Abstract
We analyzed the variation with depth in the composition of members of the domain Bacteria in samples from alkaline, hypersaline, and currently meromictic Mono Lake in California. DNA samples were collected from the mixolimnion (2 m), the base of the oxycline (17.5 m), the upper chemocline (23 m), and the monimolimnion (35 m). Composition was assessed by sequencing randomly selected cloned fragments of 16S rRNA genes retrieved from the DNA samples. Most of the 212 sequences retrieved from the samples fell into five major lineages of the domain Bacteria: α- and γ-Proteobacteria (6 and 10%, respectively), Cytophaga-Flexibacter-Bacteroides (19%), high-G+C-content gram-positive organisms (Actinobacteria; 25%), and low-G+C-content gram-positive organisms (Bacillus and Clostridium; 19%). Twelve percent were identified as chloroplasts. The remaining 9% represented β- and δ-Proteobacteria, Verrucomicrobiales, and candidate divisions. Mixolimnion and oxycline samples had low microbial diversity, with only 9 and 12 distinct phylotypes, respectively, whereas chemocline and monimolimnion samples were more diverse, containing 27 and 25 phylotypes, respectively. The compositions of microbial assemblages from the mixolimnion and oxycline were not significantly different from each other (P = 0.314 and 0.877), but they were significantly different from those of chemocline and monimolimnion assemblages (P < 0.001), and the compositions of chemocline and monimolimnion assemblages were not significantly different from each other (P = 0.006 and 0.124). The populations of sequences retrieved from the mixolimnion and oxycline samples were dominated by sequences related to high-G+C-content gram-positive bacteria (49 and 63%, respectively) distributed in only three distinct phylotypes, while the population of sequences retrieved from the monimolimnion sample was dominated (52%) by sequences related to low-G+C-content gram-positive bacteria distributed in 12 distinct phylotypes. Twelve and 28% of the sequences retrieved from the chemocline sample were also found in the mixolimnion and monimolimnion samples, respectively. None of the sequences retrieved from the monimolimnion sample were found in the mixolimnion or oxycline samples. Elevated diversity in anoxic bottom water samples relative to oxic surface water samples suggests a greater opportunity for niche differentiation in bottom versus surface waters of this lake.
Understanding the factors that control the composition and diversity of microbial communities is important to understanding how bacterial populations function to facilitate biogeochemical processes. Mono Lake is an ideal site for such studies because it has strong geochemical gradients, a simple food web, and a relatively simple microbial community. Mono Lake is an alkaline (pH 9.8), hypersaline (84 to 94 g liter−1) soda lake located east of the Sierra Nevada mountains, approximately 160 km south of Lake Tahoe, Calif. Because of the contrast between the density of saline lake water and freshwater flowing into soda lakes, the control of their water balance by local hydrologic conditions, and the absence or diminished effects of many of the mixing mechanisms found in estuaries or the open ocean (tides, fetch, and swell), soda lakes have a tendency to become meromictic in response to regional hydrologic events (29, 44). Mono Lake is no exception to this generality and has been meromictic since 1995 as a result of a high level of runoff during the El Niño winter of 1994 to 1995 that coincided with decreased diversions from tributary streams.
With prolonged meromixis, anoxic bottom water at a high pH can attain high levels of toxic inorganic compounds, including sulfide and ammonia (33). These conditions have a profound effect on the ecology of the lake (30, 44) and may be expected to affect the composition and activity of the microbial community of the lake. Stratification affects the flux of reduced metabolites into the surface layer from bottom water and lake bottom sediments. Reduced substances accumulate to high concentrations in bottom water, and biota in surface layers may be deprived of essential nutrients. High concentrations of biologically generated reduced substances (sulfide, for example) may directly affect geochemical processes (i.e., metal precipitation or speciation) and thus indirectly affect biota by creating secondary nutrient limitations. Sulfide toxicity has also been shown to affect microbial processes directly—for example, denitrification (31) and ammonia oxidation (34). Sulfide toxicity may result in the exclusion of zooplankton grazers from anoxic or hypoxic regions of the water column. In Mono Lake, this situation leads to the accumulation of bacterial biomass at depth, to an increase in average cell size (large bacterioplankton are observed in surface water before the annual bloom of the brine shrimp Artemia monica, the dominant zooplankter in Mono Lake), and to the formation of a deep chlorophyll maximum at the base of the oxycline.
