Abstract
The goal of this study was to identify genes in Bacillus cereus, a bacterium commonly associated with plant seeds and roots, that are affected by compounds originating from a host plant, tomato, or another rhizosphere resident, Pseudomonas aureofaciens. We constructed a B. cereus chromosomal DNA library in a promoter-trap plasmid, pAD123, which contains a promoterless version of the green fluorescent protein (GFP) gene, gfpmut3a. The library was screened by using fluorescence-activated cell sorting for clones showing a change in GFP expression in response to either tomato seed exudate or culture supernatant of P. aureofaciens strain 30-84. We identified two clones carrying genes that were induced by the presence of tomato seed exudate and nine clones carrying genes that were repressed by P. aureofaciens culture supernatant. A clone chosen for further study contained an open reading frame, designated lipA, that encodes a deduced protein with a lipoprotein signal peptide sequence similar to lipoproteins in B. subtilis. Expression of gusA under control of the lipA promoter increased twofold when cells were exposed to tomato seed exudate and in a concentration-dependent manner when exposed to a mixture of amino acids. When the wild type and a 10-fold excess of a lipA mutant were applied together to tomato seeds, 2 days after planting, the wild type displayed medium-dependent culturability, whereas the lipA mutant was unaffected. This study demonstrates the power of a promoter trap to identify genes in a gram-positive bacterium that are regulated by the biotic environment and resulted in the discovery of lipA, a plant-regulated gene in B. cereus.
Plants play host to a wide variety of microorganisms, including bacteria. The relationships between the bacteria and their host plants are diverse and include pathogenicity, symbiotic root nodule formation and nitrogen fixation, plant growth promotion, disease suppression, and probably other, as-yet-undiscovered, interactions. Two of the best-studied interactions between plant hosts and bacteria include the root nodule-inhabiting Rhizobium spp. and gall-forming Agrobacterium tumefaciens. Study of these systems led to the discovery that plants and bacteria communicate by using chemical signals that determine the outcome of the relationship between the organisms (32, 44, 45, 52). Since these pioneering studies, research has revealed that many relationships between plants and gram-negative bacteria are mediated by compounds that influence bacterial gene expression (3, 11, 29, 35, 50). In contrast, little attention has focused on the gram-positive bacteria, which are common in soil and on plant tissue (8, 10). To address this knowledge gap, we are investigating chemical communication between gram-positive bacteria and their plant hosts.
For more than two decades, genetic analysis of mutants has been the archetypal approach to understanding plant-microbe interactions. Nonnodulating mutants of Rhizobium spp. (24), avirulent mutants of A. tumefaciens (9), Ralstonia solanacearum (4, 18), and Pseudomonas syringae (2, 23), and antibiotic-deficient mutants of biocontrol agents such as Bacillus cereus (39) and Pseudomonas fluorescens (46) have provided insights into the mechanisms of interaction in each of these symbioses. Each of these classes of mutants had a dramatic null phenotype readily detected in a qualitative assay that could be conducted on thousands of candidates for an exhaustive mutant hunt. Although mutant analysis has been fruitful, it limits understanding of symbioses to genes that have global effects because they affect early stages of infection or are pleiotropic, as are the regulators and secretion systems that have been identified repeatedly in such screens. Genes with subtle quantitative effects are undoubtedly involved in the establishment of these symbioses, and yet most mutant screens will not detect them. To capture the full range of genes that contribute to a successful symbiosis, the arsenal of tools for identification of the genes must expand. Promoter traps have been a useful tool for identifying genes that were overlooked in traditional mutant screens (26, 36), and it seems likely that such methods will become essential to developing an accurate portrait of the complex interactions that result in the establishment of a symbiosis.
B. cereus is a gram-positive bacterium that is ubiquitous in soil and in the rhizosphere of plants (13, 34, 43). Previous work with recombinant inbred lines (RILs) of tomato demonstrated a genetic basis in tomato for the support of growth of B. cereus strains in the spermosphere after seed inoculation (40, 42). Several quantitative trait loci were identified that are highly correlated with growth of B. cereus strain UW85 on tomato seeds. Additionally, although there were differences among the RILs in their ability to support the growth of B. cereus, they did not differ in support of growth of strains of P. fluorescens or Pseudomonas aureofaciens (40). The results of these studies suggest that specific host factors are required for growth of B. cereus in the tomato spermosphere. We speculated that chemicals from the tomato seed or seedling might influence gene expression in B. cereus, and the outcome of this gene expression could affect the ability of B. cereus to grow in the tomato spermosphere.
The relationship between B. cereus and host plants is likely influenced by other plant-associated bacteria present in the system. Strains of P. aureofaciens are commonly found associated with field-grown plant roots (33) and have been isolated from the same alfalfa and soybean root segments as B. cereus (E. A. B. Emmert, L. Jacobson, G. S. Gilbert, and J. Handelsman, unpublished data). Compounds produced by rhizosphere bacteria living in close proximity to B. cereus could influence gene expression and ultimately behavior of B. cereus on plant tissue. To begin to address this possibility, we chose to investigate the effect of P. aureofaciens 30-84 (20, 33) culture supernatant on gene expression in B. cereus UW85.
We used a modification of differential fluorescence induction (48) to identify genes whose promoter activity was influenced by the presence of tomato seed exudate from a RIL capable of supporting growth of B. cereus or P. aureofaciens culture supernatant. We identified several genes regulated by biotic material.
MATERIALS AND METHODS
Strains and plasmids.
The strains and plasmids used in the present study are listed in Table 1. The promoter-trap plasmid, pAD123, is available through the Bacillus Genetic Stock Center (BGSC) at The Ohio State University http://bacillus.biosci.ohio-state.edu/; BGSC number ECE165).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | General purpose strain | 14 |
| E. coli GM2929 | dcm-6 dam-13::Tn9 | E. coli Genetic Stock Center |
| B. cereus UW85 | Wild type, lipA+ | 15 |
| P. aureofaciens 30-84 | Wheat rhizosphere isolate | 20, 33 |
| B. cereus UW85 ΔlipA | UW85 with SprlipA deletion mutant | This study |
| Clone SL7 | B. cereus UW85 containing pADSL7 | This study |
| Plasmids | ||
| pAD123 | Promoter-trap shuttle vector | 6 |
| pADSL7 | pAD123 with a DNA insert containing the lipA promoter | This study |
| pBC16 | B. cereus plasmid, Tcr | 30 |
| pUCspgusA | pUC18 containing pDG1726 Spr and gusA | This study |
| pBeloBAC11 | BAC vector, Cmr | 19 |
| pDG1726 | Spr cassette | 12 |
| pUC18 | General-use E. coli cloning vector | 53 |
| pTUC | pUC18 containing pBC16 Tcr | This study |
Spr, spectinomycin resistance; Tcr, tetracycline resistance; Cmr, chloramphenicol resistance.
Culture conditions.
Escherichia coli was grown in Luria-Bertani (LB) broth (27) or on LB agar at 37°C. B. cereus was grown in 0.5× or 0.1× tryptic soy broth (TSB; Becton Dickinson, Sparks, Md.); on 0.5×, 0.1×, or 0.01× tryptic soy agar (TSA); in MESAA1 with dl-malate as the carbon source (28); in MESAA1 containing tomato seed exudate (MESAA1E; 1 ml of MESAA1E contains 0.25 ml of 4× MESAA1, 0.1 ml of tomato seed exudate, and 0.65 ml of sterile distilled water); in MESAA1 containing 0.5 mM Ca2SO4 (MESAA1C; 1 ml of MESAA1C contains 0.25 ml of 4× MESAA1, 0.1 ml of 0.5 mM Ca2SO4, and 0.65 ml of sterile distilled water); MESAA1 containing P. aureofaciens 30-84 culture supernatant (MESAA1sup; 1 ml of MESAA1sup contains 0.5 ml of MESAA1 and 0.5 ml of P. aureofaciens MESAA1 48-h culture supernatant); or brain heart infusion broth (Becton Dickinson). B. cereus and P. aureofaciens cultures were incubated at 28°C with vigorous shaking.
