Abstract
A high-affinity phosphate transporter gene, TcPHO, was isolated from a growth-dependent subtracted cDNA library of the marine unicellular alga Tetraselmis chui. The full-length cDNA of TcPHO obtained by 5′ and 3′ rapid amplification of cDNA ends was 1,993 bp long and encoded an open reading frame consisting of 610 amino acids. The deduced amino acid sequence of TcPHO exhibited 51.6 and 49.8% similarity to the amino acid sequences of PHO89 from Saccharomyces cerevisiae and PHO4 from Neurospora crassa, respectively. In addition, hydrophobicity and secondary structure analyses revealed 12 conserved transmembrane domains that were the same as those found in PHO89 and PHO4. The expression of TcPHO mRNA was dependent on phosphate availability. With a low-phosphate treatment, the TcPHO mRNA concentration increased sharply to 2.72 fmol μg of total RNA−1 from day 1 to day 2 and remained at this high level from days 2 to 4. Furthermore, rescue treatment with either phosphate or p-nitrophenyl phosphate effectively inhibited TcPHO mRNA expression. In contrast, TcPHO mRNA expression stayed at a low level (range, 0.25 to 0.28 fmol μg of total RNA−1) under low-nitrate conditions. The expression pattern suggests that TcPHO can be used as a molecular probe for monitoring phosphorus stress in T. chui.
Phosphorus is an essential macronutrient for the growth of planktonic algae and bacteria. Traditionally, nitrogen is considered a limiting nutrient for phytoplankton in marine environments (10). However, increasing evidence indicates that phosphorus may become limiting for primary production in various regions, including the eastern Mediterranean Sea (12), the Sargasso Sea (5), the northern Red Sea (17), and the western North Atlantic Ocean (30). Furthermore, in the central Atlantic Ocean and the Red Sea, the nitrogen fixation rate of Trichodesmium spp. is also correlated with the cellular content of phosphorus (23, 27).
The conventional methods for determining phosphorus limitation are based mainly on either chemical analysis of phosphate concentration or determination of alkaline phosphatase activity. However, there are problems with experimental manipulation and data interpretation (26). Recently, molecular diagnostic indicators of limiting factors were considered as an alternative tool to identify phytoplankton physiological states in the field (20). One advantage of this approach is that the physiological status of phytoplankton in situ can be identified without incubation. Another advantage is the ability to detect species-specific activities via immunocytochemistry or in situ hybridization. For example, several stress proteins expressed under nutrient starvation conditions have been discovered by using the 14C-labeling method (14, 15), and one of these proteins, flavodoxin, has been used to demonstrate that phytoplankton in the northeast Pacific Ocean is iron stressed (16).
The proper use of a molecular probe relies on a thorough understanding of the targets in metabolic pathways. In the case of phosphate utilization, transport of phosphate across the plasma membrane by a membrane-associated transporter is the first step and an essential step for both prokaryotic and eukaryotic cells. It has been proposed that generally, low-affinity phosphate transporters with a Km of approximately 1 mM are continuously expressed proteins. On the other hand, high-affinity transporters usually have a much lower Km (range, 1 to 15 μM), and their expression is regulated by an inducible expression system when the cells are under phosphate-limiting conditions (21). In the budding yeast Saccharomyces cerevisiae, two derepressive high-affinity phosphate transporters, PHO84 and PHO89, have recently been identified. The expression of PHO84, an H+-coupled phosphate transporter, is stringently controlled by the phosphate concentration in the medium (2). This protein also functions as a receptor for ambient phosphate signals. The other high-affinity phosphate transporter in S. cerevisiae, PHO89, is believed to be an Na+-coupled transporter, because its activity is highly correlated with the external Na+ concentration. Expression of PHO89 is also controlled by the phosphate concentration, but the maximum activity occurs at pH 9.5, which is much higher than the pH reported for PHO84 activity, pH 4.5 (19). Coexistence of different kinds of cation-coupled phosphate transporters in a cell was also observed in Neurospora crassa, in which PHO5 is the H+-coupled transporter and PHO4 is the Na+-coupled transporter (28).
Little is known about proteins involved in phosphate uptake in phytoplankton. Homologs of PstS, the phosphate binding protein in Escherichia coli, were identified in two marine cyanobacterial genera, Synechococcus and Prochlorococcus (24, 25). Polyclonal antiserum raised against PstS has been used to detect the degree of phosphorus depletion in natural picoplankton in a mesocosm experiment (25). In eukaryotic phytoplankton, two membrane-associated proteins with molecular masses of >200 and 50 kDa were identified in Dunaliella tertiolecta and Phaeodacrylum tricornutum under phosphorus deficiency conditions (14). The levels of the former compound were correlated with various degrees of phosphorus limitation. However, the primary functions of this protein seem to be hydrolysis and mobilization of stored polyphosphates, which means that the protein is not directly involved in phosphate uptake (8).
