Abstract
Most soils inhibit fungal germination and growth to a certain extent, a phenomenon known as soil fungistasis. Previous observations have implicated microorganisms as the causal agents of fungistasis, with their action mediated either by available carbon limitation (nutrient deprivation hypothesis) or production of antifungal compounds (antibiosis hypothesis). To obtain evidence for either of these hypotheses, we measured soil respiration and microbial numbers (as indicators of nutrient stress) and bacterial community composition (as an indicator of potential differences in the composition of antifungal components) during the development of fungistasis. This was done for two fungistatic dune soils in which fungistasis was initially fully or partly relieved by partial sterilization treatment or nutrient addition. Fungistasis development was measured as restriction of the ability of the fungi Chaetomium globosum, Fusarium culmorum, Fusarium oxysporum, and Trichoderma harzianum to colonize soils. Fungistasis did not always reappear after soil treatments despite intense competition for carbon, suggesting that microbial community composition is important in the development of fungistasis. Both microbial community analysis and in vitro antagonism tests indicated that the presence of pseudomonads might be essential for the development of fungistasis. Overall, the results lend support to the antibiosis hypothesis.
The vast majority of natural soils suppress the germination and growth of fungi to a certain extent (29). This phenomenon, referred to as widespread soil fungistasis, was first described by Dobbs and Hinson (11). The intensity of fungistasis is dependent on the physical and chemical soil properties as well as soil microbial activity (1, 12, 29, 33, 37). The importance of the last factor was demonstrated by the relief of fungistasis by (partial) sterilization treatments and addition of antibiotics (12, 41, 43).
Fungi differ strongly in their sensitivity to fungistasis. In general, plant pathogenic fungi appear to be more sensitive than saprophytic fungi (8, 17). The sensitivity of plant pathogenic fungi to fungistasis is thought to protect them from germinating and initiating growth under unfavorable conditions (17, 29). However, this protection comes at a cost, since the viability of resting structures decreases during prolonged incubation in soils (30). The negative effects of fungistasis on the inoculum density of plant pathogenic fungi has been suggested as a mechanism to explain the commonly found correlation between fungistasis and disease suppressiveness (21, 24, 25, 29). Therefore, manipulation of fungistasis has often been mentioned as a potential measure to control plant pathogenic fungi (20, 29). The idea is to stimulate germination, and subsequent dying, of plant pathogens when no host plant is present or to suppress their germination and growth in the presence of a host plant. So far, no consistent success has been obtained with fungistasis-manipulating measures (21, 24), and a major hurdle in the successful manipulation of fungistasis is our limited knowledge of its mechanisms.
The most popular explanation for the microbial cause of fungistasis is that strong competition by soil microorganisms limits carbon availability to the germinating spores or invading hyphae (4, 19, 29, 33). Limited carbon availability in soils is the rule rather than the exception, and not only fungi but also actinomycetes (actinostasis) and nonfilamentous bacteria (bacteriostasis) are restricted in their growth in soils (19, 29). The importance of nutrients is further indicated by the partial or full alleviation of fungistasis after addition of simple energy substrates, like sugars or amino acids, to soil (29).
Other studies have, however, suggested that fungistasis can be attributed to the presence of antifungal compounds of microbiological origin (15, 26, 27, 39), and the production of fungus-inhibiting compounds has been described for a wide range of soil microorganisms (5, 7, 16).
Unfortunately, it is difficult to distinguish between these two proposed mechanisms of fungistasis, as nutrient status may be involved in both (16, 35). Yet, the involvement of antibiosis in fungistasis may imply that the microbial community composition of a soil is critical because not all microorganisms produce antifungal compounds and the spectrum of antifungal compounds produced is different for different microbial species (38). If carbon limitation is the major causative factor of fungistasis, (potential) microbial activity should be more important than microbial composition.
The aim of the present study was to obtain better insight into the relative importance of microbial activity and microbial community composition for the development of fungistasis. This was done by following the temporal dynamics of fungistasis, microbial activity, microbial numbers, and bacterial community composition in soils after partial sterilization or additions of growth substrates.
MATERIALS AND METHODS
Soils.
The study was done with two Dutch coastal dune soils: an outer dune soil from Oostvoorne (51° 08 min N, 4° 05 min E) and an inner dune soil (third dune row from the coast) from the northern part of the Wadden Island Terschelling (53° 23 min N, 5° 16 min E).
Both soils, which were sampled from the upper 10 cm between tussocks of Marram grass (Ammophila arenaria), had a sandy texture, with more than 99% of the grains being > 75 μm. The soil from Oostvoorne was sampled in February 1998 and had the following properties: pH (water), 8.9; moisture, 4.1% (wt/wt); and organic matter, 2.5 g kg−1. The properties of the soil from Terschelling, sampled in February 2000, were: pH (water), 6.9; moisture, 6.2% (wt/wt); and organic matter, 12.5 g kg−1.
In a previous study, it was shown that soils from these locations suppressed hyphal growth of a wide range of fungi (8). Originally, it was planned to do all the experiments with the soil from Oostvoorne. However, this soil appeared to cause nonbiological fungistasis for two of the test fungi, Fusarium culmorum (see Results and Discussion) and Trichoderma harzianum (data not shown). This was most likely due to the alkalinity of this soil, and therefore it was decided to do the main experiment with the less alkaline soil from Terschelling.
Experiment 1 (pilot study).
Soil sampled from the Oostvoorne site was mixed well, with addition of demineralized water to establish a moisture content of 7.5% (wt/wt). The mixed soil was divided into subportions, which were subjected to the following treatments designed to manipulate the microbial activity and community composition: microwave radiation (3 min at 850 W and 2450 Hz per 500 g of soil), acetylglucosamine addition, glucose plus ammonium sulfate addition, cellulose plus ammonium sulfate addition, and tryptic soy broth addition. Carbon sources were added at 1 g per kg of dry soil and ammonium sulfate at 0.3 g per kg of dry soil. Petri dishes (8.5-cm diameter) were filled with 55 g of moist soil from all treatments, including untreated sand. The soil was spread evenly in an 8-mm layer. The petri dishes were sealed with Parafilm to minimize water loss and incubated at 20°C.
After 0, 1, 2, 4, 8, and 12 weeks of incubation, the following measurements and analyses were performed on two randomly taken plates: hyphal extension of the fungi Chaetomium globosum (saproptroph) and Fusarium culmorum (plant pathogen), plate count of bacteria, CO2 production, pH, and inorganic N. Both fungi had been isolated from dune soils (10).