Compared to more thoroughly investigated marine and freshwater environments, relatively little is known about the microbial ecology of saline lakes. Interest in soda lake microbiology has centered primarily on the isolation and characterization of individual microorganisms with potential industrial applications (32). Little attention has been paid to the community as a whole. A recent study examined the phylogenetic diversity of bacteria isolated from soda lakes (15). However, little work has been done on the in situ phylogenetic diversity of bacterial populations in soda lakes or on ascribing aspects of the biogeochemical cycles of soda lakes to these bacteria. Since soda lakes are environments of extreme physiochemical conditions, organisms living in these lakes may possess novel adaptations to these environments—for example, transport systems, osmoregulatory compounds, or ectoenzymes. Some of these adaptations may involve enzymes or other physiological properties with potential commercial significance (24, 28).
Soda lakes are also anomalous environments in that they have high concentrations of dissolved organic carbon (DOC), dissolved inorganic phosphorus, and high standing crops of bacteria. These characteristics suggest that the microbial loop processes that have been studied more thoroughly for marine and freshwater environments (3, 4) are somehow decoupled in soda lakes, allowing DOC to accumulate. Understanding the reasons for this situation will help improve understanding of the consequences of the microbial loop for the ecology and biogeochemistry of other aquatic environments and provide insight into the processes that contribute to the storage and deposition of organic carbon (e.g., see references 14 and 45).
In a previous study (27), denaturing gradient gel electrophoresis (DGGE) was used to investigate seasonal changes in the vertical structure of Mono Lake microbial communities during a period of holomixis. This study indicated that there were significant differences in the compositions of bacterial assemblages with depth under stratified conditions but did not provide a detailed description of the phylogenetic compositions of these communities. The study presented here was conducted to compare the phylogenetic compositions and diversity of bacterial assemblages at four depths chosen to represent a range of water column redox conditions. Compared to samples from other aquatic environments, the Mono Lake microbial assemblages that we sampled displayed low diversity. We found distinctly different patterns of diversity in oxic versus anoxic waters of the lake, with surface water microbial communities being characterized by lower microbial diversity dominated by high-G+C-content gram-positive bacteria (Actinobacteria), while bottom water microbial communities had higher diversity dominated by low-G+C-content gram-positive bacteria (Bacillus and Clostridium).
MATERIALS AND METHODS
Sample collection.
The water samples used in this study were collected from the central basin of Mono Lake at station 6 (37o57.822′ N, 119o 01.305′ W; station S30 in previous reports) on 20 July 2000. Samples were collected from 2 m (oxic mixolimnion), 17.5 m (base of the oxycline), 23 m (top of the chemocline), and 35 m (monimolimnion). These depths were chosen because they represent four different zones of water column redox conditions and their DGGE banding patterns are different (data not shown, but see reference 27), indicating differences in community composition. Water samples were collected by using a Niskin bottle. Samples were screened to remove brine shrimp, placed in opaque plastic bottles, and kept in coolers until they were processed (within 4 h of collection). Vertical profiles of temperature, pressure, conductivity, photosynthetically active radiation (PAR, 400 to 700 nm; Licor 2π sensor), turbidity (WetLabs C-Star transmissometer, 10-cm path length), and in vivo fluorescence (WetLabs WetStar fluorometer) were obtained with a SeaBird SeaCat profiler. Oxygen profiles were obtained with a polarographic oxygen sensor (YSI) equipped with a Clarke-type electrode.
Bacterial counts.
Water (10 ml) for bacterioplankton enumeration was preserved with buffered formaldehyde at a 2% final concentration. Samples were stored at 4°C in the dark for no more than 1 month until enumeration was done. Bacterial cells in 0.1 to 1 ml of lake water were mixed with filtered lake water as needed to dilute and disperse cells; the mixtures were filtered onto black 0.2-μm-pore-size polycarbonate membrane filters, rinsed once with 2 ml of 10% acetic acid, stained with 2 ml of 4′,6′-diamidino-2-phenylindole (DAPI) (1 μg ml−1 final concentration), and enumerated by epifluorescence microscopy (54).