During an effort to establish a growth stage-dependent subtractive cDNA library of a prasinophyte, Tetraselmis chui, a gene homologous to the genes encoding previously described high-affinity phosphorus transporters was isolated. The mRNA expression of this gene was investigated by using a real-time quantitative reverse transcription (RT)-PCR assay, and a strong correlation between mRNA abundance and phosphate availability was confirmed. To our knowledge, this is the first time that the sequence and level of expression of a high-affinity phosphate transporter in a eukaryotic alga have been reported. This discovery not only provides a more detailed understanding of the metabolic physiology of phosphorus in eukaryotic phytoplankton but also permits production of molecular probes which potentially can be used to indicate whether phosphorus assimilation is a limiting process in marine environments.
MATERIALS AND METHODS
Culture conditions.
A unialgal culture of T. chui Butcher (Prasinophyceae) clone TA was provided by H.-M. Su of the Tungkang Marine Laboratory, Pingtung, Taiwan. The culture was grown in f/2-enriched seawater medium at 20°C with continuous illumination by using an irradiance of 145 microeinsteins m−2 s−1 (3, 9). Cell concentrations were determined with a light microscope (BX60; Olympus) at a magnification of ×100 and were used to calculate the population growth rate. To obtain a high rate of growth of algal cells, three-quarters of the culture was removed daily and replaced with fresh f/2 medium. On day 3 after inoculation, cells in the removed fraction were harvested and used for RNA extraction; this sample was designated the rapid-growth culture (RG culture). The remaining part of the culture, after proper dilution with f/2 medium, was maintained under the same conditions, but fresh medium was not added daily. When the growth rate began to decrease, cells were harvested on day 7 and used as the early-stationary-phase culture (ES culture). RNAs extracted from the two cultures were subsequently used for suppression subtractive hybridization.
In the P starvation experiment, 10-ml portions of an algal culture in the exponential phase were added to 100-ml portions of low-phosphate f/2 medium (phosphate concentration, 0.363 μM [1% of the phosphate concentration in normal f/2 medium]) in 250-ml flasks at 20°C under continuous illumination. Similarly, low-nitrate f/2 medium (nitrate concentration, 8.83 μM [1% of the nitrate concentration in normal f/2 medium]) was used in the N starvation experiment. Nutrient-replete cultures under the same growth conditions served as controls. Samples were taken at various growth stages to determine cell concentration, mRNA abundance, and alkaline phosphatase activity. On day 3 after inoculation, a nutrient rescue treatment was performed with selected flasks to raise the phosphate concentration to the normal f/2 medium level by addition of either inorganic phosphate or organic p-nitrophenyl phosphate (PNPP) (Sigma). On day 4, the same sampling and analysis procedures were performed to evaluate the effects of phosphate addition.
Extraction of total RNA.
Approximately 107 cells were harvested by centrifugation and disrupted by sonication. Next, total RNA was isolated with an RNeasy plant mini kit (Qiagen) by following the manufacturer's instructions. The isolated crude RNA was treated with DNase I (RNase free; Roche) at 37°C for 1 h and was purified by acidic phenol (pH 4.0)-chloroform-isoamyl alcohol (25:24:1) extraction. The RNA concentration was determined by spectrophotometry (U2000; Hitachi) at wavelengths of 260 and 280 nm.
Suppression subtractive hybridization.
The suppression subtractive hybridization technique described by Desai et al. (6) was used to identify differentially expressed genes in the ES culture. First, poly(A)+ RNA of both the RG and ES cultures was isolated from total RNA with a QuickPrep Micro mRNA purification kit (Amersham Pharmacia Biotech). Two sets of double-stranded cDNA fragments were generated and mutually subtracted by using a PCR-selected cDNA subtraction kit (Clontech) according to the manufacturer's instructions. Briefly, each set of double-stranded cDNA was digested with RsaI. Next, the RsaI-digested ES culture cDNA was diluted, divided into two parts, and ligated separately to either adapter 1 or adapter 2R. Subtractive hybridization was conducted by adding an excess amount of RG culture cDNA to adapter 1-ligated ES culture cDNA, and the same procedure was repeated for the adapter 2R-ligated ES culture cDNA. Subsequently, a second hybridization was performed by mixing the two reaction mixtures together with additional denatured RG culture cDNA. In this way, the cDNA fragments specific to the ES culture formed double-stranded cDNA with adapter 1 on one end and with adapter 2R on the other end. Finally, ES culture-specific cDNA was amplified by two rounds of PCR with the nested primer pairs embedded in adapters 1 and 2R. The PCR products were then ligated into pGem-T vectors (Promega) to generate a subtracted ES culture cDNA library for further screening. Similarly, RG culture-specific cDNA was generated by ligating adapter 1 and adapter 2R to cDNA from the RG culture in the process of subtractive hybridization. A more detailed explanation of the procedure has been provided previously by Armbrust (1).
