreted proteins. There is a single sortase gene srtA (LSL_1606) in the genome and genes encoding putative signal peptidase I (LSL_0876) and signal peptidase II (LSL_0825). The genome contains the second lowest total number of predicted transporters among sequenced Lactobacillus genomes (Table 3, which is published as supporting information on the PNAS web site), among which there are 23 phosphotransferase transporters.
Megaplasmid-Encoded Properties and Plasmid Profiles of L. salivarius.
There are no genes on the megaplasmid pMP118 that might be considered strictly essential for viability. For example, pMP118 harbors an additional copy of rpsN (LSL_1944, ribosomal protein S14P), which is a paralog of the chromosomal gene LSL_1422 and is homologous to rpsN2 of L. plantarum and L. johnsonii. The gene on pMP118 encoding a bile-salt hydrolase (choloylglycine hydrolase; LSL_1801) is one of only two genes encoding bile-inactivating enzymes detected in the genome. The copy number of pMP118 was estimated by PCR to be 4.7 ± 0.6 copies per chromosome equivalent in stationary phase, so gene dosage effects would contribute to amplifying the contribution of LSL_1801 to bile resistance.
The L. salivarius UCC118 chromosome harbors one copy each of ldhL and ldhD genes, whereas pMP118 encodes an additional copy of the ldhD gene (LSL_1887), whose product is 42% identical to the L. plantarum enzyme.d-lactate is an important component of cell wall precursors in L. plantarum (37), and the additional pMP118-encoded LdhD could increase the efficiency of d-lactate production, provided that the gene is expressed and the gene product is catalytically active. In addition, LSL_1901 on pMP118 encodes a bifunctional acetaldehyde/alcohol dehydrogenase, which is the only enzyme in this strain that catalyzes the formation of ethanol from acetyl-CoA via acetaldehyde. Although not essential, the presence of this additional reductive pathway on pMP118 likely would improve the redox-balancing capability of strain UCC118.
L. salivarius UCC118 elaborates a two-component Class IIb bacteriocin, Abp118, that has been characterized in ref. 12. The cloned “chromosomal” EcoRI fragment containing the Abp118 locus that we previously sequenced was actually derived from pMP118, and the gene now designated LSL_1924 contains the EcoRI site at one end of the cloned fragment. This EcoRI fragment includes LSL_1921, which encodes a presalivaricin B homolog; we previously reasoned that this protein was likely nonfunctional, because an internal deletion introduced in the gene for Abpα eliminated all antibacterial activity from the cloned fragment in L. lactis and L. plantarum (12). However, the gene LSL_1924 that was truncated in the original cloned fragment encodes a potential bacteriocin transport protein. From consideration of the complete sequence of this region, it is possible that the salivaricin gene is functional but requires specific transport or accessory proteins. Interestingly, we have recently detected the genetic locus for Abp118 production in L. salivarius strains isolated from chickens (38), suggesting this trait may be widely distributed, and its linkage to strain plasmid content is being investigated.
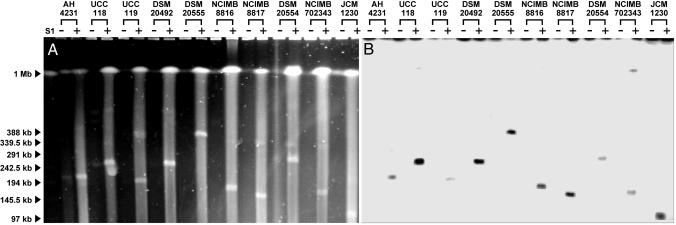
We used a combination of hybridization and S1 nuclease treatment, in combination with pulsed-field gel electrophoresis (PFGE) to investigate plasmid content of L. salivarius strains from varying sources (described in Table 4, which is published as supporting information on the PNAS web site). Nuclease S1 preferentially nicks and then linearizes megaplasmids because of their torsional stress (39). L. salivarius comprises two subspecies, salivarius and salicinius (36), and both were found to contain megaplasmids (Fig. 3), of sizes varying from 100 kb to 380 kb. All of these megaplasmids hybridized with a pMP118 repA gene probe. (Prominent bands in S1 nuclease-treated samples in Fig. 3A are chromosomal DNA, which caused nonspecific hybridization in Fig. 3B in some samples). It is noteworthy that pMP118 contains a tract of genes that show low relatedness to known or suspected conjugation genes (see Table 5, which is published as supporting information on the PNAS web site), thus representing a functional or remnant plasmid transfer locus. The ability to disseminate by conjugation would explain the apparently universal presence of pMP118-related plasmids (in strains so far tested), and this potential tra locus also might be involved in mobilization of smaller plasmids.