DNA extraction.
Water samples (125 to 900 ml; see Table 2) were pressure (ca. 50 kPa) filtered through Millipore Sterivex filter cartridges (0.22-μm-pore size) to collect microbial biomass for subsequent DNA extraction. Excess water was expelled. The cartridges then were filled (1.8 ml) with a buffer containing 50 mM Tris (pH 8.3), 40 mM EDTA, and 0.75 M sucrose; capped; frozen on dry ice; shipped to the laboratory; and stored at −70°C until processed. DNA extraction and purification were carried out essentially as described by Ferrari and Hollibaugh (19). Briefly, 40 μl of lysozyme (50 mg ml−1) was added to each cartridge, and the cartridges were incubated for 60 min at 37°C. Fifty microliters of proteinase K (20 mg ml−1) and 100 μl of a 20% (wt/vol) solution of sodium dodecyl sulfate were added to each cartridge, and the cartridges were incubated at 55°C for 2 h. DNA was purified from 800 μl of the lysate by sequential extraction with 800 μl of phenol-chloroform-isoamyl alcohol (25:24:1), chloroform-isoamyl alcohol (24:1), and finally n-butanol. The aqueous phase was removed, placed in a Centricon-100 concentrator (Amicon), mixed with 500 μl of a buffer containing 10 mM Tris and 1 mM EDTA (pH 8.0), and centrifuged at 1,000 × g for 10 min. Next, 500 μl of TE was added to the Centricon-100 concentrator, and the mixture was centrifuged for another 10 min.
TABLE 2.
Comparison of the compositions of libraries from samples collected at different depths in Mono Lakea
| Depth (m) at which samples were collected | Similarity index (%) for the library from samples collected at the following depth (m):
|
||
|---|---|---|---|
| 17.5 | 23 | 35 | |
| 2 | 67 (0.314, 0.877) | 18 (<0.001, <0.001) | 0 (<0.001, <0.001) |
| 17.5 | 16 (<0.001, <0.001) | 0 (<0.001, <0.001) | |
| 23 | 28 (0.006, 0.124) | ||
The index used was the Sorensen similarity index. Values in parentheses are the probabilities that the compositions of the libraries were different, calculated by using the LIBSHUFF program (X compared to Y, Y compared to X, where X is the library indicated in the stub and Y is the library indicated in the column head) (see Singleton et al. [60] for details).
PCR.
Small-subunit rRNA genes in samples were amplified by using the following primers: 9F, specific for the domain Bacteria (forward, 5′-GAGTTTGATCCTGGCTCAG-3′, positions 9 to 27), and universal primer 1490R (reverse, 5′-GGTTACCTTGTTACGACTT-3′, positions 1490 to 1508) (37, 69). All small-subunit rRNA gene nucleotide positions reported here are based on the Escherichia coli sequence found in the report by Brosius et al. (8). Primers were synthesized either at Operon Technologies (Oakland, Calif.) or at the University of Georgia Molecular Genetics Instrumentation Facility. PCR mixtures were prepared in a total volume of 100 μl containing PCR buffer (10 mM Tris-HCl, 50 mM KCl, 0.1% Triton X-100), 2.5 mM MgCl2, 200 μM each deoxyribonucleoside triphosphate (dATP, dCTP, dGTP, and dTTP), 1 μM each primer, and 20 to 100 ng of template DNA. PCR was performed by using the following conditions: initial denaturation at 95°C for 5 min; pause at 82°C to add Taq DNA polymerase (5 U; Promega); 25 cycles of denaturation (45 s at 94°C), annealing (45 s at 48°C), and extension (1 min at 72°C); and a final extension at 72°C for 45 min. Extracts from filters through which no water had passed served as negative controls. Reactions were run in triplicate and then combined and purified by using Wizard PCR Preps (Promega). The concentration of the resulting PCR product was estimated by using the Hoechst dye assay (52).