Screening of the subtracted cDNA library.
About 10-ng portions of denatured plasmid DNA isolated from individual clones of the subtracted ES culture library were blotted onto two identical membranes. After prehybridization in a buffer containing 50% formamide, 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.02% sodium dodecyl sulfate, 0.1% sodium lauroyl sarcosine, and 2% block reagent (Roche), each membrane was hybridized in the prehybridization buffer with either the digoxigenin-labeled ES culture-specific probe or the RG culture-specific probe at 42°C overnight. The ES and RG culture-specific probes were generated by PCR by using the subtractive hybridization products as templates with digoxigenin-11-dUTP containing a deoxynucleoside triphosphate mixture and the nested primer pairs embedded in adapters 1 and 2R. Subsequent washing was performed as described by Cheng et al. (3), and CDP-Star (Roche) was used for chemiluminescent detection.
DNA and amino acid sequence analyses.
DNA sequencing of the cloned fragments was performed by using an ABI Prism 377A DNA sequencer (Applied Biosystems). The nucleic acid and deduced amino acid sequences were analyzed by using both Lasergene (DNASTAR) and the BESTFIT program from the Wisconsin sequence analysis package of the Genetics Computer Group. Both BLASTX and BLASTP algorithms from the National Center for Biotechnology Information web site (http://www.ncbi.nlm.nih.gov) were also used for sequence analysis. A hydropathy plot and secondary structure analyses of ES99 were conducted by using TopPredII software (http://bioweb.Pasteur.fr/seqanal/interfaces/toppred.html) (4, 13, 29).
Generation of full-length cDNA of ES99.
Full-length cDNA was obtained by both 5′ and 3′ rapid amplification of cDNA ends (RACE) by using the manufacturer's protocol (Roche), with the following modification for 5′ RACE: the homopolymeric tailing was performed with dGTP instead of dATP. Thus, a forward primer of poly(dC) (5′-GCA TGC GCG CGG CCG CGG AGG CCC CCC CCC CCC CC-3′) and nested forward primer anchor-dC (5′-GCA TGC GCG CGG CCG GGA GG-3′) were used in the first and second PCRs, respectively. Subsequent PCR products were cloned into pGem-T vectors (Promega). Positive clones were identified by colony hybridization by using the standard protocol (22).
Real-time quantitative RT-PCR.
DNase I-treated total RNA (1 μg) from various nutrient treatments and known amounts of standard sense RNA were reverse transcribed into cDNA by using an oligo(dT)15 primer (Promega) and MultiScript reverse transcriptase (Applied Biosystems) at 37°C for 10 min and at 48°C for 30 min. PCRs were initiated by adding cDNA to a mixture containing 1× SYBR Green PCR master mixture (Applied Biosystems), 300 nM forward primer TetES99-for (5′-CAT TGT CAG GTC GGA GCT ACC-3′), and 300 nM reverse primer TetES99-rev (5′-TAG AGA CCA GAC GTA GTG AGG CC-3′). The reaction was then conducted with a GeneAmp 5700 sequence detection system (Applied Biosystems). The PCR conditions were 95°C for 10 min for one cycle and then 95°C for 15 s and 60°C for 1 min for 40 cycles. The fluorescence intensity of the complex formed by SYBR Green and double-stranded PCR product was continuously monitored from cycles 1 to 40. The threshold cycle (CT) at which the fluorescence intensity became higher than a preset threshold was used to calculate the ES99 mRNA concentration with a calibration curve constructed by obtaining CT values from a series consisting of known amounts of standard sense RNA.
The standard ES99 sense RNA was synthesized by using a NotI-digested ES99 gene fragment containing part of the C-terminal coding and 3′ untranslated regions as a template and T7 RNA polymerase (Roche) in a reaction mixture containing RNase inhibitor (Promega), each nucleoside triphosphate at a concentration of 1 mM, and 1× transcription buffer at 37°C for 2 h. The synthesized ES99 sense RNA was treated with DNase I and was purified by acid phenol-chloroform extraction. The concentration was determined by spectrophotometry. In addition, the size of the standard RNA was confirmed by electrophoresis on a 2% agarose gel containing 1.2 M formaldehyde.
Finally, the specificity of the quantitative RT-PCR products was confirmed by performing a melting temperature analysis with a GeneAmp 5700 sequence detection system (Applied Biosystems) at temperatures ranging from 65 to 95°C for 20 min and was also examined by electrophoresis on a 3% agarose gel containing 0.5× Tris-boric acid-EDTA buffer.