Experiment 2 (main study).
The main study was done with soil from Terschelling at its field moisture content of 6.2% (wt/wt). After mixing, the soil was divided into subportions that received the following treatments: none (untreated soil), partial sterilization by microwave (4 min at 700 W and 2450 Hz per 600 g of soil) (microwaved), autoclaving (twice 30 min each at 120°C per 4 kg of soil) followed by 1% (wt/wt) inoculation with nonsterile soil from Terschelling (autoclaved-inoculated), autoclaving followed by mixing in open air to allow air infection (autoclaved-open), and autoclaving followed by closed incubation (autoclaved-closed). After the treatments had been applied, portions of 675 g of well-mixed soil were transferred to glass jars. The jars were sealed with Parafilm to minimize water loss and incubated at 20°C. For the autoclaved-closed treatment only, soil (675-g portions) was autoclaved inside the jars.
At several times during the incubation (0, 1, 2, 3, 5, 7, and 10 weeks for untreated, microwaved, and autoclaved-inoculated soil; 0, 5, and 10 weeks for autoclaved-closed and autoclaved-open soil), the contents of two randomly chosen jars per treatment were pooled and mixed and subjected to measurements and analyses of hyphal extension for four fungi, Chaetomium globosum (same strain as in experiment 1), Fusarium culmorum (same strain as experiment 1), F. oxysporum (potential plant pathogen, dune soil isolate), and Trichoderma harzianum (saprotroph, dune soil isolate); bacterial and fungal CFU, with whole-cell fatty acid-based identification of dominant bacterial colony types; in vitro inhibition of fungi by dominant bacterial colony types; bacterium-specific PCR-denaturing gradient gel electrophoresis and sequence analysis of 16S ribosomal DNA (rDNA) fragments; CO2 production; pH; and inorganic N content. All measurements and analyses were done three times.
Hyphal extension (experiments 1 and 2).
Hyphal extension of test fungi was measured by the method described by De Boer et al. (8). This method tests the ability of a fungus to invade a nutrient-poor medium, such as soil, from a nutrient-rich spot. Briefly, an agar disk (potato dextrose agar, 1-cm diameter) from the growing margin of the fungal colony was inverted and placed centrally on top of soil in a petri dish. After 3 weeks of incubation at 20°C, the extension of the mycelium was determined with a binocular microscope, and the area of hyphal extension was calculated. The surface growth on the soil samples was compared with that on nutrient-free, acid-washed beach sand, moisture content 7.5% (wt/wt). In experiment 1, petri dishes containing soil could be used immediately for this test, whereas in experiment 2, petri dishes were filled with sand that had been incubating in glass jars.
Plate and microscopic counts (experiments 1 and 2).
Enumeration of bacterial and fungal CFU was performed with 1/10-strength tryptic soy broth-agar for bacteria and water-agar for fungi as described previously (8).
For experiment 2, an estimate of total soil bacteria was obtained by performing epifluorescence microscopy counts of 4′,6′-diamidino-2-phenylindole-stained cells in soil smears (31).
FAME analysis (experiment 2).
Bacterial CFU were grouped on the basis of colony morphology. Representatives of the most dominant groups were identified with a commercially available gas chromatograph software system for the analysis of whole-cell fatty acid methyl ester (FAME) profiles (Microbial ID, Inc., Newark, Del.). Culturing, harvesting, chromatography, and data analysis were performed as described by Janse (23).
In vitro antagonism (experiment 2).
Bacteria that were representative of dominant colony types and were identified by FAME and partial sequencing of the in vitro 16S rRNA gene were tested for antagonism against the fungi F. culmorum and T. harzianum. The test was done on water-agar as described by De Boer et al. (9) to simulate the nutrient-poor conditions in soil. Briefly, bacteria were streaked on a 1.5-cm zone in the center of a petri dish containing water-agar and incubated for 1 week at 20°C. The fungal inocula (potato dextrose agar disks) were placed at a distance of 1 cm from the bacterial zone. The effects of the bacteria on mycelium formation was recorded after 2 weeks of incubation at 20°C.
DNA extraction and PCR-denaturing gradient gel electrophoresis (experiment 2).
DNA was extracted from 0.25 g (wet weight) soil samples, and the Mobio soil DNA extraction kit according to the manufacturer's specifications (Mobio Laboratories, Solana Beach, Calif.) except that bead beating (twice for 30 s each) was used instead of Vortex mixing. Samples were eluted in 50 μl of 10 mM Tris (pH 8.0) and diluted to a final concentration of 50 ng μl−1.
The V6 to V8 region of the 16S rRNA gene was amplified from soil DNA with the primers 968f-GC and 1401r (18) and the following thermocycling program: 120 s at 94°C for one cycle; 30 s at 92°C, 60 s at 55°C, and 45 s at 72°C plus 1 s per cycle for 35 cycles; and 300 s at 68°C for one cycle. Each amplification reaction mix consisted of 30 nM each primer, 1 μl of template DNA (50 pg), 1 U of Expand High Fidelity DNA polymerase (Boehringer, Mannheim, Germany), and the manufacture's recommended buffer conditions. PCR products were examined by standard 1.5% (wt/vol) agarose-0.5× TBE (Tris-borate-EDTA) gel electrophoresis with ethidium bromide staining, to confirm product integrity and estimate yield.
Approximately 0.5 μg of PCR product was used for denaturing gradient gel electrophoresis analysis, with the method of Muyzer et al. (34) as modified below. Gels contained 6% (wt/vol) polyacrylamide (37:1 acrylamide/bisacrylamide) and 0.5× TAE (Tris-acetate-EDTA) and were 1.5-mm thick and measured 20 by 20 cm. The linear gradient used was from 45% to 65% denaturant, where 100% denaturing acrylamide is defined as containing 7 M urea and 40% (vol/vol) formamide (34). To ensure well-polymerized slots, a 10-ml top gel containing no denaturants was added before polymerization was complete. All denaturing gradient gel electrophoresis analyses were run with a D-Gene system (Bio-Rad Laboratories, Hercules, Calif.) at a constant temperature of 60°C.
Electrophoresis was for 10 min at 200 V, after which the voltage was lowered to 80 V for an additional 16 h. Gels were stained in MilliQ (Millipore B.V., Etten-Leur, The Netherlands) water containing 0.5 mg of ethidium bromide per liter and destained twice in MilliQ water prior to UV transillumination. Gel images were digitally captured with the ImaGo system (B & L, Maarssen, The Netherlands). Comparisons of denaturing gradient gel electrophoresis profiles were done with Pearson's index, taking both band number and intensity into account after signal normalization, and dendrogram construction was performed with the ImageMaster Elite Database program (version 2.0) (Amersham Pharmacia Biotech) as described by Duineveld et al. (13).