Antibiotic selection for E. coli strains containing pAD123 or its derivatives was done with either 10 μg of chloramphenicol or 100 μg of ampicillin/ml. Selection for B. cereus containing pAD123 or its derivatives was done with 10 μg of chloramphenicol/ml. Selection for the chromosomal gusA reporter strain was done with 75 μg of spectinomycin/ml.
Electroporation of E. coli and B. cereus.
E. coli was electroporated by using the protocol in the operating manual for the GenePulser transfection apparatus (Bio-Rad Laboratories, Hercules, Calif.).
B. cereus was grown overnight in 10 ml of 0.5× TSB. A total of 2 ml of overnight culture was used to inoculate 500 ml of brain heart infusion broth. When the culture reached an optical density at 600 nm of 0.3, it was centrifuged at 8,000 × g for 10 min at 4°C, and the pellet was resuspended in 40 ml of cold electroporation buffer (0.5 mM MgCl2, 272 mM sucrose, 0.2 mM K2HPO4, 50 μM KH2PO4), filter sterilized, and stored at 4°C. Cells were centrifuged at 6,000 × g for 6 min at 4°C. The pellet was resuspended in 0.5 ml of cold electroporation buffer, and the cell suspension was placed on ice. DNA (0.1 to 1.0 μg) in 1 to 20 μl of water was mixed with 100 μl of the cell suspension, incubated on ice for 5 min, and transferred to a chilled cuvette with a 0.2-cm interelectrode gap (Bio-Rad). Cells were electroporated at 25 μF, 200 Ω, and 1.2 kV in a GenePulser electroporation apparatus (Bio-Rad). Cells were transferred to 2 ml of LB medium in an 18-mm test tube, incubated at 37°C with shaking for 1 h, and spread onto 0.5× TSA containing the appropriate antibiotics. All plasmids isolated from E. coli DH5α were electroporated into, and isolated from, E. coli GM2929 prior to B. cereus electroporation.
Construction of the promoter-trap library.
The promoter trap library was constructed as described previously (6), with the following modifications. Chromosomal DNA from B. cereus UW85 was isolated by using the Easy-DNA kit (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol, with the addition of vortexing the cells for 3 min in the presence of 0.1-mm-diameter silica beads before the addition of solution A. B. cereus UW85 chromosomal DNA was partially digested with Sau3A, and 1- to 3-kb fragments were purified by using the QIAquick gel purification system (Qiagen, Valencia, Calif.) prior to ligation into pAD123 (6). The ligation mixture was introduced into E. coli DH5α by electroporation, and cells were plated on LB agar containing 10 μg of chloramphenicol and 100 μg of ampicillin/ml. Approximately 33,000 colonies were scraped from the plates after overnight incubation, and plasmids were purified by using the Qiagen QIAprep Spin Miniprep kit. The library was electroporated into E. coli GM2929, plated and purified as for E. coli DH5α, and then introduced into B. cereus UW85 by electroporation. Transformants were selected on 0.5× TSA containing 10 μg of chloramphenicol/ml. Transformation plates were scraped and pooled into two groups in 0.5× TSB containing 10 μg of chloramphenicol/ml and 10% glycerol, and aliquots were frozen and stored at −80°C.
Preparation of tomato seed exudate.
RIL 37 tomato (31, 42) was field grown, and seeds were separated from the fruit in the laboratory as described previously (41). Wisconsin 55 tomato seed was purchased from Jung Seed Company (Randolph, Wis.).
A total of 50 mg of tomato seeds were surface disinfected by a brief rinsing with 95% ethanol, followed by shaking at 25°C for 5 min in 100 ml of diluted bleach solution (99 ml of deionized water and 1 ml of 6% hypochlorite solution). Seeds were rinsed three times for 5 min each in 100 ml of sterile deionized water at 25°C with shaking. Each 50-mg portion of disinfected seeds was transferred to a single well of a 24-well plate containing 1.5 ml of sterile 0.5 mM Ca2SO4 (1× exudate). The plate was sealed with parafilm, wrapped in foil, and placed on a shaker at 25°C, with gentle shaking. After 24 h of incubation, 10 μl was removed and spotted on 0.1× TSA. The agar plate was incubated overnight at 28°C. Exudate that did not show bacterial growth on 0.1× TSA was collected after 48 h of incubation, filter sterilized by using a 0.22-μm (pore-size) nylon presterilized syringe filter, and used in experiments. Exudate was prepared immediately before use for all flow cytometry experiments and the quantitative β-glucuronidase (GUS) assays.
Preparation of P. aureofaciens culture supernatant.
P. aureofaciens strain 30-84 (20, 33) was grown in MESAA1 medium for 48 h with vigorous shaking. Cells were pelleted by centrifugation (6,000 × g for 6 min), and culture supernatant was filter sterilized and stored at −20°C. For screening, thawed supernatant was diluted with an equal volume of MESAA1 medium (MESAA1sup).
Screening of the promoter-trap library by using a modified differential fluorescence induction technique.
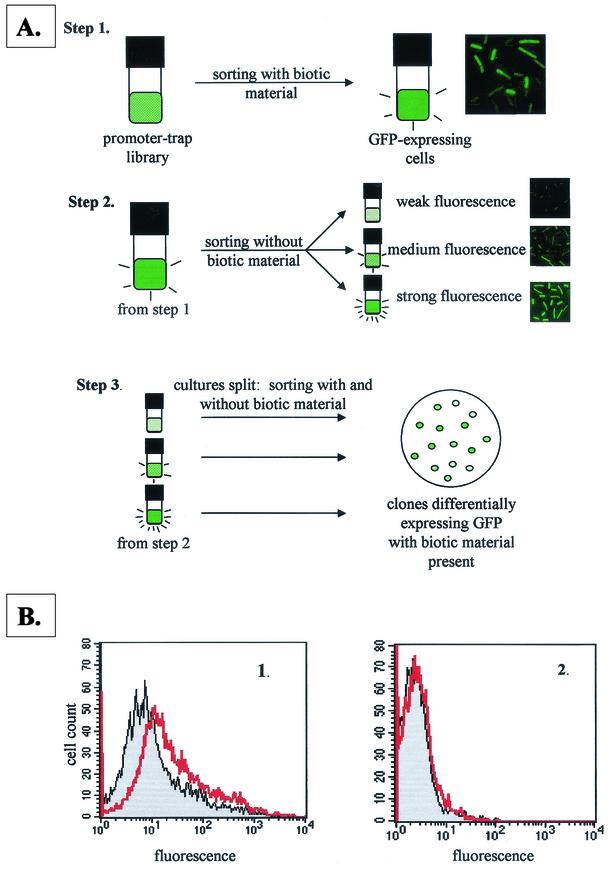
Figure 1A depicts the screening strategy. Sorting was performed by using a FACStarPlus apparatus (Becton Dickinson) fitted with an argon laser emitting at 488 nm. Bacteria were detected by forward scatter, side scatter, and fluorescence. Data were collected by using logarithmic amplifiers. The flow rate was adjusted for data collection of 1,500 events/s. All culture media contained 10 μg of chloramphenicol/ml. One sample from each group of the stored library was thawed on ice and used to inoculate 50 ml of MESAA1. The library was incubated for 16 h, and 0.5-ml samples were pelleted in 1.5-ml microcentrifuge tubes by spinning for 30 s at 14,000 × g. The supernatant was discarded, and the pellet was resuspended in twice the original volume of MESAA1E (exudate sort) or MESAA1sup (supernatant sort). Cells were incubated for 4 h and sorted, retaining green fluorescent protein (GFP)-expressing cells. The GFP-expressing cells represented ca. 4% of the total cell population. The sorted cells were grown overnight in 1 ml of MESAA1. After 15 h, the cells were pelleted by centrifugation for 30 s at 14,000 × g, the supernatant was discarded, and the pellet was resuspended in 2 ml of fresh MESAA1C (exudate sort) or MESAA1 (supernatant sort). Cells were incubated for 4 h and sorted into three groups based on fluorescence intensity: weak fluorescence (0 to 33% of maximum), medium fluorescence (33 to 66% of maximum), and strong fluorescence (66 to 99% of maximum). The groups were incubated overnight in 1 ml of MESAA1. After 15 h, the individual cultures were split into two 0.5-ml aliquots, and each aliquot was centrifuged as described above. One aliquot per group was resuspended in 1 ml of MESAA1C or MESAA1. The other aliquot was resuspended in 1 ml of MESAA1E or MESAA1sup. All cultures were incubated for 4 h. For each group, the MESAA1C or MESAA1 culture was sorted, and fluorescence intensity data for the population were collected. Immediately after the first sort, the MESAA1E or MESAA1sup culture was sorted. Any portion of the population that exhibited more or less fluorescence than the cells grown in MESAA1C or MESAA1 was collected in 1 ml of 0.5× TSB and plated for single colonies on 0.5× TSA within 1 h after sorting. The plates were incubated overnight. Individual colonies were restreaked on the same medium for isolated colonies. For the exudate and supernatant sorts, 25 isolates from each group (weak, medium, and strong) were picked at random for flow cytometry to identify individual clones showing differential fluorescence in response to exudate or supernatant.