Alkaline phosphatase activity analysis.
About 105 cells from T. chui cultures were pelleted by centrifugation at 8,000 × g for 10 min. Next, the cells were resuspended in 990 μl of f/2 medium containing an additional 135 μl of Tris-glycine buffer (50 mM, pH 8.5) and 13.5 μl of 1 mM MgCl2. Alkaline phosphatase activity was determined by adding 50 μl of 10 mM PNPP (Sigma) as the substrate. Following incubation for 90 min at 20°C in the dark, the absorbance of p-nitrophenol (PNP) was determined with a spectrophotometer (U2000; Hitachi) at 400 nm. The concentration of PNP was determined from the absorbance by using a molar extinction coefficient of 18,000 cm−1 (17, 31).
Nucleotide sequence accession number.
The nucleotide sequence of ES99 (TcPHO) has been deposited in the GenBank database under accession number AF520588.
RESULTS
Subtracted cDNA library construction and screening.
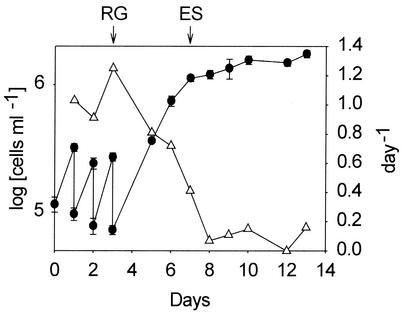
By using the daily dilution method we successfully maintained the culture of T. chui at the exponential phase with high growth rates (range, 0.9 to 1.3 day−1) and a population density of 2.5 × 105 cells ml−1 (Fig. 1). When the daily addition of fresh medium was stopped after day 3, the cell number continued to increase and reached 1.1 × 106 cells ml−1 on day 7, but the growth rate decreased to 0.4 day−1. By using mRNA obtained from the RG and ES cultures, a PCR-based subtraction cDNA library of T. chui was constructed in order to enrich genes specifically expressed in the ES culture. A total of 34 cDNA fragments were selected from the ES culture library for further dot blot hybridization screening. cDNA fragments that could be hybridized with the ES culture probe but not with the RG culture probe were considered ES culture-specific clones, and 22 such clones were obtained. Sequence analysis and a database search of these cDNA clones revealed that most of them were gene fragments exhibiting low levels of similarity to genes in the GenBank database. The only exception was cDNA fragment ES99, which was 476 bp long and had an identity score of 41% with the N. crassa phosphate permease gene, PHO4 (18).
FIG. 1.
Time courses for cell number (•) and growth rate (▵) for the T. chui cultures used for construction of subtracted cDNA libraries. The cultures were diluted daily with fresh medium between days 1 and 3 after inoculation. The arrows indicate the times at which samples of total RNA were collected from the RG and ES cultures. The error bars indicate the standard errors of the means for cell number (n = 3).
cDNA and the putative peptide structure of TcPHO.
The full-length cDNA of ES99 obtained by RACE was 1,993 bp long. It contained an 1,833-bp coding region, as well as 91- and 69-bp stretches of 5′ and 3′ untranslated regions, respectively. The open reading frame in the coding region encoded a polypeptide containing 610 amino acids (Fig. 2a). Furthermore, the DNA sequence in the coding region was confirmed by RT-PCR amplification by using a high-fidelity DNA polymerase (Advantage cDNA polymerase mixture; Clontech) with primers located at amino acid positions 1 to 7 and 605 to 610. A deduced amino acid sequence comparison revealed that the ES99 sequence exhibited 38% identity and 49.8% similarity with the sequence of PHO4 from N. crassa (18). In addition, the ES99 sequence exhibited 40% identity and 51.6% similarity with the sequence of PHO89 from S. cerevisiae (7, 19) (Fig. 2b). A hydropathy plot and secondary structure analysis of the ES99 polypeptide by using TopPredII indicated that there were 12 transmembrane regions connected by 11 hydrophilic loops (Fig. 2a). Based on these similarities, ES99 cDNA should encode a high-affinity membrane-associated sodium phosphate transporter protein, which was tentatively designated TcPHO.
FIG. 2.
(a) Deduced amino acid sequence encoded by TcPHO from T. chui. Twelve predicted transmembrane domains are underlined, and two conserved repeats are shaded. The numbers on the right indicate amino acid positions. The GenBank accession number of the sequence is AF520588. (b) Phylogenic relationship between TcPHO and other high-affinity phosphate transporters whose physiological functions have been experimentally established. Alignment of amino acid sequences and phylogenetic analysis were performed by using the Lasergene software (DNASTAR, Inc.) according to an algorithm described by Hein (11). The sequences included are the sequences of PHO84 (GenBank accession number NP_013583) and PHO89 (NP_009855) from S. cerevisiae; PHO4 (JQ0116) and PHO5 (AAA74899) from N. crassa; and AtPT1 (AAB17265), AtPT2 (AAB88291), and AtPHT2 (AAM53960) from Arabidopsis thaliana. Scale bar = 100 nucleotide substitutions.