Clone libraries.
The PCR product (50 ng) was ligated into pGEM-T Easy Vector (Promega) and transformed into competent E. coli JM109 cells. The transformed cells were plated on Luria-Bertani plates containing 100 μg of ampicillin ml−1, 80 μg of 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) ml−1, and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) as recommended by the manufacturer and incubated overnight at 37°C.
Sequencing.
Clones containing an insert were selected randomly from each library for sequencing. We used only one library from each sample for the analysis presented here; however, 10 other libraries were constructed from different samples and used to identify DGGE bands. Although clones from these other libraries were selected for sequencing because they gave DGGE bands that matched bands of interest from the samples, rather than being selected randomly, they contained the same sequences with approximately the same frequency distribution as that which we determined by random sampling.
All sequences were obtained with an automatic sequencer operated at the Molecular Genetics Instrumentation Facility. A total of 239 clones (55 from 2 m, 61 from 17.5 m, 57 from 23 m, and 66 from 35 m) were selected, purified, and screened by sequencing in one direction through a hypervariable region (V3) of the 16S rRNA gene by using primer 356F (forward, 5′-CCTACGGGAGGCAGCAG-3′), yielding readable sequences of ∼600 bp. Ten of the sequences were ambiguous and were discarded. The remaining sequences were tested for chimeras by using the Ribosomal Database Project CHIMERA-CHECK program. Chimeras were also detected by generating phylogenetic trees with different regions of the sequences. Seventeen of these clones were identified as containing chimeric sequences and were excluded from further analysis. The remaining 212 cloned sequences (51 from the oxic mixolimnion, 2 m; 59 from the base of the oxycline, 17.5 m; 46 from the top of the chemocline, 23 m; and 56 from the monimolimnion, 35 m) were used in phylogenetic analyses. Forty of the these clones, representing all of the major phylotypes, were also sequenced with plasmid primers Sp6 and T7 to obtain nearly complete sequences of the 16S rRNA gene.
Phylogenetic analyses.
Sequences were assigned to major groups, and reference sequences were chosen, based on BLAST (1) similarities. For further phylogenetic analyses, sequences were aligned by using the PILEUP tool of the Wisconsin package, version 10.0 (Genetics Computer Group Inc.). Phylogenetic trees were constructed by using positions 411 to 854 (Proteobacteria), 491 to 882 (high-G+C-content gram-positive organisms), 541 to 972 (low-G+C-content gram-positive organisms), and 540 to 993 (Cytophaga-Flexibacter-Bacteroides [CFB]). Phylogenetic trees were inferred, and bootstrap analysis (100 replicates) was performed with the PHYLIP package (17) by using evolutionary distances (i.e., Jukes-Cantor distances) and the neighbor-joining method. Another phylogenetic tree was constructed by using the nearly full-length sequences (positions 110 to 1404). Sequence similarity values cited below are based on GAP analysis (Wisconsin package, version 10.0) of the nearly full-length sequences representing each phylotype, when they were available, or the shorter, “screening” sequences when they were not.
Statistical analysis and sequence population diversity.
For the purposes of assigning sequences to distinct phylotypes, sequences with similarities of greater than 97% were considered to represent the same phylotypes. In most studies in which similar analyses were performed, the discriminator values used were greater (i.e., 98 to 99% similarity) (36, 47, 59). We chose to use a relatively conservative value to be inclusive (i.e., we tended to “lump” rather than “split”) in consideration of possible errors in the sequences due to PCR (70) and cloning (51, 65). We also wanted to avoid focusing on microdiversity (11, 20). As a result, the taxa (phylotypes) that we define for the purposes of this analysis may not represent individual “species” of bacteria, although 97% 16S rRNA gene sequence similarity was proposed previously as a discriminator of species versus strain differences (25, 66).