FIG.1.
(A) Fluorescence-activated cell sorting screening method adapted from Valdivia and Falkow (48). Biotic material was either P. aureofaciens 30-84 culture supernatant or RIL 37 tomato seed exudate. (B) Flow cytometry data showing the effect of 0.1× RIL 37 exudate on GFP expression in clone SL7 (containing the lipA promoter). (Panel 1) Fluorescence intensity of the SL7 cell population without (gray shaded peak) and with (red unshaded peak) RIL 37 exudate present. The fluorescence intensity (GFP expression) increases in the presence of exudate. (Panel 2) Fluorescence intensity of the cell population of B. cereus containing pAD123, a negative control. The exudate did not affect the fluorescence intensity of the cell population.
Flow cytometry to verify differential GFP expression.
Each individual clone was grown overnight in 1 ml of MESAA1. Cultures were split and centrifuged for 30 s at 14,000 × g. One pellet was resuspended in 1 ml of MESAA1C (exudate clone) or MESAA1 (supernatant clone), and the other was resuspended in 1 ml of MESAA1E (exudate clone) or MESAA1sup (supernatant clone). Cultures were incubated for 4 h. Samples were analyzed by using a FACSCalibur flow cytometer (Becton Dickinson) with an air-cooled argon laser at 488 nm; forward and side scatter data were collected by using logarithmic amplifiers. Data were analyzed by using the CELLQuest data acquisition and general analysis software (Becton Dickinson). Purified plasmid was electroporated into B. cereus UW85, and flow cytometry was repeated to confirm that fluorescence changed in response to the addition of exudate or supernatant.
Sequence analysis of clones.
Plasmid DNA was isolated from the B. cereus clones by using a QIAprep Spin Miniprep protocol (Qiagen) according to the manufacturer's instructions, except that cells were resuspended in 300 μl of buffer P1 and 50 μl of 0.1-mm silica beads was added to each tube. The tubes were vortexed at high speed for 3 min, the beads were allowed to settle, and 250 μl of supernatant was transferred to a fresh 1.5-ml microcentrifuge tube. The instructions provided by the manufacturer were then followed.
The plasmids were electroporated into E. coli DH5α. Purified plasmid from DH5α (Qiagen QIAprep Spin Miniprep) was used for sequencing, performed with the ABI PRISM BigDye Terminators v3.0 cycle sequencing kit (Perkin-Elmer), by using primers flanking the multiple cloning site of pAD123 (left, 5′-ACC TGA CGT CTA AGA ACC CAT TAT T-3′; right, 5′-GGG ACA ACT CCA GTG AAA AGT TCT T-3′). Sequencing gels were run at the University of Wisconsin-Madison Biotechnology Center, on an ABI 377 automated DNA sequencer. Each sequencing reaction produced ca. 700 bases of readable sequence. The sequences were analyzed with BLASTN and BLASTX algorithms on the National Center for Biotechnology Information website (www.ncbi.nlm.nih.gov/BLAST/) (1).
Construction of a chromosomal lipA-gusA reporter strain.
A chromosomal gusA fusion to the B. cereus UW85 lipA promoter was constructed by using a pUC18-based suicide plasmid, and this resulted in a functional wild-type gene with an additional wild-type promoter fused to the gusA reporter gene on the chromosome.
pUC18 was modified to contain a spectinomycin resistance cassette (12) and gusA from mTn5SSgusA40 (51) and was designated pUCspgusA. A 600-bp PCR-amplified DNA fragment containing the lipA promoter region (forward, 5′-GCC AGG ATC CGC ATG ATA CAT AGT CAA C-3′; reverse, 5′-CAA ACT GCA GGG CAG CAA AAT CTT CAC C-3′) was directionally cloned into the modified pUC18 vector. The plasmid was introduced into B. cereus UW85 by electroporation, and integrants were selected by plating onto 0.5× TSA containing 100 μg of spectinomycin/ml. Integration was confirmed by Southern analysis (data not shown).
Quantitative GUS assays.
Enzyme assays were conducted on cultures with 4-methylumbelliferyl-β-glucuronide (MUG) as a substrate for the GUS enzyme (17). Assays were performed in 96-well opaque black microtiter dishes.
Cultures of B. cereus containing the chromosomal lipA-gusA fusion were grown overnight in MESAA1, and 100 μl was subcultured into 2 ml of the same medium. Cultures were incubated for 4 h with shaking, split, centrifuged, resuspended in either 2 ml of MESAA1 without exudate (or amino acid mixture) or 2 ml of MESAA1 containing exudate (or amino acid mixture), and incubated for 4 h. Then, 1 ml of each culture was centrifuged for 2 min at 14,000 × g, the supernatant was removed, and the pellet was frozen at −80°C. Pellets were thawed at room temperature and resuspended in 0.2 ml of resuspension buffer (50 mM Na2HPO4 [pH 7.0], 10 mM Na2EDTA, [pH 8.0]). Cells were permeabilized by adding 8 μl of 0.1% sodium dodecyl sulfate and 16 μl of CHCl3, followed by vortexing at high speed for 10 s. Next, 10 μl of permeabilized cells was added to 100 μl of MUG assay buffer prewarmed to 37°C (50 mM Na2HPO4 [pH 7.0], 1 mM Na2EDTA [pH 8.0], 5 mM dithiothreitol, 1 mM MUG). At each of four time points (6, 12, 18, and 24 min), 20 μl of the reaction was added to 180 μl of stop buffer (0.2 M Na2CO3). Product accumulation was measured in a Wallac Victor2 1420 multilabel counter (Perkin-Elmer) with a 355-nm lamp filter and a 460-nm emission filter by using 4-methylumbelliferone (MU) to generate a standard curve. The BCA protein assay kit (Pierce, Rockford, Ill.) was used to determine protein concentrations in calculating enzyme activities. Enzyme assays were repeated at least three times with independent cultures.