Sequence analysis.
Denaturing gradient gel electrophoresis bands selected for sequence analysis (see Fig. 3A) were excised, taking care that only the innermost portion of each band was removed to avoid contamination from other bands and background. DNA was eluted from denaturing gradient gel electrophoresis bands for subsequent reamplification and doubled-stranded sequencing of purified PCR products as described by Duineveld et al. (13). Sequence comparisons were performed with the FASTA and BLAST programs (2, 36) and did not include the regions corresponding to the primer binding sites.
Soil respiration (experiments 1 and 2).
Portions (40 g) of soil were weighed into screw-cap bottles (315 ml), which were closed and incubated for 72 h at 20°C. The concentration of CO2 in the headspace was subsequently measured with a gas chromatograph (Carlo Erba GC 6000) equipped with a hot wire detector and a Molsieve 5 Å column, operated at 80°C with helium as the carrier gas.
Soil analyses (experiments 1 and 2).
Moisture and pH were determined as described by Maly et al. (31). Organic matter was determined as weight loss of dry soil after 24 h at 430°C.
Data analysis.
For experiment 1, data for each treatment were pooled for the incubation periods 0 to 2 weeks and 4 to 12 weeks, as there were only two replicates per time interval. These data were analyzed with one-way analysis of variance. Differences between treatment means were inspected with Tukey's honestly significantly difference at the 5% level. For experiment 2, significant differences (5% level) between the mean of the untreated soil sample and that of the treated soil samples were inspected for each time interval with the two-sample t test.
Relationships between CO2 production and mycelial extension were identified by calculation of correlations (Pearson).
Nucleotide sequence accession numbers.
Sequences recovered in this study have been deposited in the EMBL database under accession numbers AJ428134 to AJ428157 and AJ428179 to AJ428182.
RESULTS
Experiment 1.
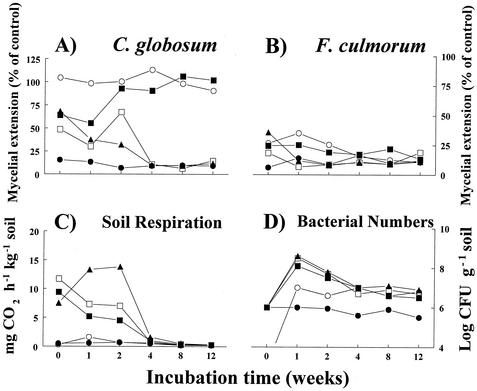
Fusarium culmorum and Chaetomium globosum responded differently to microwave and substrate addition treatments of the soil from Oostvoorne (Fig. 1A and B). The treatments had little effect on hyphal extension of F. culmorum. Only during the initial part of the incubation was a small but significant (P < 0.05) relief of fungistasis observed for the microwave and tryptic soy broth treatments. Hyphal extension of C. globosum was significantly (P < 0.05) greater in all treatments during the first 2 weeks of incubation. Thereafter, both the microwave- and tryptic soy broth-treated soils maintained complete relief of fungistasis during the whole incubation period, whereas fungistasis in the glucose- and acetylglucosamine-treated soils returned to levels typical of the untreated soil.
FIG. 1.
Effect of initial treatments (microwave and nutrient addition) of Oostvoorne dune soil on temporal dynamics of the hyphal extension ability of Chaetomium globosum (A) and Fusarium culmorum (B), soil respiration (C), and bacterial CFU (D). Hyphal extension from potato dextrose agar disks into soil was measured at all indicated time intervals and is given as a percentage of the extension measured into acid-purified beach sand. Symbols: •, control; ○, microwave; ▪, tryptic soy broth; □, acetylglucosamine; ▴, glucose.
CO2 production was significantly increased by the C additions during the initial part (0 to 2 weeks) of the incubation (Fig. 1C). Thereafter, CO2 production was as low as that in the untreated dune soil. Microwave treatment had no significant effect on CO2 production.
Bacterial CFU increased rapidly in the carbon addition treatments and were highest after 1 week of incubation (Fig. 1D). Although the CFU decreased steadily during the incubation period, they remained an order of magnitude higher than those of the untreated soil. The initial number of CFU of the microwave treatment was below the detection limit of 103 (g of soil)−1. The numbers increased rapidly during the first week but not to the levels seen for the carbon additions. In the later part of the incubation, the CFU numbers of the microwave treatment were similar to those of the carbon addition treatments.
Experiment 2: hyphal extension.
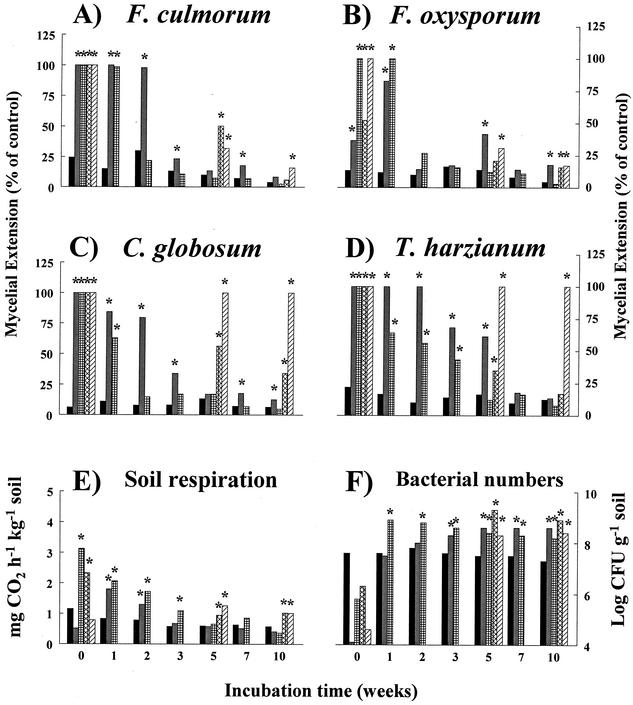
Initially, the microwave and autoclave treatments had a strong stimulating effect on hyphal extension of all fungi (Fig. 2A to D). F. culmorum hyphal extension at time zero was even a factor of 1.5 to 2 higher on sterilized sand than on the acid-purified sand (not shown in Fig. 2 because of the scale). In the inoculated, autoclaved soil, the hypha-extending ability returned within 2 weeks (F. culmorum, F. oxysporum, and C. globosum) or 5 weeks (T. harzianum) to the levels in the untreated soil. In the microwave-treated soil, hyphal extension also decreased again, albeit more slowly than in the autoclaved-inoculated treatment.