Fig. 3.
Megaplasmids of varying size are found in L. salivarius. (A) PFGE of genomic DNA of 10 L. salivarius strains. (B) Corresponding Southern hybridization with the pMP118 repA probe. (+) or (−) indicate treatment with S1 nuclease. Arrowheads to left indicate λ DNA concatamers used as size standard. Detailed strain information is provided in Supporting Text. The estimated size of megaplasmids are as follows: AH4231, 210 kb; UCC118, 242 kb; UCC119, 195 kb; DSM20492, 240 kb; DSM20555, 380 kb; NICMB8816, 180 kb; NCIMB8817, 145 kb; DSM20554, 260 kb; NCIMB702343, 160 kb; JCM1230, 100 kb.
The megaplasmid pMP118 is the largest plasmid from lactic acid bacteria in current nucleotide databases. However, plasmids >100 kb in Lactobacillus spp. have been reported. Lactacin F production by L. acidophilus strain 88 was shown by conjugation analysis to be linked to a 110-kb plasmid, pPM68 (40). A plasmid of 150 kb was identified in L. gasseri by PFGE and was suggested to be linear on the basis of electrophoretic behavior (41). However, its size and conformation were not confirmed. Given that many plasmid-profile studies of lactic acid bacteria (29) predated PFGE technology or did not employ conditions required to separate undigested replicons from the chromosome (39), it is possible that megaplasmids have a wider distribution in these bacteria than was previously recognized. The sequence of pMP118 reported here, in the context of the whole genome, uniquely illustrates the contribution of a megaplasmid to diverse metabolic and phenotypic properties of L. salivarius by both integrating with and extending chromosomally encoded features. It also provides a definitive platform for investigating the replication and dissemination of very large plasmids in this species.
The circular chromosome of L. salivarius UCC118 is the smallest Lactobacillus chromosome so far sequenced, 57.5 kb smaller than that of L. sakei and 165.5 kb smaller than that of L. johnsonii (Tables 6 and 7, which are published as supporting information on the PNAS web site). The presence of genes located on pMP118 confers numerous additional metabolic capabilities. The contribution of large numbers of pMP118 genes of unknown function to phenotype obviously cannot be evaluated. However, the fact that pMP118 contains a repertoire of genes that likely confer metabolic flexibility, seen in the context of significant megaplasmid size variation in other strains, strongly suggests that the multireplicon genome architecture of L. salivarius bestows on the species a dynamic and flexible genetic complement. This architecture could be in response to dietary fluctuations in host species, flexible niches in the GI tract, or adaptation to different hosts. In conclusion, the sequence of the L. salivarius UCC118 genome has provided a unique insight into the contribution of chromosomal and megaplasmid-encoded genes to the biology of this organism.
Materials and Methods
Bacterial Strains.
Bacterial strains are listed in Table 4.
Sequencing, Assembly, and Analysis.
The genome of L. salivarius UCC118 was sequenced by a standard shotgun strategy (see Supporting Text, which is published as supporting information on the PNAS web site, for details). The final assembly of the genome was confirmed by concordance of pulsed-field gel restriction patterns (ApaI, AsiSI, BglI, BssHII, MluI, RsrII, SacII, and SmaI) with the restriction map generated in silico. Standard procedures were used for genome annotation and analysis (Supporting Text). The genome sequence was integrated in the ERGO database (42), where a functional annotation was carried out, followed by manual verification and curation. Predicted proteins were functionally classified on the basis of Clusters of Orthologous Groups (43). The Transport Classification Database (TCDB; ref. 44) was used to categorize transport proteins. Genomes were aligned by using the Artemis Comparison Tool (45). The annotated genome sequence has been deposited in GenBank under accession nos. CP000233 (chromosome) and CP000234 (pMP118).
Metabolic Reconstruction.
A metabolic reconstruction was performed by using ERGO predictions, and Pathway Tools (46) based on EC numbers and enzymatic nomenclature. In addition, Pathway Hole Filler, a method using Bayesian statistics and blast comparisons, helped in identifying missing enzymes in the reconstructed pathways (47). Kyoto Encyclopedia of Genes and Genomes maps (48) were used to record and visualize this overview.