Isolation of the full gene sequence of lipA from a BAC library of B. cereus UW85.
PCR was used to generate a 600-bp fragment of the lipA gene by using pADSL7 as a template (forward, 5′-CCC GCA TGA TAC ATA GTC AAC-3′; reverse, 5′-CTC TGG CAG CAA AAT CTT CAC C-3′). The PCR product was labeled with digoxigenin (DIG) according to the manufacturer's protocol (Roche Molecular Biochemicals) and used in a Southern hybridization to probe purified plasmid DNA from a subset of a bacterial artificial chromosome (BAC) library of B. cereus UW85 (37). The DIG-labeled lipA probe was hybridized to the BAC clones by using a standard protocol (Roche Molecular Biochemicals).
One BAC plasmid, containing a 98-kb chromosomal DNA insert, was used to subclone a 6.0-kb fragment containing lipA sequence into pAD123. This subclone was partially sequenced by using primers flanking the multiple cloning site of pAD123 (described in the previous section) and a primer designed to sequence the 3′ portion of lipA (5′-GGT GAA GAT TTT GCT GCC-3′). PCR primers were designed to amplify a 1.3-kb DNA fragment containing the full lipA sequence (forward, 5′-GGA GGG TAC CGC ATG ATA CAT AGT CAA C-3′; Reverse, 5′-AGT TCT GCA GTG GAA ATG ACG GCG TTT-3′).
Construction of a chromosomal lipA knockout by using marker exchange mutagenesis.
Using PCR (forward 1, 5′-GGA GGG TAC CGC ATG ATA CAT AGT CAA C-3′; reverse 1, 5′-GGG AGG ATC CAA GGC AGC AAA ATC TTC A-3′; forward 2, 5′-CCT TGG ATC CTG AAG ATT TTG CTG CCT T-3′; reverse 2, 5′-AGT TCT GCA GTG GAA ATG ACG GCG TTT-3′), a unique BamHI site was introduced into the lipA coding sequence at codon 150. This modified lipA sequence was cloned into pTUC. A spectinomycin resistance cassette (12) was cloned into the PCR-generated BamHI site in the lipA sequence and resulted in a suicide plasmid containing a disrupted lipA gene sequence.
The suicide plasmid was introduced into B. cereus UW85 by electroporation, and integrants were selected by plating on 0.5× TSA containing 100 μg of spectinomycin/ml. Individual integrants were grown for 48 h with subculturing every 12 h in 2 ml of 0.5× TSB and plated for single colonies on 0.5× TSA. Single colonies were tested for spectinomycin resistance and tetracycline sensitivity (7.5 μg/ml). All potential mutants were verified by Southern analysis (data not shown).
Assays for bacterial growth on seeds.
B. cereus UW85 and the lipA mutant were grown in 0.5× TSB for 4 days to sporulation. Cultures were diluted 50-fold in sterile distilled water, and tomato seeds were inoculated with 100 μl/seed and air dried under vacuum as described previously (41). Seeds were used immediately after they were dried. When the wild type and mutant were coinoculated onto seeds, cultures were mixed by volume and diluted 50-fold prior to seed coating. In each experiment, at least two independent cultures were used to coat seeds, resulting in replication within each treatment.
Each seed was placed on a 1.5-cm water agar disk and watered with 10 μl of sterile distilled water. Fifteen water agar disks were placed in a sterilized petri dish, and the dishes were sealed with parafilm and wrapped in aluminum foil. Petri dishes were placed at 24°C. At each time point (day 0 or the day of inoculation, day 1, day 2, day 3, or day 4), six disks from each replicate were sampled per treatment. Disks were placed in 9 ml of sterile water blanks and sonicated for 30 s (Branson Ultrasonics Corp., Danbury, Conn.). Samples were diluted 10-fold in sterile distilled water and spread on 0.01×, 0.1×, or 0.5× TSA with or without 75 μg of spectinomycin/ml by using an Autoplate 3000 (Spiral Biotech, Bethesda, Md.). Plates were incubated at 28°C overnight, and colonies were enumerated. The numbers of CFU per sample were calculated by using formulas in the Autoplate 3000 user guide.
Sequence accession numbers.
The amino acid sequence of PrsA (P24327), the nucleotide sequence of a DNA fragment containing lipA (AY169384), the DNA fragment containing partial tspX sequence (AY169383) and partial sequences of the DNA inserts of P. aureofaciens supernantant-regulated clones B32 (AY169382), H11 (AY169374), H12 (AY169375), H17 (AY169376), H21 (AY169377), H23 (AY169378), H28 (AY169379), H29 (AY169380), and H33 (AY169381) are available in the GenBank database.
RESULTS
Identification of clones carrying regulated genes.
Using the modified differential fluorescence induction technique (Fig. 1A), we identified 11 clones that showed a decrease in GFP expression in the presence of P. aureofaciens 30-84 culture supernatant (Table 2). The addition of supernatant to MESAA1 medium does not impair the growth of B. cereus (data not shown).
TABLE 2.
Sequence similarity of B. cereus genes regulated by P. aureofaciens supernatant
| Clone | Insert size (kb) | No. of sequenced basesa | Sequence similarity (most similar protein, significance)b |
|---|---|---|---|
| B32 | 2.1 | 572 | B. anthracis acetyltransferase GNAT family, 1e−24 (NP_657892) |
| H11c | 1.3 | 754 | B. anthracis hypothetical protein, 8e−71 (NP_657557); B. subtilis HutP, 4e−40 (NP_391813) |
| H12 | 1.0 | 755 | B. anthracis thiolase, 3e−73 (NP_658060) |
| H17 | 1.5 | 768 | No sequence similarity, no apparent ORF |
| H21 | 2.6 | 766 | L. monocytogenes possible transcriptional regulator, 4e−42 (NP_463894) |
| H23 | 3.0 | 757 | B. anthracis hypothetical protein, 2e−14 (NP_655241) |
| H28 | 2.8 | 774 | No sequence similarity, no apparent ORF |
| H29 | 1.2 | 770 | B. anthracis amino acid permease, 4e−45 (NP_657178) |
| H33 | 0.7 | 543 | B. anthracis hypothetical protein, 0.014 (NP_655295) |
That is, the number of base pairs of the DNA insert adjacent to the gfpmut3a reporter that were sequenced.
That is, the most similar protein(s) in GenBank. The accession number is indicated in parentheses. ORF, open reading frame.
Clone H11 was isolated three times, independently.
We identified two promoter-trap clones (SL7 and SL12) in which GFP expression increased in the presence of exudate from RIL 37 tomato seed (data for clone SL7 is shown in Fig. 1B). The increase in GFP expression does not appear to be due to an increase in growth of B. cereus in the presence of exudate. B. cereus UW85 did not grow in 0.1× exudate alone, and the growth of B. cereus UW85 containing pAD123 was not affected by the addition of 0.1× exudate to MESAA1 (data not shown). Sequence analysis of the 600-bp DNA insert of pADSL12 from clone SL12 showed similarity to TspC, a tail specific protease in Bacillus anthracis (E = 3e−27; GenBank accession number NP_653631). Insertional inactivation of this gene (tspX) on the B. cereus UW85 chromosome did not result in noticeable phenotypic differences from wild type in culture or during growth on plants (data not shown).