FIG. 2.
Effect of initial sterilization treatments of Terschelling dune soil on temporal dynamics of hyphal extension ability of several soil-borne fungi (A to D), soil respiration (E), and bacterial plate counts (F). Hyphal extension from potato dextrose agar disks into soil was measured at all indicated time intervals and is given as a percentage of the extension measured into acid-purified beach sand. *, mean extension, respiration, or bacterial number is significantly higher (two-sample t test, P < 0.05) than that of the nonsterilized dune soil. Bar patterns (from left to right within each group): solid black bar, untreated soil; checkerboard bar, microwaved; checked bar, autoclaved plus 1% nonsterile dune soil inoculum; trellis bar, autoclaved and open incubation; and hatched bar, autoclaved and closed incubation.
Air-infected, autoclaved soil (autoclaved-open soil) was fully fungistatic, i.e., not significantly different from untreated soil, after 10 weeks of incubation for T. harzianum and F. culmorum, but not fully fungistatic for F. oxysporum and C. globosum. Closed-incubated, autoclaved soil was not fungistatic to T. harzianum and C. globosum after 10 weeks of incubation and was almost completely fungistatic to the Fusarium species.
The mean hyphal extension on acid-purified sand was 33, 27, 63 (i.e., the complete petri dish), and 63 cm2 for F. culmorum, F. oxysporum, C. globosum, and T. harzianum, respectively.
Experiment 2: soil respiration and microbial counts.
CO2 production increased immediately in the autoclaved-inoculated and autoclaved-open treatments compared to the untreated soil (Fig. 2E). The microwaved treatment had a significantly lower CO2 production than the control at the start of the experiment, followed by a significantly higher production during the next 2 weeks of incubation. The autoclaved-closed treatment also had a low initial CO2 production. CO2 production for the microwaved and autoclaved-inoculated treatments was back to the level of the untreated soil within 3 and 5 weeks, respectively. In both the autoclaved-closed and autoclaved-open treated soils, CO2 production was still higher than the untreated soil after 5 and 10 weeks of incubation. There was no significant difference in CO2 production between the autoclaved-closed and autoclaved-open soil for these time intervals.
After the initial decrease, numbers of bacterial CFU became significantly higher than in the untreated soil for all treatments and remained so during prolonged incubation (Fig. 2F). Microscopic counts (data not shown) revealed that, for the untreated soil only, about 4% of the total number of soil bacteria was counted on agar plates. In the microwaved, autoclaved-open, and autoclaved-closed treated soils, there were no such differences between microscopic counts and plate counts for the whole incubation period. The autoclaved-inoculated treatment also started with similar microscopic and plate counts during the first 2 weeks, but then the plate numbers decreased to 19% of the microscopic numbers at week 10. Counts of fungal CFU indicated that all treatments were colonized by fungi, but the numbers never became higher than in the untreated soil (data not shown).
Experiment 2: bacterial community composition.
The development of the culturable part of the bacterial community differed strongly between the treatments (Table 1). In the microwaved treatment, Bacillus sp. dominated initially. However, within 3 weeks of incubation, bacilli were a minor component of the total CFU. Stenotrophomonads (formerly Xanthomonas maltophilia) and xanthomonads gained in dominance and remained the dominant group throughout the rest of the incubation. Cytophaga species proliferated during the initial part of the incubation in the autoclaved-inoculated treatment, but streptomycetes became dominant as of week 3. Only Bacillus spp. were cultivated from the autoclaved-closed treated soil, whereas the autoclaved-open treated soil had a diverse composition of mainly gram-negative bacterial species. Pseudomonas spp. and Stenotrophomonas/Xanthomonas spp were present in all treatments except the autoclaved-closed incubation. Many of the bacteria cultivated from the untreated soil grew too slowly on tryptic soy broth-agar to allow FAME analysis. Of the identifiable (by FAME and colony morphology) colonies, streptomycetes were dominant.
TABLE 1.
Effect of initial sterilization treatments of Terschelling dune soil (experiment 2) on culturable microbial community compositiona
| Soil | Genus (closest match) | % of isolates belonging to group
|
|||
|---|---|---|---|---|---|
| Wk 1 | Wk 3 | Wk 5 | Wk 10 | ||
| Untreated dune soil | Bacillus/Paenibacillus | 5 | ND | ND | ND |
| Cellulomonas | 5 | ND | ND | ND | |
| Cytophaga | 5 | ND | ND | ND | |
| Micrococcus | 2 | ND | ND | ND | |
| Pseudomonas | 2 | ND | ND | ND | |
| Stenotrophomonas/Xanthomonas | 4 | ND | ND | ND | |
| Streptomyces | 18 | ND | ND | ND | |
| Unidentified | 59 | ND | ND | ND | |
| Microwaved | Arthrobacter | 14 | |||
| Bacillus/Paenibacillus | 67 | ||||
| Burkholderia | 14 | ||||
| Pseudomonas | 25 | 9 | 14 | ||
| Stenotrophomonas/Xanthomonas | 19 | 61 | 72 | 71 | |
| Unidentified | 19 | 15 | |||
| Autoclaved and inoculated | Bacillus/Paenibacillus | 25 | 38 | ||
| Cytophaga | 45 | 4 | |||
| Pseudomonas | 10 | 15 | 13 | 13 | |
| Stenotrophomonas/Xanthomonas | 13 | 5 | 25 | ||
| Streptomyces | 50 | 38 | 38 | ||
| Unidentified | 32 | 26 | 11 | ||
| Open incubation | Cytophaga | ND | ND | 25 | |
| Pseudomonas | ND | ND | 53 | 8 | |
| Sphingobacterium | ND | ND | 10 | 3 | |
| Stenotrophomonas/Xanthomonas | ND | ND | 8 | ||
| Unidentified | ND | ND | 37 | 28 | |
| Autoclaved and closed incubation | Bacillus/Paenibacillus | ND | ND | 100 | 100 |
Based on whole-cell fatty acid analysis (FAME) of morphologically different colony types. ND, not determined.