Effect of nutrient depletion on TcPHO mRNA expression.
In the real-time quantitative RT-PCR analysis, the CT had a linear relationship with the logarithm of the TcPHO sense RNA concentration in the standard solution. The slope of the standard curve was −3.58, which was close to the theoretical value, −3.32 (data not shown). The variation among replicates was also small, and the coefficients of variation ranged from 0.08 to 0.71%.
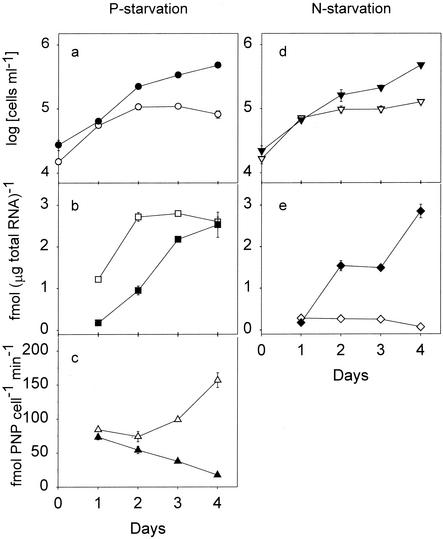
In the P starvation experiment, cells in the nutrient-replete and low-phosphate treatment preparations grew exponentially for the first 2 days after inoculation, with the growth rates ranging from 0.85 to 1.30 day−1. The growth rate of the nutrient-replete culture declined to 0.4 day−1 after day 2, and the concentration reached 4.8 × 105 cells ml−1 by day 4. In contrast, growth completely stopped after day 2 in the low-phosphate culture, and by day 4 the concentration was only 8.2 × 104 cells ml−1 (Fig. 3a).
FIG. 3.
Effects of nutrient depletion on cell number, TcPHO mRNA level, and alkaline phosphatase activity in T. chui. (a to c) P starvation experiment. (a) Time course for cell number. (b) TcPHO mRNA expression level. (c) Alkaline phosphatase activity. (d and e) N starvation experiment. (d) Time course for cell number. (e) TcPHO mRNA expression level. Solid symbols, data from nutrient-replete cultures; open symbols, data from low-phosphate or low-nitrate cultures. The error bars indicate the standard errors of the means (n = 3). For data points without an error bar the error bar is smaller than the symbol.
In the nutrient-replete culture, the TcPHO mRNA level gradually increased with the age of the culture. The highest mRNA level, 2.53 fmol μg of total RNA−1, was observed on day 4. However, in the low-phosphate culture, the TcPHO mRNA level increased sharply to 2.72 fmol μg of total RNA−1 between days 1 and 2 and remained at this high level from day 2 to day 4 (Fig. 3b).
The variation in alkaline phosphatase activity was not correlated with the variation in the TcPHO mRNA level. In the nutrient-replete culture, the alkaline phosphatase activity decreased from day 1 to day 4. On the other hand, the alkaline phosphatase activity in the low-phosphate culture decreased slightly from day 1 to day 2 but began to increase from day 2 and reached the highest level, 157 fmol of PNP cell−1 min−1, on day 4 (Fig. 3c).
In the N starvation experiment, cells in the nutrient-replete culture showed a growth curve similar to that of their counterparts in the P starvation experiment. However, the low-nitrate culture stopped growing completely after day 2, and the cell number remained at 1.3 × 105 cells ml−1 until day 4 (Fig. 3d). Again, the TcPHO mRNA expression level in the nutrient-replete culture increased with culture age. In contrast, the level of TcPHO mRNA in the low-nitrate culture remained low (0.25 to 0.28 fmol μg of total RNA−1) from day 1 to day 3 and dropped to 0.07 fmol μg of total RNA−1 on day 4 (Fig. 3e).
Nutrient rescue effects on TcPHO expression.