The gene identified in the pADSL7 DNA insert encodes a deduced protein with a lipoprotein signal peptide sequence (LALSAC) similar to lipoproteins in B. subtilis (47); therefore, we designated the gene lipA. From sequence analysis of the region of DNA surrounding lipA, the gene does not appear to be part of an operon. Forty nucleotides upstream of lipA a putative open reading frame of 111 amino acids was detected in the opposite orientation. Sequence similarity to a B. anthracis hypothetical protein (E = 7e−22; GenBank accession number NP_656196) was identified ca. 450 nucleotides downstream of lipA in the same orientation.
Quantitative GUS assays.
To quantify the increase in lipA promoter activity in the presence of RIL 37 exudate, we constructed a chromosomal lipA-gusA fusion, in which lipA activity increased twofold in the presence of 0.1× RIL 37 exudate (data not shown). The addition of 0.05× or 0.2× RIL 37 exudate resulted in a <2-fold induction of lipA promoter activity (1.2- and 1.4-fold, respectively); therefore, 0.1× exudate was used as the optimal concentration in further experiments.
In contrast to RIL 37 exudate, exudate prepared from Wisconsin 55 tomato seed did not influence the activity of the lipA promoter, indicating that lipA induction is influenced by either the genotype or cultural history of the tomato seed (data not shown). The addition of 0.0001× Wisconsin 55 exudate to 0.1× RIL 37 exudate abolished induction of lipA promoter activity (data not shown).
Exudates used in the experiments described above contained the accumulated compounds released from seeds over 48 h. To determine when the compound(s) responsible for induction of lipA promoter activity were released from the seed, RIL 37 exudate of various ages was used in the GUS assay. Exudate was collected after 24 h and stored at −20°C. The liquid covering the tomato seeds was replaced with fresh 0.5 mM CaSO4, and exudate was again collected after 24 h. The first 24-h fraction was compared to the second 24-h fraction for induction of lipA promoter activity by using the GUS assay. Sufficient accumulation of inducing compounds had occurred after 24 h to induce lipA promoter activity ∼2-fold, whereas the exudate collected in the second 24 h period only slightly induced lipA promoter activity (data not shown).
Characterization of inducing compounds in exudate.
Seed exudates contain many small molecules that include amino acids, sugars, organic acids, peptides, and volatiles (21, 25, 49). Using the quantitative GUS assay, a solution of amino acids induced lipA promoter activity in a concentration-dependent manner (1× amino acid mix = 96.4 ± 1.9 nmol of MUG cleaved/min/μg of protein, 0.1× amino acid mix = 71.4 ± 2.9 nmol of MUG cleaved/min/μg of protein, 0.01× amino acid mix = 48.4 ± 5.9 nmol of MUG cleaved/min/μg of protein, no amino acid mix = 10.3 ± 0.4 nmol of MUG cleaved/min/μg of protein). The solution contained amino acids not already present in MESAA1 medium (28).
To determine whether amino acids were the component in seed exudate responsible for lipA induction, we determined the free amino acid content of RIL 37 and Wisconsin 55 tomato seed exudates (Table 3). This information was used to reconstruct the amino acid mixtures present in the exudates, and these mixtures were used in the GUS assay. Both mixtures failed to induce lipA promoter activity (data not shown), suggesting that compounds other than, or in combination with, amino acids in RIL 37 exudate induce lipA promoter activity.
TABLE 3.
Free amino acid and ammonia content of RIL 37 exudate, Wisconsin 55 exudate, and the 12-amino-acid mixturea
| Amino acid or ammonia | RIL 37
|
Wisconsin 55
|
1× Amino acid mixture
|
|||
|---|---|---|---|---|---|---|
| μM | Mol% | μM | Mol% | mM | Mol% | |
| l-Aspartic acid | 0.24 | 7.6 | 2.52 | 5.5 | 0.013 | 0.1 |
| l-Threonine | 0.09 | 2.6 | 2.82 | 5.6 | 0.024 | 0.2 |
| l-Serine | 0.18 | 4.5 | 2.40 | 4.1 | 0.022 | 0.2 |
| l-Asparagine | ND | ND | 1.80 | 3.9 | 1.19 | 11.1 |
| l-Glutamic acid | 0.27 | 9.6 | 3.24 | 7.9 | ND | ND |
| l-Glutamine | 0.21 | 7.3 | 2.10 | 5.0 | 0.60 | 6.2 |
| l-Proline | ND | ND | 1.68 | 3.2 | 0.88 | 7.1 |
| l-Glycine | 0.63 | 11.3 | 2.10 | 2.6 | 1.93 | 10.2 |
| l-Alanine | 0.27 | 5.7 | 3.39 | 5.1 | ND | ND |
| l-Valine | ND | ND | 0.27 | 0.6 | ND | ND |
| l-Cysteine | ND | ND | 0.06 | 0.2 | 0.19 | 3.2 |
| l-Methionine | ND | ND | ND | ND | 0.82 | 8.6 |
| l-Isoleucine | 0.03 | 0.9 | 1.77 | 3.8 | 1.08 | 10.0 |
| l-Leucine | 0.03 | 0.9 | 3.09 | 6.6 | ND | ND |
| l-Tyrosine | 0.03 | 1.3 | 1.11 | 3.4 | 0.55 | 7.0 |
| l-Phenylalanine | 0.09 | 3.5 | 1.77 | 4.8 | 0.85 | 9.9 |
| l-Tryptophan | ND | ND | ND | ND | 0.67 | 9.6 |
| Ammonia | 6.30 | 25.6 | 95.04 | 26.5 | 0.25 | 0.3 |
| l-Lysine | 0.12 | 4.2 | 1.77 | 4.2 | 0.003 | 0.0 |
| l-Histidine | ND | ND | 0.42 | 1.1 | 0.70 | 7.6 |
| l-Arginine | 0.36 | 15 | 2.07 | 5.9 | 0.71 | 8.8 |
Analysis was performed by Scientific Research Consortium (St. Paul, Minn.) on a Beckman Instruments model 6300 dedicated amino acid analyzer. ND, none detected.
Neither a mixture of sugars (17.7 mM glucose, 3.3 mM fructose, 4.6 mM maltose, and 9.3 mM xylose [17]), nor a mixture of volatiles (0.1% methanol and 1% ethanol), nor an organic acid (0.1% [vol/vol] lactic acid) induced lipA promoter activity in the GUS assay (data not shown).
Construction and characterization of the lipA mutant of B. cereus UW85.
To determine the effect of lipA on the behavior of B. cereus, a lipA knockout mutant of B. cereus UW85 was constructed by marker-exchange mutagenesis. The mutant grew at a rate similar to that of the wild type in complex medium (0.1× TSB), defined medium (MESAA1), and defined medium containing 0.1× RIL 37 tomato seed exudate (MESAA1E) (data not shown). The mutant sporulated, produced the antibiotic zwittermicin A, and survived for 30 days at 4 and 25°C in culture (MESAA1 and 0.1× TSB) and in soil similarly to wild type (data not shown). The lipA mutant and wild type were tested for secretion of functional α-amylase and general protease activity in culture supernatant, and no differences between the mutant and wild type were observed (data not shown).