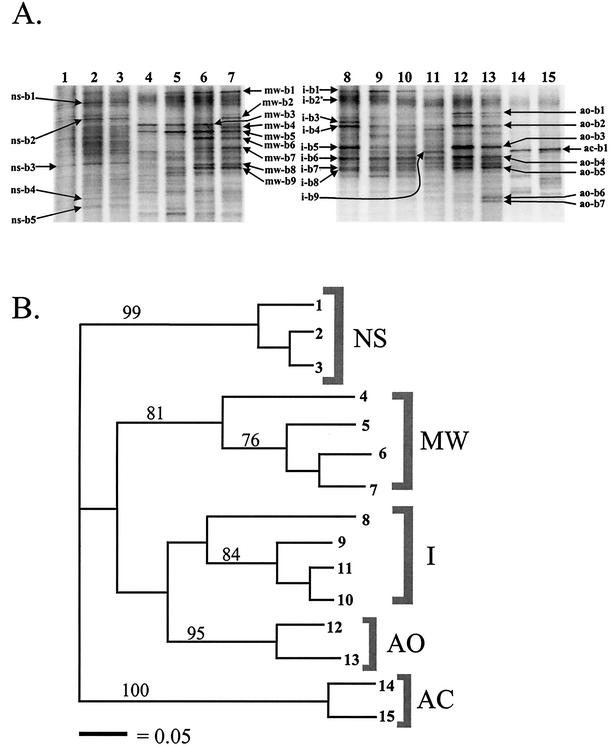
Bacterial 16S rDNA fingerprints indicated clear differences in the dominant bacterial populations in soils of the different treatments (Fig. 3A). Also, a succession in dominant bacterial populations over the course of the experiment was observed. Despite this succession, denaturing gradient gel electrophoresis patterns within a treatment over time showed greater similarity to each other than did those between treatments (Fig. 3B).
FIG. 3.
(A) Denaturing gradient gel electrophoresis analysis of bacterial 16S rDNA fragments amplified from dune soil subjected to different sterilization treatments. Arrows point to bands that were excised for sequence analysis (see Table 2), and the band designation reflects the treatment from which it was recovered. Lanes: 1 to 3, nonsterilized soil (NS) at 0, 5, and 10 weeks, respectively; 4 to 7, microwave-treated soil (MW) at 1, 2, 5, and 10 weeks, respectively; 8 to 11, autoclaved soil plus 1% nonsterile dune soil inoculum (I) at 1, 2, 5, and 10 weeks, respectively; 12 and 13, autoclaved soil allowing air infections (AO) at 5 and 10 weeks, respectively; 14 and 15, autoclaved soil, closed incubation (AC) at 5 and 10 weeks, respectively. (B) Dendrogram analysis of the denaturing gradient gel electrophoresis gel shown in A. Numbers on the right refer to lane designations in A. Only bootstrap values above 70 are shown.
Dominant denaturing gradient gel electrophoresis bands were excised for sequence analysis, and the closest database matches with recovered band sequences are shown in Table 2. Some of the recovered sequences were consistent with data concerning the dominant culturable bacteria isolated from these soils. For instance, sequences showing affinity with the genera Xanthomonas, Stenotrophomonas, Pseudomonas, and Bacillus were seen as dominant denaturing gradient gel electrophoresis bands in some treatments where they were also observed as common isolates. In addition, a number of populations were detected by the PCR-denaturing gradient gel electrophoresis method that were not observed in our culture-dependent survey. These included the genera Variovorax, Brevibacillus, and Rhodanobacter. The relative importance of streptomycetes among isolates was not found for denaturing gradient gel electrophoresis analysis.
TABLE 2.
Comparison of band sequences with database entries
| Banda | Nearest databank entryb | % identity | Accession no. of nearest databank entry |
|---|---|---|---|
| ns-b1 | Uncultured eubacterium strain WD2124 | 96.2 | AJ292676 |
| ns-b2 | Bacillus macroides | 93.8 | AF157696 |
| ns-b3 | Variovorax paradoxus strain E4C | 98.2 | AF209469 |
| ns-b4 | No legible sequence | ||
| ns-b5 | No legible sequence | ||
| mw-b1/i-b1 | Ultramicrobacterium sp. strain MY14 | 94.2 | AB008503 |
| mw-b2 | Matsuebacter sp. strain 9 | 97.5 | AB024305 |
| mw-b3 | Brevibacillus centrosporus | 99.0 | D78458 |
| mw-b4 | Unidentified activated-sludge bacterium AI051 | 89.5 | U45691 |
| mw-b5 | Brevibacillus sp. strain HC6 | 98.2 | AF252328 |
| mw-b6 | Xanthomonas axonopodis | 97.7 | AF123091 |
| mw-b7 | Variovorax paradoxus | 91.2 | AF209469 |
| mw-b8 | Rhodanobacter lindanoclas | 97.7 | AF039167 |
| mw-b9 | Rhodanobacter lindanoclas | 97.0 | AF039167 |
| i-b2 | No legible sequence | ||
| i-b3 | Janthinobacterium lividum | 99.0 | AF174648 |
| i-b4/ao-b2 | Janthinobacterium lividum | 100 | AF174648 |
| i-b5/ao-b3 | Pseudomonas lundensis | 99.0 | AB021395 |
| i-b6/ao-b4 | Stenotrophomonas maltophilia | 95.4 | AF417866 |
| i-b7 | Pseudomonas brassicacearum | 94.5 | AF100321 |
| i-b8 | Pseudomonas brassicacearum | 94.8 | AF100321 |
| i-b9 | Sphingomonas capsulata | 89.3 | D16147 |
| ao-b1 | Herbaspirillum sp. strain BA17 | 97.0 | AF364861 |
| ao-b5 | Psychrophilic marine bacterium PS03 | 97.2 | AF20021 |
| ao-b6 | Uncultured Alcaligenes sp. | 95.0 | AF312672 |
| ao-b7 | Uncultured Alcaligenes sp. | 94.3 | AF312672 |
| ac-b1 | Bacillus sp. isolate P54-2 | 94.1 | AJ297719 |
See Fig. 3A.
When independent database entries gave the same BLAST value, only the entry from the best-characterized source is given (i.e., full species characterization over genus description, bacterial isolate over environmental clone).
Experiment 2: in vitro antagonism.
Bacteria that were identified as Pseudomonas spp. were strongly antagonistic against both Fusarium culmorum and T. harzianum (Table 3). None of the other representatives of dominant groups was antagonistic against T. harzianum. Bacilli and xanthomonads also did not show any antagonistic activity against F. culmorum, and stenotrophomonads showed variable antagonistic activity against this fungus. Of all the strains tested, 42% and 17% were antagonistic against F. culmorum and T. harzianum, respectively.