We then assumed that the relative abundance of phylotypes defined as described above reflects the relative abundance of the bacteria possessing those sequences in the population of bacteria in our sample. It is not necessary to make this assumption for the purposes of the subsequent analyses, but the analyses are devoid of ecological significance without it. We recognize that PCR biases, differential extraction efficiencies, multiple copies of the rrn operon per genome, and similar artifacts (16, 56, 67, 70) may affect the distribution of sequences in our libraries. We also recognize the possibility that not all of the bacteria present in Mono Lake when we sampled are represented in our libraries, as highly divergent sequences may not have been amplified by the primers used. While the assumption made above is a major one, analysis of the same and similar samples by DGGE with a different set of PCR primers produced qualitatively similar results (27; J. T. Hollibaugh, unpublished data), and the distributions determined by direct counting of readily identifiable organisms (a Synechococcus-like organism and the chloroplast of Picocystis salinarum) agreed well with the distributions obtained from analyses of the clone libraries.
Coverage (C) was calculated by using the following formula: C = 1 − (n1/N), where n1 is the number of phylotypes that occurred only once in the clone library and N is the total number of clones analyzed (48). Rarefaction curves (26) were produced by using software available online at http://www.uga.edu/∼strata/software.html. The phylogenetic compositions of libraries were compared by using the Sorensen similarity index, Cs = 2j/(a + b), where j is the number of phylotypes common to both samples and a and b are the numbers of phylotypes in libraries A and B, respectively (43). The diversity of the libraries and the total number of species in our samples were estimated by using the approach presented by Curtis et al. (10), which assumes a lognormal distribution