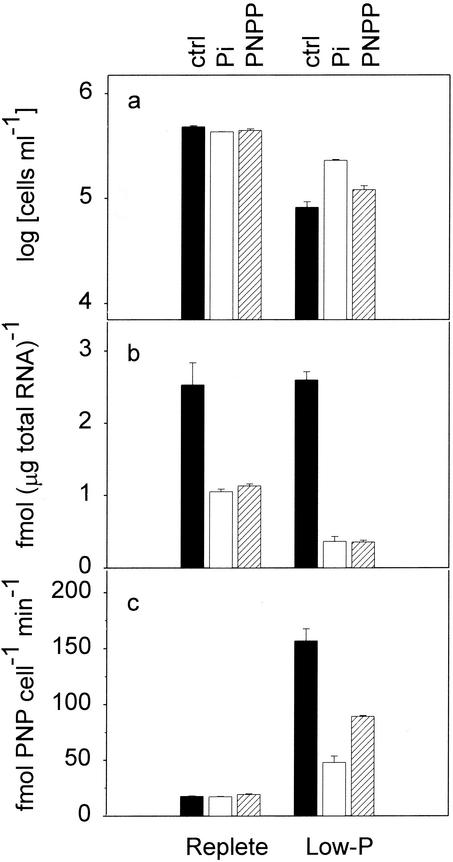
In the P starvation experiment, rescue treatment in which the phosphorus concentration was restored to the full-strength f/2 medium level on day 3 had no effect on cell number in the nutrient-replete culture (Fig. 4a). In the low-phosphate cultures, addition of phosphate resulted in increased cell concentrations compared to the concentration in the no-addition control. However, addition of organic phosphate (PNPP) caused no obvious increase in the cell concentration. The concentration in the PNPP-supplemented culture was 1.2 × 105 cells ml−1, which was only slightly higher than the 8.2 × 104 cells ml−1 in the no-addition control on day 4.
FIG. 4.
Effects of phosphate addition (rescue effects) on cell growth (a), TcPHO mRNA expression (b), and alkaline phosphatase activity (c) in T. chui. Phosphate was added on day 3 after inoculation, and various measurements were obtained on day 4. The additional phosphate was provided as either inorganic phosphate (Pi) (open bars) or organic phosphate (PNPP) (cross-hatched bars). The culture which received no phosphorus on day 3 served as the control (ctrl) (solid bars). The error bars indicate the standard errors of the means (n = 3).
In both the nutrient-replete and low-phosphate cultures, addition of phosphate effectively inhibited TcPHO mRNA expression. In the nutrient-replete cultures, the TcPHO mRNA levels dropped from 2.53 to 1.05 and 1.13 fmol μg of total RNA−1 in the culture to which phosphate was added and in the culture to which PNPP was added, respectively. In the low-phosphate cultures, the TcPHO mRNA level dramatically decreased from 2.60 to 0.37 fmol μg of total RNA−1 when phosphate and PNPP were added (Fig. 4b).
In the nutrient-replete cultures, nutrient rescue had no effect on alkaline phosphatase activity. The activities in the no-addition control and in the two cultures to which phosphate was added remained low (around 17 fmol of PNP cell−1 min−1) (Fig. 4c). However, in the low-phosphate cultures, phosphate addition decreased the alkaline phosphatase activity. The activities were 157 fmol of PNP cell−1 min−1 in the no-addition control and 48 and 89 fmol of PNP cell−1 min−1 in the cultures to which phosphate and PNPP were added, respectively.
DISCUSSION
A phylogenetic analysis based on amino acid similarity revealed that TcPHO is closely related to high-affinity sodium phosphate transporters from S. cerevisiae (PHO89) and N. crassa (PHO4) (Fig. 2b). Although the level of similarity is not especially high, TcPHO does have the 12 membrane-spanning domains found in PHO4 and PHO89 (Fig. 2a). The largest hydrophilic loop in TcPHO is also located between membrane-spanning domains VII and VIII and is on the side of the membrane opposite the N and C termini. This hydrophilic loop, if it is proven to be extracellular, should be suitable for production of an antiserum against TcPHO. Furthermore, two conserved short internal repeats with the sequence G-A-N-D-V-A-N were identified in TcPHO. One repeat is located in the hydrophilic loop between membrane-spanning domains I and II, while the other is located in the hydrophilic loop before domain VIII (Fig. 2a). Similar repeats are present in PHO4 and in PHO89 at similar positions (20).
The results of the nutrient starvation experiments clearly indicated that TcPHO expression is tightly linked to the level of ambient phosphate. The amount of TcPHO mRNA increased immediately after algal cells were inoculated into the low-phosphate medium, and TcPHO expression was effectively inhibited by the addition of external phosphate (Fig. 3b and 4b). Both orthophosphate and organic phosphate (PNPP) had similar inhibitory effects on TcPHO transcription in these rescue experiments. In contrast, the level of TcPHO expression remained low in the low-nitrate medium in which there was a great excess of phosphate (Fig. 3e). This evidence indicates that TcPHO expression responds solely to the external concentration of phosphate. A similar result was obtained for S. cerevisiae. Northern blot analysis showed that PHO89 was transcribed only under phosphorus-limited conditions (19).