TABLE 3.
Antagonism on water-agar of representatives of dominant colony-forming bacteria in the soil from Terschelling (experiment 2, all treatments) against two fungia
| Bacterial group | No. of isolates tested | No. of isolates showing in vitro antagonismb against:
|
|
|---|---|---|---|
| Fusarium culmorum | Trichoderma harzianum | ||
| Bacillus | 8 | 0 | 0 |
| Paenibacillus | 6 | 0 | 0 |
| Pseudomonas | 7 | 7 | 6 |
| Stenotrophomonas | 6 | 3 | 0 |
| Xanthomonas | 4 | 0 | 0 |
| Cytophaga | 2 | 2 | 0 |
| Other | 14 | 8 | 2 |
Identified by whole-cell fatty acid analysis and/or partial sequencing of 16S rDNA.
Prolonged inhibition (>2 weeks) of fungal extension (see text).
DISCUSSION
Several results in this study indicate that soil microbial community composition can be a major determinant of soil fungistasis. The most pronounced result in this respect was the prolonged relief of suppression of hyphal growth of C. globosum in the soil from Oostvoorne (experiment 1) after partial sterilization or tryptic soy broth addition. The CO2 production in these treatments dropped to the level of the untreated soil within 3 weeks, whereas the number of bacteria remained an order of magnitude higher than in the untreated soil. This indicates that intense competition for nutrients, i.e., low availability of carbon together with high carbon demand, must have occurred during the incubation of the microwave- and tryptic soy broth-treated soils but that this was not sufficient to induce fungistasis. These results strongly suggest that the changes in the bacterial community which were induced by these treatments resulted in a relative dominance of bacterial populations that produced smaller amounts of substances inhibiting C. globosum.
In contrast to observations for the soil from Oostvoorne, hyphal growth of C. globosum as well as that of all other fungi tested was significantly correlated with CO2 production in the microwave-treated soil from Terschelling (Table 4). The same was seen for the autoclaved-inoculated treatment. Although these results might suggest that hyphal growth was limited by carbon availability, several observations in experiment 2 indicate that carbon availability alone was not sufficient to explain the development of fungistasis in the soil from Terschelling.
TABLE 4.
Pearson correlations between soil respiration (CO2 production) and hyphal
| Fungus | Treatmenta | Expt | No. of data | Correlation coefficientb (r) |
|---|---|---|---|---|
| C. globosum | ACG | 1 | 8 | 0.954* |
| Glucose | 1 | 8 | 0.877* | |
| TSB | 1 | 8 | 0.126 | |
| Microwave | 1 | 8 | 0.355 | |
| Microwave | 2 | 7 | 0.967* | |
| Inoculation | 2 | 7 | 0.782* | |
| All | 2 | 25 | 0.607* | |
| F. culmorum | ACG | 1 | 8 | 0.032 |
| Glucose | 1 | 8 | 0.534 | |
| TSB | 1 | 8 | 0.640 | |
| Microwave | 1 | 8 | 0.761* | |
| Microwave | 2 | 7 | 0.985* | |
| Inoculation | 2 | 7 | 0.741 | |
| All | 2 | 25 | 0.747* | |
| F. oxysporum | Microwave | 2 | 7 | 0.789* |
| Inoculation | 2 | 7 | 0.826* | |
| All | 2 | 25 | 0.785* | |
| T. harzianum | Microwave | 2 | 7 | 0.865* |
| Inoculation | 2 | 7 | 0.968* | |
| All | 2 | 25 | 0.609* |
ACG, acetylglucosamine addition; TSB, tryptic soy broth addition.
*, significant correlation at P < 0.05.
First, suppression of hyphal growth returned more rapidly in the autoclaved-inoculated treatment than in the microwaved treatment, even though carbon availability, as indicated by CO2 production, appeared to be higher in the autoclaved-inoculated treatment during the first weeks of incubation. Second, the suppression of hyphal growth by the microwaved soil remained significantly lower than that of the untreated control for a prolonged period (7 or 10 weeks), whereas CO2 production was similar from week 3 on. Third, mycelial extension of C. globosum and T. harzianum remained much higher in the closed-incubated autoclaved soil compared to the open-incubated autoclaved soil even though the soil respiration did not differ.
The most likely explanation for all these observations is that the distinct microbial communities present after the different treatments produced different amounts or varieties of antifungal compounds. That this apparent production of antifungal compounds correlated with available carbon limitation is not surprising, as microbial production of antibiotics can be induced by interspecific competition for substrates (14, 40).
The higher hyphal growth in the control, i.e., acid-washed, autoclaved beach sand, compared to the dune sand also indicated that nutrient limitation itself was not sufficient to cause fungistasis. It might, however, be argued that nutrients can be withdrawn from the potato dextrose agar disks by soil microorganisms in the nonsterilized dune sands, thereby resulting in a decrease of hyphal extension (29). Therefore, in an additional experiment, we compared hyphal extension from potato dextrose agar discs as performed in the present experiment with hyphal extension from potato dextrose agar disks that were separated from the soil by a sterile stainless steel disk and found no differences (W. de Boer and P. J. A. Klein Gunnewiek, unpublished data). This indicates that nutrient withdrawal is not likely to explain the suppression of hyphal extension.
Other studies have also contended that competition for nutrients cannot fully explain the extent of soil fungistasis, invoking microbial community composition to partially explain differences in fungistasis levels (15, 20, 22, 42). However, no attempts were made to monitor changes within the microbial community. In this study (experiment 2), we showed that all fungistasis-relieving treatments resulted in a large shift in the bacterial community composition, and differences between treatments remained apparent throughout the incubation period. This observation could implicate bacterial community composition as a determining factor in the development of fungistasis, and the variable speed with which suppression of hyphal growth returned for the different treatments is in agreement with this assertion. The bacilli that dominated the autoclaved-closed treatment apparently had no suppressive effect on C. globosum and T. harzianum. Also, the absence of fungus-inhibiting activities by bacilli and paenibacilli on water-agar indicated that these bacteria are probably not involved in fungistasis development.
The fact that fungistasis in the autoclaved-inoculated treatment returned very rapidly to the level of the untreated control may indicate that important microbial components were introduced that were no longer present after the microwaved, autoclaved-open, and autoclaved-closed treatments. For instance, actinomycetes, which are known for their production of antifungal compounds (3, 5), may have been eliminated from these treatments. However, actinomycetes were not essential to the development of fungistasis, as prolonged incubation of microwaved and autoclaved-open treated soils also resulted in strong fungistasis despite no detection of actinomycetes in these soils.