When inoculated onto separate RIL 37 tomato seeds placed on water agar disks, the lipA mutant and wild type showed the same growth pattern and reached the same detectable population sizes (on the second day of the assay when plated on 0.1× TSA, the values were 6.02 ± 0.09 average log CFU/sample for the wild type and 5.98 ± 0.09 average log CFU/sample for the lipA mutant). When the lipA mutant was applied to the seeds in a 10-fold excess in a mixture with the wild type, the detectable population of the wild type decreased on the second day of the assay compared to the first day when the samples were cultured on 0.1× TSA, whereas the detectable population of the mutant increased (Fig. 2). To determine whether this decrease of the wild-type population was due to death of wild-type cells or reduced culturability, on the second day of the assay the bacteria isolated from the seed were spread on various strengths of TSA. The culturability of the wild type was lowest on 0.01× TSA and higher on 0.1× and 0.5× TSA (Fig. 3). Detection of the lipA mutant was not affected by medium type (Fig. 3). There appears to be a physiological difference between the wild-type and mutant cells that results in medium-dependent culturability of the wild-type cells on the second day of growth on tomato seeds.
FIG. 2.
Growth on seed. Tomato seeds were inoculated with the wild type and a 10-fold excess of the lipA mutant at the start of the assay (day 0). Detectable populations of the wild type (gray bars) and the lipA mutant (black bars) were determined by plating on 0.1× TSA and are presented as the average log CFU/sample. Values represent the average of five experiments. Error bars represent the standard error of the mean.
FIG. 3.
Medium-dependent culturability of the wild type (gray bars) on day 2 of growth on tomato seeds. The lipA mutant (black bars) was unaffected. At the start of the assay, the lipA mutant was inoculated in a 10-fold excess with the wild type. Samples were plated on either 0.01×, 0.1×, or 0.5× TSA. Detectable populations are represented as the average log CFU/sample. Values are the average of three experiments with six samples per experiment. Error bars represent the standard error of the mean.
A small decrease in the detectable populations of the wild type on day 2 of the assay was observed when the seeds were inoculated with equal amounts of the wild type and mutant. No decrease in the detectable populations of the wild type was observed when the seeds were inoculated with the mutant and a 10-fold excess of the wild-type (Table 4). The extent of the decrease in detectable populations of the wild type on day 2 of the assay when the seeds were incoulated with the wild type and a 10-fold excess of the mutant varied between experiments (Fig. 2), with the most dramatic decrease below the limit of detection of the assay (Table 4).
TABLE 4.
B. cereus growth as determined by tomato seed assay with inoculation at various ratios of wild type and lipA mutant
| Ratio in inoculum (wild type:lipA mutant)a | Avg log CFU/sampleb ± SE at:
|
|||
|---|---|---|---|---|
| Time zero
|
48 h
|
|||
| Wild type | lipA mutant | Wild type | lipA mutant | |
| 1:10 | 4.06 ± 0.08 | 4.84 ± 0.05 | 1.26 ± 0.00 | 5.95 ± 0.02 |
| 1:1 | 4.66 ± 0.08 | 4.73 ± 0.05 | 4.10 ± 0.91 | 5.88 ± 0.05 |
| 10:1 | 5.09 ± 0.06 | 4.14 ± 0.09 | 5.79 ± 0.05 | 5.32 ± 0.05 |
Ratios were mixed by volume.
Calculated from an average of six samples per treatment ± the standard error of the mean after being plated on 0.1 × TSA. The detection limit of the assay was log10 1.26.
DISCUSSION
As a model system for increasing our understanding of the relationship between gram-positive plant-associated bacteria and other organisms in their environment, we have chosen to study the influence of chemicals exuded from tomato seeds and a rhizosphere-colonizing strain of P. aureofaciens on gene expression in B. cereus. Using this system, we identified nine clones carrying genes regulated by P. aureofaciens supernatant and two carrying genes regulated by tomato seed exudate. We characterized lipA, a tomato seed exudate-regulated gene in B. cereus.
The choice of biotic factors that might serve as gene regulators was based on previous work involving the interactions between B. cereus, its host plant, and other rhizosphere bacteria. In experiments with field-grown soybean and alfalfa plants, we routinely isolated strains of P. aureofaciens from the same root segments as B. cereus, suggesting that signaling between B. cereus and P. aureofaciens could be biologically relevant (Emmert et al., unpublished). For the present study, we used supernatant from the well-characterized P. aureofaciens strain 30-84, originally isolated from the rhizosphere of a wheat plant (20, 33). Further characterization of the compounds in P. aureofaciens culture supernatant responsible for repression of B. cereus gene expression will be aided by the availability of mutants that lack the ability to produce secondary metabolites (5). Sequence analysis of the P. aureofaciens supernatant-regulated clones identified similarity to genes encoding products involved in processes such as nutrient acquisition, as well as everal sequences with similarity to hypothetical proteins of unknown function (Table 2).
The choice of plant material was guided by previous work with RILs of tomato that identified several quantitative trait loci that were highly correlated with growth of B. cereus strains on tomato seeds (42). The RILs that differed in support of B. cereus growth did not differ in support of growth of several Pseudomonas species (40), indicating that host factors that are species specific for bacteria are required for B. cereus growth in the tomato spermosphere. To determine whether chemical compounds from the tomato seed or seedling influenced gene expression in B. cereus, we used tomato seed exudate from RIL 37, a line capable of supporting growth of B. cereus. RIL 37 exudate induced lipA promoter activity twofold, and exudate from RIL 29 and RIL 83, lines not supportive of growth, induced lipA promoter activity to the same extent as exudate from RIL 37 (data not shown), suggesting that induction of lipA promoter activity is not correlated with the ability of RILs to support B. cereus growth. Exudate from a genetically distinct tomato, Wisconsin 55, did not induce lipA promoter activity. We speculated that differences in seed exudate chemistry between RIL 37 and Wisconsin 55 were responsible for induction of lipA promoter activity through an inducing compound(s) in RIL 37 exudate, an inhibiting compound(s) in Wisconsin 55 exudate, or a specific ratio of compounds present in RIL 37 exudate. Our results suggest that although a mixture of amino acids induces lipA promoter activity, it is likely that compounds in the exudates other than amino acids are responsible for influencing lipA promoter activity. Further chemical analysis of RIL 37 and Wisconsin 55 exudates will be necessary to identify the compound or mixture of compounds that affect lipA promoter activity.
From sequence analysis of lipA, we deduced that this gene encodes a protein with a signal peptide sequence similar to lipoproteins in B. subtilis (47). More specifically, BLASTX sequence analysis of lipA identified sequence similarity to PrsA of B. subtilis (39% identity). PrsA is a major lipoprotein in B. subtilis and is a molecular chaperone homologous to peptidyl prolyl cis-trans isomerases (38), important for the proper folding of secreted proteins (16, 22). Inactivation of prsA in B. subtilis is lethal (22), whereas inactivation of lipA in B. cereus is not, suggesting that they may have different roles, possibly acting as molecular chaperones for different subsets of secreted proteins. Bacillus species are known to secrete large quantities of various proteins, and it is possible that these secreted proteins play a role in the response of B. cereus to plant hosts. Comparative analysis of functional proteins secreted by the wild type and lipA mutant may clarify the physiological role of LipA in B. cereus.
It appears that the role of LipA in the ability of B. cereus to grow in the presence of tomato seeds and seedlings is also complex. Our data demonstrate that wild-type B. cereus, when inoculated onto tomato seeds with a 10-fold excess of the lipA mutant, enters an altered physiological state that results in medium-dependent culturability on the second day of the growth on seed assay.
The influence of the physiological state of B. cereus cells on culturability has been observed in broth culture (E. A. B. Emmert and J. Handelsman, unpublished data). The ability to grow on rich medium (0.1× TSA) and defined medium (MESAA1 agar) was compared between late-exponential-phase and stationary-phase B. cereus grown in brain heart infusion broth. At stationary phase, the detectable populations of B. cereus on MESAA1 agar were 200-fold lower than on 0.1× TSA. During late exponential phase, the detectable populations on MESAA1 agar were at least 29,000-fold lower than on 0.1× TSA. These results indicate that culturability of B. cereus is related to the growth stage of the bacteria. Although growth in liquid medium is not directly comparable to growth on tomato seeds, the possibility exists that the medium-dependent culturability of the wild type on the second day of the assay is related to the growth stage of the cells.