Although the variation in alkaline phosphatase activity was correlated with the level of TcPHO mRNA expression in the low-phosphate medium, the increase in the TcPHO mRNA level occurred prior to the increase in alkaline phosphatase activity (Fig. 3b and c). Apparently, when T. chui senses that the ambient phosphorus level is beginning to decrease, it first scavenges trace amounts of orthophosphate from the environment by using a high-affinity transporter, such as TcPHO. Only after the external orthophosphate is almost exhausted does T. chui activate its alkaline phosphatase to utilize organic phosphate. This hypothesis can be used to explain results observed in phosphate-replete cultures (Fig. 3b and c). The slowly decreasing alkaline phosphatase activity suggests that the phosphate concentrations were not low enough to activate alkaline phosphatase. However, as the phosphate was gradually consumed as the culture aged, TcPHO expression increased to meet the needs for growth.
In a recent review, Palenik and Wood (20) suggested that the high-affinity phosphate transporter system is a good candidate for production of molecular probes in marine research if this system can be identified in phytoplankton. Based on structural similarities and a physiological response to a phosphate deficiency, TcPHO is clearly part of the high-affinity transporter system in algae and should be an ideal molecular indicator for phosphorus limitation in T. chui. Obviously, it is desirable to extend this approach to evaluate phosphate limitation in all eukaryotic phytoplankton in marine ecosystems. To do this, information about high-affinity phosphate transporter genes in other phytoplankton groups is required in order to produce proper probes or antisera. In addition, alkaline phosphatase activity has been shown to vary with both phytoplankton species and irradiance (32). How these factors influence the expression of high-affinity phosphate transporter genes also needs to be investigated in order to avoid possible confusion in data interpretation.
Acknowledgments
We thank C. C. Lin and D. N. Lin for their help with DNA sequencing.
This study was supported by research grants NSC90-2313-B-019-017 and NSC89-2313-B-019-078 from the National Science Council of the Republic of China.
REFERENCES
- 1.Armbrust, E. V. 1999. Identification of a new gene family expressed during the onset of sexual reproduction in the centric diatom Thalassiosira weissflogii. Appl. Environ. Microbiol. 65:3121-3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bun-ya, M., M. Nishimura, S. Harashima, and Y. Oshima. 1991. The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell. Biol. 11:3229-3238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cheng, L.-C., S.-P. L. Hwang, and J. Chang. 1997. Gene sequence and expression of an analog of proliferating cell nuclear antigen (PCNA) in the alga Tetraselmis chui and detection of the encoded protein with anti-rat PCNA monoclonal antibody. Appl. Environ. Microbiol. 63:4010-4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claros, M. G., and G. von Heijne. 1994. TopPredII: an improved software for membrane protein structure predictions. Comput. Applic. Biosci. 10:685-686. [DOI] [PubMed] [Google Scholar]
- 5.Cotner, J. B., Jr., and R. G. Wetzel. 1992. Uptake of dissolved inorganic and organic phosphorus compounds by phytoplankton and bacterioplankton. Limnol. Oceanogr. 37:232-243. [Google Scholar]
- 6.Desai, S., J. Hill, S. Trelogan, L. Diatchenko, and P. D. Siebert. 2000. Identification of differentially expressed genes by suppression subtractive hybridization, p. 81-112. In S. P. Hunt and F. J. Livesey (ed.), Functional genomics. Oxford University Press, New York, N.Y.