Pseudomonads and stenotrophomonads/xanthomonads were always present when fungistasis reappeared in the different treatments and were absent from the nonfungistatic autoclaved-closed treated soil. These groups are known for their production of antifungal compounds (14, 28, 32). The strong antifungal activity of Pseudomonas isolates against both F. culmorum and T. harzianum on water-agar indicated that they did produce inhibiting compounds under nutrient-limiting conditions. On the contrary, none of the stenotrophomonad/xanthomonad isolates inhibited T. harzianum on water-agar, and only a few strains inhibited F. culmorum. Therefore, the results suggest that the presence and activity of pseudomonads could be essential for the development of fungistasis in these soils.
According to the FAME analysis, the dominant pseudomonads were most closely related to P. putida, P. chlororaphis, or P. fluorescens, and this was confirmed by partial 16S rDNA sequence analysis. The presence and activity of pseudomonads were also indicated to be an important causal factor of fungal suppression by compost (20). However, the in vitro antagonism test indicated that in addition to pseudomonads, other bacteria can also contribute to fungistasis, based upon their production of antifungal compounds under nutrient-limiting conditions.
Toyota et al. (41) hypothesized that microbial diversity may be related to fungistasis. Their hypothesis was based on the observation that addition of a single microbial species to sterile soil aggregates never resulted in the same level of suppression against colonization by F. oxysporum f. sp. raphani as observed for nonsterilized aggregates. It may well be that, in addition to the nutrient status, the complexity of interactions also determines the quantity and quality of antifungal metabolites. For instance, it has been reported that many bacteria which did not normally produce antibiotics could be induced to do so when exposed to other strains or to supernatants from other bacterial cultures (7).
In most studies, fungistasis has been assayed by measuring the repression of spore germination (15, 19, 26, 29, 33) and not by the restriction of hyphal extension, as was done here. In addition to hyphal extension measurements, spore germination tests (spores on nitrocellulose filters on top of soil) were done for a number of fungi as part of the main experiment. The patterns observed for the relief and reappearance of inhibition of spore germination mirrored the results obtained for hyphal extension (W. de Boer and P. Verheggen, unpublished results). It has been suggested by Lockwood (29) that the same mechanism mediates both repression of spore germination and inhibition of hyphal extension, but that spore germination is more sensitive.
Dobbs and Gash (12) have already provided a demonstration that sterilization did not fully remove the fungistasis of some soils. They named this phenomenon residual mycostasis and attributed it to abiotic factors mostly linked to alkalinity. The sensitivity to this residual mycostasis was shown to be species dependent. In the present study, we found a strong residual mycostasis for F. culmorum in the soil from Oostvoorne.
In conclusion, our results indicate that microbial community composition is an important factor determining soil fungistasis and that the presence and antifungal activity of pseudomonads may be essential in this respect. Therefore, this study supports the antibiosis hypothesis as the principal explanation for fungistasis. Although the nutrient status of the soil is likely to play a role by inducing the production of antifungal compounds, it is the microbial community composition and the interactions within that community that determine the quality and quantity of these antifungal compounds.
Footnotes
This is publication 3050 NIOO-KNAW of The Netherlands Institute of Ecology.
REFERENCES
- 1.Alabouvette, C. 1999. Fusarium wilt suppressive soils: an example of disease-suppressive soils. Australas. Plant Pathol. 28:57-64. [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrade, O. A., D. E. Mathre, and D. C. Sands, 1994. Natural suppression of take-all disease of wheat in Montana soils. Plant soil 164:9-18. [Google Scholar]
- 4.Arora, D. K., A. B. Filonow, and J. L. Lockwood. 1983. Exudation from 14C-labeled fungal propagules in the presence of specific microorganisms. Can. J. Microbiol. 29:1487-1492. [Google Scholar]
- 5.Behal, V. 2000. Bioactive products from Streptomyces. Adv. Appl. Microbiol. 47:113-156. [PubMed] [Google Scholar]
- 6.Brüggemann, J., J. R. Stephen, Y.-J. Chang, S. J. Macnaughton, G. A. Kowalchuk, E. Kline., and D. C. White. 2000. Competitive PCR-denaturing gradient gel electrophoresis analysis of bacterial mixtures: an internal standard and an appraisal of template enumeration accuracy. J. Microbiol. Methods 40:111-123. [DOI] [PubMed] [Google Scholar]
- 7.Burgess, J. G., E. M. Jordan, M. Bregu, A. Mearns-Spragg, and K. G. Boyd. 1999. Microbial antagonism: a neglected avenue of natural products research. J. Biotechnol. 70:27-32. [DOI] [PubMed] [Google Scholar]
- 8.De Boer, W., P. J. A. Klein Gunnewiek, and J. Woldendorp. 1998. Suppression of hyphal growth of soil-borne fungi by dune soils from vigorous and declining stands of Ammophila arenaria. New Phytol. 138:107-116. [Google Scholar]
- 9.De Boer, W., P. J. A. Klein Gunnewiek, P. Lafeber, J. D. Janse, B. E. Spit, and J. W. Woldendorp. 1998. Antifungal properties of chitinolytic dune soil bacteria. soil Biol. Biochem. 30:193-203. [Google Scholar]
- 10.De Rooij-Van der Goes, P. C. E. M., W. H. Van der Putten, and C. Van Dijk. 1995. Analysis of nematodes and soil-borne fungi from Ammophila arenaria (Marram grass) in Dutch coastal foredunes by multivariate techniques. Eur. J. Plant Pathol. 101:149-162. [Google Scholar]
- 11.Dobbs, C. G., and W. H. Hinson. 1953. A widespread fungistasis in soil. Nature (London) 172:197-199. [DOI] [PubMed] [Google Scholar]
- 12.Dobbs, C. G., and M. J. Gash. 1965. Microbial and residual mycostasis in soils. Nature (London) 207:1354-1356. [Google Scholar]
- 13.Duineveld, B. M., G. A. Kowalchuk, A. Keizer, J. Van Elsas, and J. A. Van Veen. 2001. Analysis of the bacterial communities in the rhizosphere of chrysanthemum via denaturing gradient gel electrophoresis of PCR amplified 16S ribosomal RNA and DNA fragments. Appl. Environ. Microbiol. 67:172-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellis, R. J., T. M. Timms-Wilson, and M. J. Bailey. 2000. Identification of conserved traits in fluorescent pseudomonads with antifungal activity. Environ. Microbiol. 2:274-284. [DOI] [PubMed] [Google Scholar]
- 15.Fradkin, A., and Z. A. Patrick. 1985. Interactions between conidia of Cochliobolus sativus and soil bacteria as affected by physical contact and exogenous nutrients. Can. J. Plant Pathol. 7:7-18. [Google Scholar]
- 16.Fravel, D. R. 1988. Role of antibiosis in the biocontrol of plant diseases. Annu. Rev. Phytopathol. 26: 75-91. [Google Scholar]
- 17.Garrett, S. D. 1970. Pathogenic root-infecting fungi. Cambridge University Press, Cambridge, UK.