of species abundance and uses the relative abundance of phylotypes in clone libraries and the total abundance of organisms in the samples (here, DAPI abundance estimates times sample volume) to estimate the total number of phylotypes present. Sequences identified as originating from chloroplast 16S rRNA genes were not included in any of these analyses.
The statistical significance of differences in the compositions of pairs of libraries was tested by using the LIBSHUFF program (60). Sequences (∼500 bp) were aligned as described above, and evolutionary distances were calculated by using the Jukes-Cantor algorithm. The distance matrix was then used to determine the significance of differences in the compositions of pairs of libraries.
Nucleotide sequence accession numbers.
The sequences of the cloned inserts were deposited in GenBank (see Fig. 2 for accession numbers). Only one representative of sequences that were more than 99% similar was deposited.
FIG. 2.
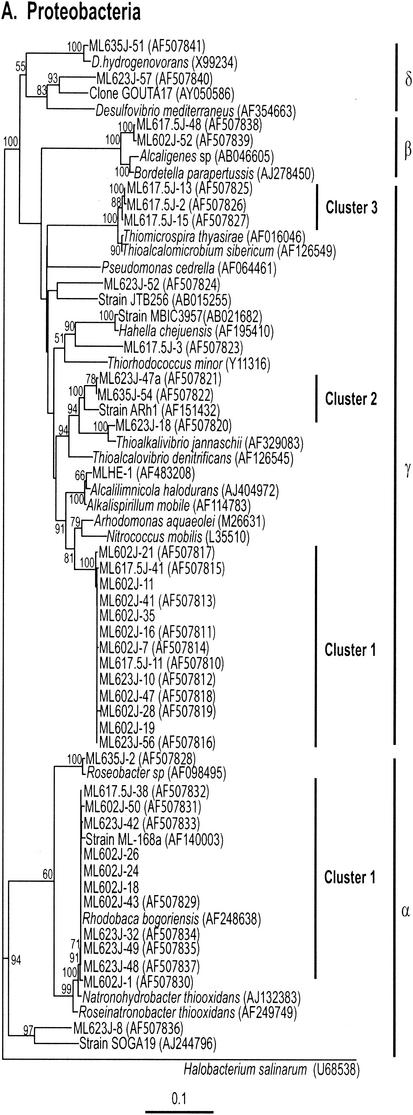
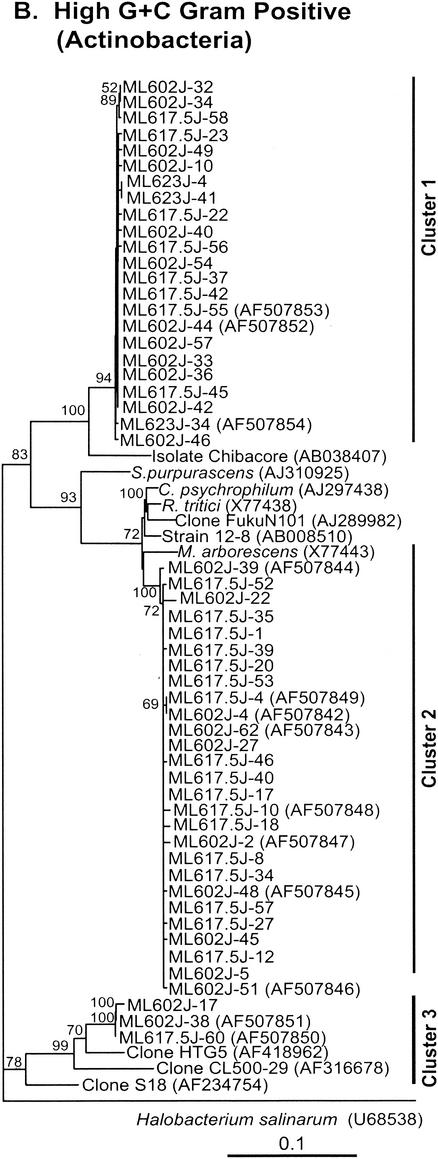
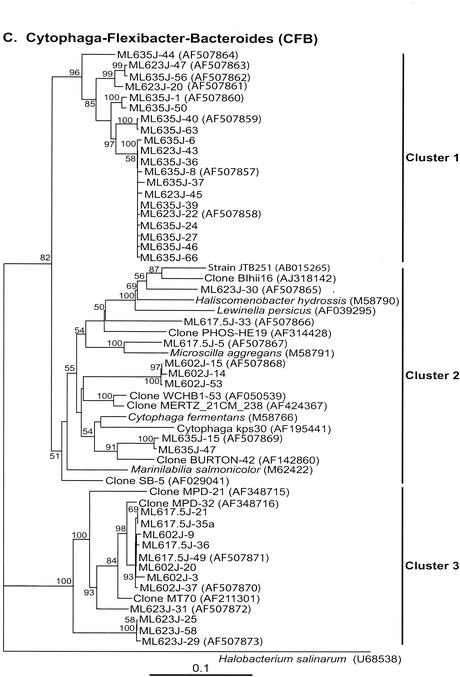
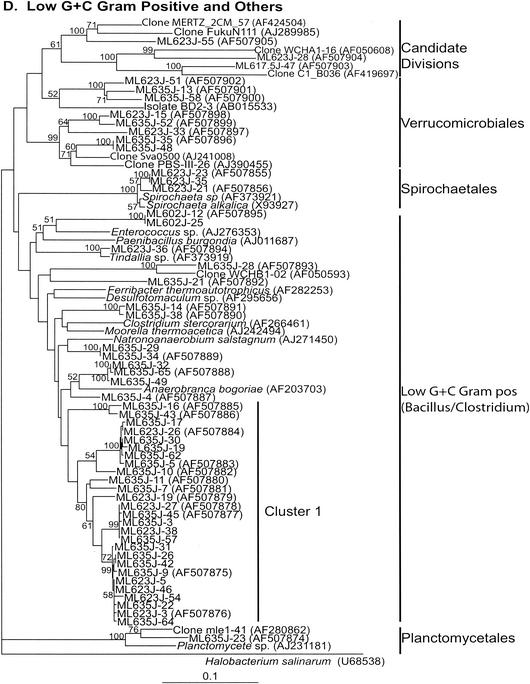
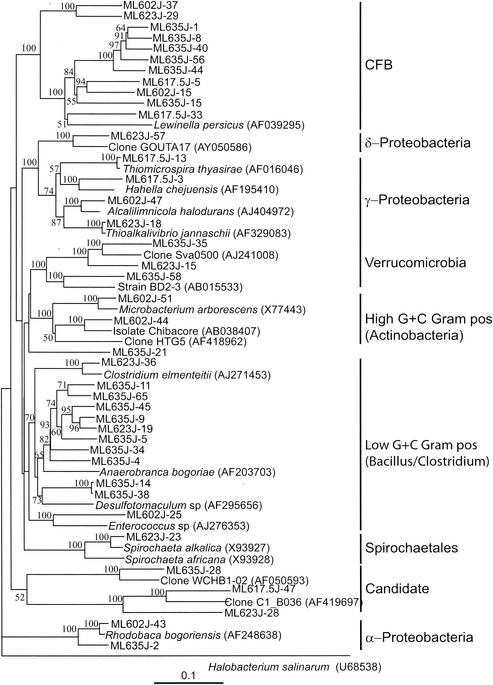
Neighbor-joining trees (partial sequences, ∼400 bp) showing phylogenetic relationships of 16S rRNA gene sequences cloned from Mono Lake to closely related sequences from GenBank. Clones from this study are coded as follows, with ML623J-31 as an example: ML, Mono Lake; 6, station 6; 23, sample depth in meters; J, month in which samples were collected (July); and 31, number assigned to the clone in that library (thus, ML623J is the library identifier). Scale bars indicate Jukes-Cantor distances. Bootstrap values of >50% (for 100 iterations) are shown. The trees are unrooted, with Halobacterium salinarum as the outgroup. GenBank accession numbers are given in parentheses. pos, positive.
RESULTS
Water column profiles.
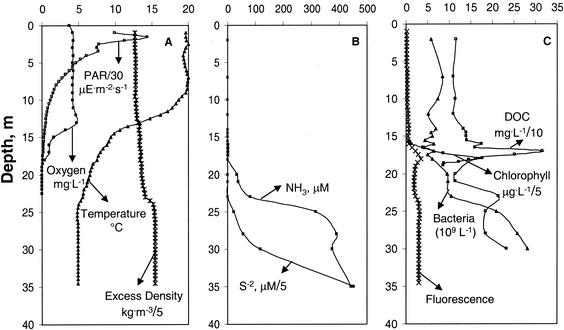