Copy Number Determination.
The copy number of pMP118 relative to the chromosome was estimated by quantitative PCR. A range of gene fragments were amplified first to find equal PCR efficiencies. The genes lspD (LSL_1838) and a pseudogene (LSL_1320) on pMP118 and chromosome, respectively, were selected. The ratio of plasmid to chromosome was calculated by using the formula 2(Ct chromosomal gene − Ct plasmid gene), where Ct is the crossing threshold value.
PFGE and Hybridization.
Nuclease S1 treatment and PFGE was carried out as described in ref. 39. Southern hybridization followed standard protocols (49). The probe was a 765-bp amplicon corresponding to the repA gene of pMP118 (LSL_1739).
Supplementary Material
Acknowledgments
We thank numerous Alimentary Pharmabiotic Centre colleagues for help with this work, T. Walunas of Integrated Genomics for assistance with ERGO, Kyoto Encyclopedia of Genes and Genomes/Pathway Solutions for permission to use images of Kyoto Encyclopedia of Genes and Genomes reference pathways, D. Walsh for HPLC analysis, K. Hayes for annotation assistance, and Prof. M. H. Saier, Jr., for making the Transport Classification Database available. This research was supported by Science Foundation Ireland through a Centre for Science, Engineering, and Technology award to the Alimentary Pharmabiotic Centre and by grants from the Higher Education Authority PRTLI1 and PRTLI3 programmes, the Department of Agriculture and Food FIRM 01/R&D/C/159 program, and the Irish Research Council for Science, Engineering, and Technology EMBARK postdoctoral program (to C.C.).
Abbreviations
- GI
gastrointestinal
- IS
insertion sequence
- PFGE
pulsed-field gel electrophoresis.
Footnotes
References
- 1.Klaenhammer T. R. J. Nutr. 2000;130:415S–416S. doi: 10.1093/jn/130.2.415S. [DOI] [PubMed] [Google Scholar]
- 2.Klaenhammer T. R., Russell W. M. Encyclopedia Food Microbiology. Vol. 2. Amsterdam: Elsevier; 2000. pp. 1151–1157. [Google Scholar]
- 3.Tannock G. W. Appl. Environ. Microbiol. 2004;70:3189–3194. doi: 10.1128/AEM.70.6.3189-3194.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vaughan E. E., Heilig H. G., Ben-Amor K., de Vos W. M. FEMS Microbiol. Rev. 2005;29:477–490. doi: 10.1016/j.femsre.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Molin G., Jeppsson B., Johansson M. L., Ahrne S., Nobaek S., Stahl M., Bengmark S. J. Appl. Bacteriol. 1993;74:314–323. doi: 10.1111/j.1365-2672.1993.tb03031.x. [DOI] [PubMed] [Google Scholar]
- 6.Ahrne S., Nobaek S., Jeppsson B., Adlerberth I., Wold A. E., Molin G. J. Appl. Microbiol. 1998;85:88–94. doi: 10.1046/j.1365-2672.1998.00480.x. [DOI] [PubMed] [Google Scholar]
- 7.Heilig H. G., Zoetendal E. G., Vaughan E. E., Marteau P., Akkermans A. D., de Vos W. M. Appl. Environ. Microbiol. 2002;68:114–123. doi: 10.1128/AEM.68.1.114-123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dunne C., Murphy L., Flynn S., O’Mahony L., O’Halloran S., Feeney M., Morrissey D., Thornton G., Fitzgerald G., Daly C., et al. Antonie Van Leeuwenhoek. 1999;76:279–292. [PubMed] [Google Scholar]
- 9.Dunne C., O’Mahony L., Murphy L., Thornton G., Morrissey D., O’Halloran S., Feeney M., Flynn S., Fitzgerald G., Daly C., et al. Am. J. Clin. Nutr. 2001;73:386S–392S. doi: 10.1093/ajcn/73.2.386s. [DOI] [PubMed] [Google Scholar]