- 18.Heuer, H., M. Krsek, P. Baker, K. Smalla, and E. Wellington. 1997. Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63:3233-3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ho, W. C., and W. H. Ko. 1986. Microbiostasis by nutrient deficiency shown in natural and synthetic soils. J. Gen. Microbiol. 132:2807-2815. [Google Scholar]
- 20.Hoitink, H. A. J., and M. J. Boehm. 1999. Biocontrol within the context of soil microbial communities: A substrate-dependent phenomenon. Annu. Rev. Phytopathol. 37:427-446. [DOI] [PubMed] [Google Scholar]
- 21.Hornby, D. 1983. Suppressive soils. Annu. Rev. Phytopathol. 21:65-85. [Google Scholar]
- 22.Hu, S., A. H. C. Van Bruggen, R. J. Wakeman, and N. J. Grunwald. 1997. Microbial suppression of in vitro growth of Pythium ultimum and disease incidence in relation to soil C and N availability. Plant soil 195:43-52. [Google Scholar]
- 23.Janse, J. D. 1991. Pathovar discrimination within Pseudomonas syringae subsp. Savastanoi with whole cell fatty acid analysis and pathogenicity as criteria. Syst. Appl. Microbiol. 14:79-84. [Google Scholar]
- 24.Knudsen, I. M. B., K. Debosz, J. Hockenhull, D. F. Jensen, and S. Elmholt. 1999. Suppressiveness of organically and conventionally managed soils towards brown foot rot of barley. Appl. soil Ecol. 12:61-72. [Google Scholar]
- 25.Larkin, R. P., D. L. Hopkins, and F. N. Martin. 1996. Suppression of Fusarium wilt of watermelon by nonpathogenic Fusarium oxysporum and other microorganisms recovered from a disease-suppressive soil. Phytopathology 86:812-819. [Google Scholar]
- 26.Liebman, J. A., and L. Epstein. 1992. Activity of fungistatic compounds from soil. Phytopathology 82:147-153. [Google Scholar]
- 27.Liebman, J. A., and L. Epstein. 1994. Partial characterization of volatile fungistatic compound(s) from soil. Phytopathology 84:442-446. [Google Scholar]
- 28.Ligon, J. M., D. S. Hill, P. E. Hammer, N. R. Torkewitz, D. Hofmann, H.-J. Kemf, and K.-H. Van Pée. 2000. Natural products with antifungal activity from Pseudomonas biocontrol bacteria. Pest Manag. Sci. 56:688-695. [Google Scholar]
- 29.Lockwood, J. L. 1977. Fungistasis in soils. Biol. Rev. 52:1-43. [Google Scholar]
- 30.Lockwood, J. L. 1986. Soiborne plant pathogens: Concepts and connections. Phytopathology 76:20-27. [Google Scholar]
- 31.Malý, S., G. W. Korthals, C. Van Dijk, W. Van der Putten, and W. De Boer. 2000. Effect of vegetation manipulation of abandonded arable land on soil microbial properties. Biol. Fertil. soils 31:121-127. [Google Scholar]
- 32.Minkwitz, A., and G. Berg. 2001. Comparison of antifungal activities and 16S ribosomal DNA sequences of clinical and environmental isolates of Stenotrophomonas maltophilia. J. Clin. Microbiol. 39:139-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mondal, S. N., and M. Hyakumachi. 1998. Carbon loss and germinability, viability, and virulence of chlamydospores of Fusarium solani f.sp. phaseoli after exposure to soil at different pH levels, temperatures, and matric potentials. Phytopathology 88:148-155. [DOI] [PubMed] [Google Scholar]
- 34.Muyzer, G., E. C. De Waal, and A. C. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes for 16S rRNA. Appl. Environ. Microbiol. 55:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paulitz, T. C. 1990. Biochemical and ecological aspects of competition in biological control, p. 713-724. In R. Baker and P. E. Dunn (ed.), New directions in biological control: alternatives for suppressing agricultural pests and diseases. Alan R. Liss, Inc., New York, N.Y.
- 36.Pearson, W. R., and D. J. Lipman. 1988. Improved tools for biological sequence analysis. Proc. Natl. Acad. Sci. USA 85:2444-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qian, P., and L. F. Johnson. 1987. Chemical and physical soil characteristics related to lysis of oospores of Pythium ultimum. Phytopathology 77:1062-1066. [Google Scholar]
- 38.Riley, M. A. 1998. Molecular mechanisms of bacteriocin evolution. Annu. Rev. Genet. 32:255-278. [DOI] [PubMed] [Google Scholar]
- 39.Romine, M., and R. Baker. 1973. soil fungistasis: Evidence for an inhibitory factor. Phytopathology 63:756-759. [Google Scholar]
- 40.Slattery, M., I. Rajbhandari, and K. Wesson. 2001. Competition-mediated antibiotic induction in the marine bacterium Streptomyces tenjimariensis. Microb. Ecol. 41:90-96. [DOI] [PubMed] [Google Scholar]
- 41.Toyota, K., K. Ritz, and I. A. Young. 1996. Microbiological factors affecting colonisation of soil aggregates by Fusarium oxysporum f. sp. raphani. soil Biol. Biochem. 28:1513-1521. [Google Scholar]
- 42.Van Os, G. J., and J. H. Van Ginkel. 2001. Suppression of Pythium root rot in bulbous Iris in relation to biomass and activity of the soil microflora. soil Biol. Biochem. 33:1447-1454. [Google Scholar]
- 43.Wiseman, B. M., S. M. Neate, K. O. Keller, and S. Smith. 1996. Suppression of Rhizoctonia solani anastomosis Group 8 in Australia and its biological nature. soil Biol. Biochem. 28:727-732. [Google Scholar]