The presence of the lipA mutant appears to influence the culturability of wild-type cells, because only when the wild type was inoculated onto tomato seeds with sufficient quantities of the lipA mutant was this phenomenon observed. This might be due to the spatial distribution of the wild type and mutant cells when inoculated onto the tomato seeds. When a 10-fold excess of wild-type cells is inoculated onto a seed with the mutant, the wild-type cells are more likely to be surrounded by other wild-type cells, and medium-dependent culturability is not observed. When a 10-fold excess of mutant cells is inoculated onto a seed with wild-type cells, the wild-type cells are more likely to be surrounded by mutant cells, and medium-dependent culturability is observed. The mutant cells might produce one or more molecules that, in sufficient concentration, alter the physiological state of the wild type, preferentially obtain nutrients that are then not available to the wild type, and result in its altered physiological state—or possibly even be unresponsive to inhibitory compounds exuded from the seed that the wild type is sensitive to. Based on the available data, we can only speculate as to how exuded plant compounds and the lipA mutant could affect the wild type in our assay. Further study of the role of LipA in the physiology of B. cereus, combined with identification of the compounds in the tomato seed exudates responsible for induction and/or repression of lipA promoter activity, will be necessary to fully explain the role that LipA plays in interactions with plants.
The subtlety of the interaction reported here coincides with the emerging picture of the complex and dynamic relationship of seed exudate to the bacteria that colonize the seed. For example, canavanine, an arginine analog secreted by alfalfa seeds that is highly toxic to B. cereus in vitro, inhibits the growth of B. cereus more at a distance from the seed than directly on the seed surface (7). Since simple diffusion kinetics suggest that the concentration would be highest at the seed surface, some other factor in exudate most likely moderates the activity of canavanine at the seed surface. Canavanine toxicity might be modulated by arginine, which protects cells from canavanine toxicity when supplied at sufficiently high concentrations, and might protect B. cereus at the seed surface. We also have presented here evidence suggesting that amino acids antagonize each others' activity, and the ratio of amino acids or presence of other factors in the seed exudate determines whether lipA is induced.
In these experiments we have shown that the modified differential fluorescence induction method is useful for identifying genes in B. cereus regulated by biotic material. Using this technique we discovered lipA, a previously unidentified gene in B. cereus that may play a role in the ability of this bacterium to interact with host plants. Our screening of the library for exudate-regulated genes was not exhaustive, and the potential exists for finding many other interesting plant-regulated genes. Further use of this method with B. cereus will bring a greater understanding of the influence of plant hosts and of other plant-associated microorganisms on the gene expression and ecological behavior of gram-positive bacteria.
Acknowledgments
We thank Caitilyn Allen for helpful discussions and Laurie Luther for help in preparing the manuscript. We thank K. Schell at the Flow Lab at the University of Wisconsin-Madison Comprehensive Cancer Center for assistance with flow cytometry and cell sorting and data analysis.
A.K.D. was supported by NIH grants 5 T32 GM07215 and NIH 5 T32 GM08349 and by a U.S. EPA Office of Research and Development, National Center for Environmental Research, Science to Achieve Results (STAR) Fellowship U-915617-01. This work was supported by Hatch Project 4038 from the College of Agricultural and Life Sciences at the University of Wisconsin-Madison.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, D., and D. Mills. 1983:Tn5. transposon mutagenesis of 2 phytopathogenic pseudomonas-syringae pathovars. Phytopathology 73:824-826. [Google Scholar]
- 3.Beaulieu, C., and F. van Gijsegem. 1990. Identification of plant-inducible genes in Erwinia chrysanthemi 3979. J. Bacteriol. 172:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher, C., P. Barberis, A. Trigalet, and D. Demery. 1985. Transposon mutagenesis of Pseudomonas solanacearum: isolation of Tn5 induced avirulent mutants. J. Gen. Microbiol. 131:2449-2457. [Google Scholar]
- 5.Chancey, S. T., D. W. Wood, and L. S. Pierson III. 1999. Two-component transcriptional regulation of N-acyl homoserine lactone production in Pseudomonas aureofaciens. Appl. Environ. Microbiol. 65:2294-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunn, A. K., and J. Handelsman. 1999. A vector for promoter-trapping in Bacillus cereus. Gene 226:297-305. [DOI] [PubMed] [Google Scholar]
- 7.Emmert, E. A. B., J. L. Milner, J. C. Lee, K. L. Pulvermacher, H. A. Olivares, J. Clardy, and J. Handelsman. 1998. Effect of canavanine from alfalfa seed on the population biology of Bacillus cereus. Appl. Environ. Microbiol. 64:4683-4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Felske, A., A. Wolterink, R. van Lis, and A. D. L. Akkermans. 1998. Phylogeny of the main bacterial 16S rRNA sequences in Drentse A grassland soils (The Netherlands). Appl. Environ. Microbiol. 64:871-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garfinkel, D. J., R. B. Simpson, L. W. Ream, F. F. White, M. P. Gordon, and E. W. Nester. 1981. Genetic analysis of crown gall: fine structure map of the T-DNA by site-directed mutagenesis. Cell 27:143-153. [DOI] [PubMed] [Google Scholar]
- 10.Gilbert, G. S., J. L. Parke, M. K. Clayton, and J. Handelsman. 1993. Effects of an introduced bacterium on bacterial communities on roots. Ecology 74:840-854. [Google Scholar]
- 11.Goldstein, A. H., K. Braverman, and N. Osorio. 1999. Evidence for mutualism between a plant growing in a phosphate-limited desert environment and a mineral phosphate solubilizing (MPS) rhizobacterium. FEMS Microbiol. Ecol. 30:295-300. [DOI] [PubMed] [Google Scholar]
- 12.Guérout-Fleury, A., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 13.Halverson, L. J., M. K. Clayton, and J. Handelsman. 1993. Population biology of Bacillus cereus UW85 in the rhizosphere of field-grown soybeans. Soil Biol. Biochem. 25:485-493. [Google Scholar]
- 14.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 165:557-580. [DOI] [PubMed] [Google Scholar]
- 15.Handelsman, J., S. Raffel, E. H. Mester, L. Wunderlich, and C. R. Grau. 1990. Biological control of damping-off of alfalfa seedlings with Bacillus cereus UW85. Appl. Environ. Microbiol. 56:713-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jacobs, M., J. B. Anderssen, V. Kotinen, and M. Sarvas. 1993. Bacillus subtilis PrsA is required in vivo as an extracytoplasmic chaperone for secretion of active enzymes synthesized either with or without pro-sequence. Mol. Microbiol. 8:957-966. [DOI] [PubMed] [Google Scholar]
- 17.Jefferson, R. A., and K. J. Wilson. 1988. The GUS gene fusion system, p. B14/1-B14/33. In S. Gelvin, R. Schilperoort, and D. P. Verma (ed.), Plant molecular biology manual. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 18.Kelman, A. 1954. The relationship of pathogenicity of Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology 44:693-695. [Google Scholar]
- 19.Kim, U.-J., B. W. Birren, T. Slepak, V. Mancino, C. Boysen, H.-L. Kang, M. I. Simon, and H. Shizuya. 1996. Construction and characterization of a human bacterial artifical chromosome library. Genomics 34:213-218. [DOI] [PubMed] [Google Scholar]
- 20.Kluyver, A. J. 1956. Pseudomonas aureofaciens nov. spec. and its pigments. J. Bacteriol. 72:406-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koller, D., A. M. Mayer, A. Poljakoff-Mayber, and S. Klein. 1962. Seed germination. Annu. Rev. Plant Physiol. 13:437-464. [Google Scholar]
- 22.Kotinen, V. P., and M. Sarvas. 1993. The PrsA lipoprotein is essential for protein secretion in Bacillus subtilis and sets a limit for high-level secretion. Mol. Microbiol. 8:727-737. [DOI] [PubMed] [Google Scholar]
- 23.Lindgren, P. B., R. C. Peet, and N. J. Panopoulos. 1986. A gene cluster of Pseudomonas syringae pv. Phaseolicola controls pathogenicity on bean plants and hypersensitivity on non-host plants. J. Bacteriol. 168:512-522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Long, S. R., W. J. Buikema., and F. M. Ausubel. 1982. Cloning of Rhizobium meliloti nodulation genes by direct complementation of Nod− mutants. Nature 298:485-488. [Google Scholar]
- 25.Lugtenberg, B. J. J., L. V. Kravchenko, and M. Simons. 1999. Tomato seed and root exudate sugars: composition, utilization by Pseudomonas biocontrol strains and role in rhizosphere colonization. Environ. Microbiol. 1:439-446. [DOI] [PubMed] [Google Scholar]
- 26.Mahan, M. J., D. M. Heitoff, R. I. Sinsheimer, and D. A. Low. 2000. Assessment of bacterial pathogenesis by analysis of gene expression in the host. Annu. Rev. Genet. 34:139-164. [DOI] [PubMed] [Google Scholar]
- 27.Miller, J. H. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, N.Y.