- 10.McCarthy J., O’Mahony L., O’Callaghan L., Sheil B., Vaughan E. E., Fitzsimons N., Fitzgibbon J., O’Sullivan G. C., Kiely B., Collins J. K., Shanahan F. Gut. 2003;52:975–980. doi: 10.1136/gut.52.7.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheil B., McCarthy J., O’Mahony L., Bennett M. W., Ryan P., Fitzgibbon J. J., Kiely B., Collins J. K., Shanahan F. Gut. 2004;53:694–700. doi: 10.1136/gut.2003.027789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Flynn S., van Sinderen D., Thornton G. M., Holo H., Nes I. F., Collins J. K. Microbiology. 2002;148:973–984. doi: 10.1099/00221287-148-4-973. [DOI] [PubMed] [Google Scholar]
- 13.Siezen R. J., van Enckevort F. H., Kleerebezem M., Teusink B. Curr. Opin. Biotechnol. 2004;15:105–115. doi: 10.1016/j.copbio.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 14.Klaenhammer T. R., Barrangou R., Buck B. L., Azcarate-Peril M. A., Altermann E. FEMS Microbiol. Rev. 2005;29:393–409. doi: 10.1016/j.femsre.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 15.Kleerebezem M., Boekhorst J., van Kranenburg R., Molenaar D., Kuipers O. P., Leer R., Tarchini R., Peters S. A., Sandbrink H. M., Fiers M. W., et al. Proc. Natl. Acad. Sci. USA. 2003;100:1990–1995. doi: 10.1073/pnas.0337704100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pridmore R. D., Berger B., Desiere F., Vilanova D., Barretto C., Pittet A.-C., Zwahlen M.-C., Rouvet M., Altermann E., Barrangou R., et al. Proc. Natl. Acad. Sci. USA. 2004;101:2512–2517. doi: 10.1073/pnas.0307327101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altermann E., Russell W. M., Azcarate-Peril M. A., Barrangou R., Buck B. L., McAuliffe O., Souther N., Dobson A., Duong T., Callanan M., et al. Proc. Natl. Acad. Sci. USA. 2005;102:3906–3912. doi: 10.1073/pnas.0409188102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chaillou S., Champomier-Verges M. C., Cornet M., Crutz-Le Coq A. M., Dudez A. M., Martin V., Beaufils S., Darbon-Rongere E., Bossy R., Loux V., Zagorec M. Nat. Biotechnol. 2005:1527–1533. doi: 10.1038/nbt1160. [DOI] [PubMed] [Google Scholar]
- 19.Boekhorst J., Siezen R. J., Zwahlen M. C., Vilanova D., Pridmore R. D., Mercenier A., Kleerebezem M., de Vos W. M., Brussow H., Desiere F. Microbiology. 2004;150:3601–3611. doi: 10.1099/mic.0.27392-0. [DOI] [PubMed] [Google Scholar]
- 20.Bolotin A., Quinquis B., Renault P., Sorokin A., Ehrlich S. D., Kulakauskas S., Lapidus A., Goltsman E., Mazur M., Pusch G. D., et al. Nat. Biotechnol. 2004;22:1554–1558. doi: 10.1038/nbt1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Flynn S. Ph.D. thesis. Ireland: University College Cork; 2001. [Google Scholar]
- 22.Ochman H. Curr. Biol. 2002;12:R427–R428. doi: 10.1016/s0960-9822(02)00916-8. [DOI] [PubMed] [Google Scholar]
- 23.Bentley S. D., Parkhill J. Annu. Rev. Genet. 2004;38:771–792. doi: 10.1146/annurev.genet.38.072902.094318. [DOI] [PubMed] [Google Scholar]
- 24.Ng W. V., Ciufo S. A., Smith T. M., Bumgarner R. E., Baskin D., Faust J., Hall B., Loretz C., Seto J., Slagel J., et al. Genome Res. 1998;8:1131–1141. doi: 10.1101/gr.8.11.1131. [DOI] [PubMed] [Google Scholar]
- 25.del Solar G., Giraldo R., Ruiz-Echevarria M. J., Espinosa M., Diaz-Orejas R. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lobry J. R., Sueoka N. Genome Biol. 2002;3:RESEARCH0058. doi: 10.1186/gb-2002-3-10-research0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Bueno M., Valdivia E., Galvez A., Maqueda M. Curr. Microbiol. 2000;41:257–261. doi: 10.1007/s002840010130. [DOI] [PubMed] [Google Scholar]
- 28.Eppinger M., Baar C., Raddatz G., Huson D. H., Schuster S. C. Nat. Rev. Microbiol. 2004;2:872–885. doi: 10.1038/nrmicro1024. [DOI] [PubMed] [Google Scholar]