- 7.Feldmann, H., M. Aigle, G. Aljinovic, G. B. Andre, M. C. Baclet, C. Barthe, A. Baur, A. M. Becam, N. Biteau, and E. Boles. 1994. Complete DNA sequence of yeast chromosome II. EMBO J. 13:5795-5809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Graziano, L. M., J. La Roche, and R. J. Geider. 1996. Physiological responses to phosphorus limitation in batch and steady-state cultures of Dunaliella tertiolecta (Chlorophyta): a unique stress protein as an indicator of phosphate deficiency. J. Phycol. 32:825-838. [Google Scholar]
- 9.Guillard, R. R. L., and J. H. Ryther. 1962. Studies on marine planktonic diatoms. I. Cyclotella nana Hustedt and Detonula confervacae (Cleve) Gran. Can. J. Microbiol. 8:229-239. [DOI] [PubMed] [Google Scholar]
- 10.Hecky, R. E., and P. Kilham. 1988. Nutrient limitation of phytoplankton in freshwater and marine environments: a review of recent evidence on the effects of environment. Limnol. Oceangr. 33:796-822. [Google Scholar]
- 11.Hein, J. J. 1990. Unified approach to alignment and phylogenesis. Methods Enzymol. 183:626-645. [DOI] [PubMed] [Google Scholar]
- 12.Krom, M. D., N. Kress, and S. Brenner. 1991. Phosphorus limitation of primary productivity in the eastern Mediterranean Sea. Limnol. Oceanogr. 36:424-432. [Google Scholar]
- 13.Kyte, J., and R. F. Doolittle. 1982. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157:105-132. [DOI] [PubMed] [Google Scholar]
- 14.La Roche, J., R. J. Geider, L. M. Graziano, H. Murray, and K. Lewis. 1993. Induction of specific proteins in eukaryotic algae grown under iron-, phosphorus-, or nitrogen-deficient conditions. J. Phycol. 26:767-777. [Google Scholar]
- 15.La Roche, J., H. Murray, M. Orellana, and J. Newton. 1995. Flavodoxin expression as an indicator of iron limitation in the marine diatoms. J. Phycol. 31:520-530. [Google Scholar]
- 16.La Roche, J., P. W. Boyd, R. M. L. Mckay, and R. J. Geider. 1996. Flavodoxin as an in situ marker for iron stress in phytoplankton. Nature 382:802-805. [Google Scholar]
- 17.Li, H., M. J. W. Veldhuis, and A. F. Post. 1998. Alkaline phosphatase activities among planktonic communities in the northern Red Sea. Mar. Ecol. Prog. Ser. 173:107-115. [Google Scholar]
- 18.Mann, B. J., B. J. Bowman, J. Grotelueschen, and R. L. Metzenberg. 1989. Nucleotide sequence of pho-4+, encoding a phosphate-repressible phosphate permease of Neurospora crass. Gene 83:281-289. [DOI] [PubMed] [Google Scholar]
- 19.Martinez, P., and B. L. Persson. 1998. Identification, cloning and characterization of a derepressible Na+-coupled phosphate transporter in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:628-638. [DOI] [PubMed] [Google Scholar]
- 20.Palenik, B., and A. M. Wood. 1998. Molecular markers of phytoplankton physiological status and their application at the level of individual cells, p. 187-205. In K. E. Cooksey (ed.), Molecular approaches to the study of the ocean. Chapman & Hall, London, United Kingdom.
- 21.Persson, B. L., J. Petersson, U. Fristedt, R. Weinander, A. Berhe, and J. Pattison. 1999. Phosphate permeases of Saccharomyces cerevisiae: structure, function and regulation. Biochim. Biophys. Acta 1422:255-272. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sañudo-Wilhelmy, S. A., A. B. Kustka, C. J. Gobler, D. A. Hutchins, M. Yang, K. Lwiza, J. Burns, D. G. Capone, J. A. Raven, and E. J. Carpenter. 2001. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411:66-69. [DOI] [PubMed] [Google Scholar]
- 24.Scanlan, D. J., N. H. Mann, and N. G. Carr. 1993. The response of the picoplanktonic marine cyanobacterium Synechococcus species WH7803 to phosphate starvation involves a protein homologous to the periplasmic phosphate-binding protein of Escherichia coli. Mol. Microbiol. 10:181-191. [DOI] [PubMed] [Google Scholar]
- 25.Scanlan, D. J., N. J. Silman, K. M. Donald, W. H. Wilson, N. G. Carr, I. Joint, and N. H. Mann. 1997. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl. Environ. Microbiol. 63:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scanlan, D. J., and W. H. Wilson. 1999. Application of molecular techniques to addressing the role of P as a key effector in the marine ecosystems. Hydrobiologia 401:149-175. [Google Scholar]
- 27.Stihl, A., U. Sommer, and A. F. Post. 2001. Alkaline phosphatase activities among populations of the colony-forming diazotrophic cyanobacterium Trichodesmium spp. (Cyanobacteria) in the Red Sea. J. Phycol. 37:310-317. [Google Scholar]
- 28.Versaw, W. K., and R. L. Metzenberg. 1995. Repressible cation-phosphate symporters in Neurospora crassa. Proc. Natl. Acad. Sci. USA 92:3884-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.von Heijne, G. 1992. Membrane protein structure prediction hydrophobicity analysis and positive-inside rule. J. Mol. Biol. 225:487-494. [DOI] [PubMed] [Google Scholar]
- 30.Wu, J., W. Sunda, E. A. Boyle, and D. M. Karl. 2000. Phosphate depletion in the western North Atlantic Ocean. Science 289:759-762. [DOI] [PubMed] [Google Scholar]
- 31.Wynne, D. 1981. The role of phosphatases in the metabolism of Peridinium cinctum from Lake Kinneret. Hydrobiologia 83:93-99. [Google Scholar]
- 32.Wynne, D., and G.-Y. Rhee. 1988. Changes in alkaline phosphatase activity and phosphate uptake in P-limited phytoplankton, induced by light intensity and spectral quality. Hydrobiologia 160:173-178. [Google Scholar]