- 28.Milner, J. L., S. J. Raffel, B. J. Lethbridge, and J. Handelsman. 1995. Culture conditions that influence accumulation of zwittermicin A by Bacillus cereus UW85. Appl. Microbiol. Biotechnol. 43:685-691. [DOI] [PubMed] [Google Scholar]
- 29.Osbourn, A. E., C. E. Barber, and M. J. Daniels. 1987. Identification of plant-induced genes of the bacterial pathogen Xanthomonas campestris pathovar campestris using a promoter-probe plasmid. EMBO J. 6:23-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Palva, A., G. Vigren, M. Simonen, H. Rintala, and P. Laamanen. 1990. Nucleotide sequence of the tetracycline resistance gene of pBC16 from Bacillus cereus. Nucleic Acids Res. 18:1635.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paran, I., I. Goldman, S. D. Tanksley, and D. Zamir. 1995. Recombinant inbred lines for genetic mapping in tomato. Theor. Appl. Genet. 90:542-548. [DOI] [PubMed] [Google Scholar]
- 32.Peters, N. K., J. W. Frost, and S. R. Long. 1986. A plant flavone, luteolin, induces expression of Rhizobium meliloti nodulation genes. Science 233:977-980. [DOI] [PubMed] [Google Scholar]
- 33.Pierson, L. S., III, and E. A. Pierson. 1996. Phenazine antibiotic production in Pseudomonas aureofaciens: role in rhizosphere ecology and pathogen suppression. FEMS Microbiol. Lett. 136:101-108. [Google Scholar]
- 34.Raffel, S. J., E. V. Stabb, J. L. Milner, and J. Handelsman. 1996. Genotypic and phenotypic analysis of zwittermicin A-producing strains of Bacillus cereus. Microbiology 142:3425-3436. [DOI] [PubMed] [Google Scholar]
- 35.Rainey, P. B. 1999. Adaptation of Pseudomonas fluorescens to the plant rhizosphere. Environ. Microbiol. 1:243-257. [DOI] [PubMed] [Google Scholar]
- 36.Rainey, P. B., and G. M. Preston. 2000. In vivo expression technology strategies: valuable tools for biotechnology. Curr. Opin. Biotechnol. 11:440-444. [DOI] [PubMed] [Google Scholar]
- 37.Rondon, M. R., S. J. Raffel, R. M. Goodman, and J. Handelsman. 1999. Toward functional genomics in bacteria: analysis of gene expression in Escherichia coli from a bacterial artificial chromosome library of Bacillus cereus. Proc. Natl. Acad. Sci. USA 96:6451-6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rudd, K. E., H. J. Sofia, E. V. Koonin, G. Plunkett III, S. Lazar, and P. E. Rouviere. 1995. A new family of peptidyl-prolyl isomerases. Trends Biochem. Sci. 20:12-14. [DOI] [PubMed]
- 39.Silo-Suh, L. A., B. J. Lethbridge, S. J. Raffel, H. He, J. Clardy, and J. Handelsman. 1994. Biological activities of two fungistatic antibiotics produced by Bacillus cereus UW85. Appl. Environ. Microbiol. 60:2023-2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon, H. M., K. P. Smith, J. A. Dodsworth, B. Guenthner, J. Handelsman, and R. M. Goodman. 2001. Influence of tomato genotype on growth of inoculated and indigenous bacteria in the spermosphere. Appl. Environ. Microbiol. 67:514-520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith, K. P., J. Handelsman, and R. M. Goodman. 1997. Modeling dose-response relationships in biological control: partitioning host responses to the pathogen and biocontrol agent. Phytopathology 87:720-729. [DOI] [PubMed] [Google Scholar]
- 42.Smith, K. P., J. Handelsman, and R. M. Goodman. 1999. Genetic basis in plants for interactions with disease-suppressive bacteria. Proc. Natl. Acad. Sci. USA 96:4786-4790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stabb, E. V., L. M. Jacobson, and J. Handelsman. 1994. Zwittermicin A-producing strains of Bacillus cereus from diverse soils. Appl. Environ. Microbiol. 60:4404-4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stacey, G., J. Sanjuan, S. Luka, T. Dockendorff, and R. W. Carlson. 1995. Signal exchange in the Bradyrhizobium-soybean symbiosis. Soil. Biol. Biochem. 27:473-483. [Google Scholar]
- 45.Stachel, S. E., E. Messens, M. van Montagu, and P. Zambryski. 1985. Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624-629. [Google Scholar]
- 46.Thomashow, L. S., and D. M. Weller. 1988. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J. Bacteriol. 170:3499-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tjalsma, H., V. P. Kotinen, S. Pragai, H. Wu, R. Meima, G. Benema, S. Bron, M. Sarvas, and J. M. van Dijl. 1999. The role of lipoprotein processing by signal peptidase II in the gram-positive eubacterium Bacillus subtilis. J. Biol. Chem. 274:1698-1707. [DOI] [PubMed] [Google Scholar]
- 48.Valdivia, R. H., and S. Falkow. 1997. Fluorescence-based isolation of bacterial genes expressed within host cells. Science 277:2007-2011. [DOI] [PubMed] [Google Scholar]
- 49.Vanèura, V., and G. Stotzky. 1975. Gaseous and volatile exudates from germinating seeds and seedlings. Can. J. Bot. 54:518-532. [Google Scholar]
- 50.van Overbeek, L. S., and J. D. van Elsas. 1995. Root exudate-induced promoter activity in Pseudomonas fluorescens mutants in the wheat rhizosphere. Appl. Environ. Microbiol. 61:890-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wilson, K. J., S. Sessitch, J. C. Corbo, K. E. Giller, A. D. L. Akkermans, and R. A. Jefferson. 1995. β-Glucuronidase (GUS) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology 141:1691-1705. [DOI] [PubMed] [Google Scholar]
- 52.Winans, S. C. 1992. Two-way chemical signaling in Agrobacterium-plant interactions. Microbiol. Rev. 56:12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]