- 29.Wang T. T., Lee B. H. Crit. Rev. Biotechnol. 1997;17:227–272. doi: 10.3109/07388559709146615. [DOI] [PubMed] [Google Scholar]
- 30.Cerdeno-Tarraga A. M., Patrick S., Crossman L. C., Blakely G., Abratt V., Lennard N., Poxton I., Duerden B., Harris B., Quail M. A., et al. Science. 2005;307:1463–1465. doi: 10.1126/science.1107008. [DOI] [PubMed] [Google Scholar]
- 31.O’Sullivan D., Twomey D. P., Coffey A., Hill C., Fitzgerald G. F., Ross R. P. Mol. Microbiol. 2000;36:866–875. doi: 10.1046/j.1365-2958.2000.01901.x. [DOI] [PubMed] [Google Scholar]
- 32.Hols P., Kleerebezem M., Schanck A. N., Ferain T., Hugenholtz J., Delcour J., de Vos W. M. Nat. Biotechnol. 1999;17:588–592. doi: 10.1038/9902. [DOI] [PubMed] [Google Scholar]
- 33.Kilstrup M., Hammer K., Ruhdal Jensen P., Martinussen J. FEMS Microbiol. Rev. 2005;29:555–590. doi: 10.1016/j.femsre.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 34.Peant B., LaPointe G., Gilbert C., Atlan D., Ward P., Roy D. Microbiology. 2005;151:1839–1851. doi: 10.1099/mic.0.27852-0. [DOI] [PubMed] [Google Scholar]
- 35.Sturme M. H., Nakayama J., Molenaar D., Murakami Y., Kunugi R., Fujii T., Vaughan E. E., Kleerebezem M., de Vos W. M. J. Bacteriol. 2005;187:5224–5235. doi: 10.1128/JB.187.15.5224-5235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogosa M., Wiseman R. F., Mitchell J. A., Disraely M. N., Beaman A. J. J. Bacteriol. 1953;65:681–699. doi: 10.1128/jb.65.6.681-699.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goffin P., Deghorain M., Mainardi J. L., Tytgat I., Champomier-Verges M. C., Kleerebezem M., Hols P. J. Bacteriol. 2005;187:6750–6761. doi: 10.1128/JB.187.19.6750-6761.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Mulcahy G., Van Sinderen D., O’Toole P. W. in LAB8 Symposium on Lactic Acid Bacteria. Egmond aan Zee, Netherlands: Fed. Eur. Microbiol. Soc; 2005. [Google Scholar]
- 39.Barton B. M., Harding G. P., Zuccarelli A. J. Anal. Biochem. 1995;226:235–240. doi: 10.1006/abio.1995.1220. [DOI] [PubMed] [Google Scholar]
- 40.Muriana P. M., Klaenhammer T. R. Appl. Environ. Microbiol. 1987;53:553–560. doi: 10.1128/aem.53.3.553-560.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roussel Y., Colmin C., Simonet J. M., Decaris B. J. Appl. Bacteriol. 1993;74:549–556. [PubMed] [Google Scholar]
- 42.Overbeek R., Larsen N., Walunas T., D’Souza M., Pusch G., Selkov E., Jr, Liolios K., Joukov V., Kaznadzey D., Anderson I., et al. Nucleic Acids Res. 2003;31:164–171. doi: 10.1093/nar/gkg148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatusov R. L., Fedorova N. D., Jackson J. D., Jacobs A. R., Kiryutin B., Koonin E. V., Krylov D. M., Mazumder R., Mekhedov S. L., Nikolskaya A. N., et al. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Busch W., Saier M. H., Jr Crit. Rev. Biochem. Mol. Biol. 2002;37:287–337. doi: 10.1080/10409230290771528. [DOI] [PubMed] [Google Scholar]
- 45.Carver T. J., Rutherford K. M., Berriman M., Rajandream M. A., Barrell B. G., Parkhill J. Bioinformatics. 2005;21:3422–3423. doi: 10.1093/bioinformatics/bti553. [DOI] [PubMed] [Google Scholar]
- 46.Karp P. D., Paley S., Romero P. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 47.Green M. L., Karp P. D. BMC Bioinformatics. 2004;5:76. doi: 10.1186/1471-2105-5-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kanehisa M. Trends Genet. 1997;13:375–376. doi: 10.1016/s0168-9525(97)01223-7. [DOI] [PubMed] [Google Scholar]
- 49.Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Woodbury, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.