Fig. 1 shows the vertical distribution of water column variables at Mono Lake station 6 when our samples were collected. A steep thermocline was evident between depths of 9 and 24 m, with a mixolimnion temperature of 20°C and a monimolimnion temperature of 5°C (Fig. 1A). A chemocline was present over a depth range of 23 to 26 m. There was a slight increase in oxygen concentration at 12 m, and then the oxygen concentration decreased rapidly below this depth to the limit of detection at 17 m. The chemocline and monimolimnion were anoxic. Sulfide and ammonia concentrations began to increase at 18 to 20 m, reaching their highest levels at 35 m (Fig. 1B). Chlorophyll concentration and fluorescence were low in the mixolimnion, and both showed a distinct peak at the base of the oxycline that corresponded to a high DOC concentration (Fig. 1C). Bacterial abundance was 6 × 109 cells liter−1 in the mixolimnion, and there was a slight increase (10 × 109 cells liter−1) at the base of the oxycline. Bacterioplankton abundance was fivefold higher in the monimolimnion than in the mixolimnion (Fig. 1C).
FIG. 1.
Depth distributions of selected physicochemical and biological variables in the Mono Lake water column at station 6 on 20 July 2000. (A) PAR, oxygen, temperature, and excess density. μE, microeinsteins. (B) Ammonia (NH3) and sulfide (S2−). (C) Bacterial abundance, DOC, fluorescence, and chlorophyll. Note that some variable values are scaled as indicated (by numbers following slashes) to fit axis ranges.
Phylogenetic analyses.
Partial sequences of the 16S rRNA genes fell into the following groups.
(i) Chloroplast-like sequences
A total of 18 (29%) of the sequences retrieved from the 17.5-m sample, 5 (11%) retrieved from the 23-m sample, and 1 each (2%) retrieved from the 2- and 35-m samples were from the chloroplast 16S rRNA gene of the chlorophyte alga P. salinarum. Two other sequences retrieved from the libraries of the 2- and 17.5-m samples were related (96% similarity) to the chloroplast 16S rRNA gene of the diatom Odontella sinensis.
(ii) α-Proteobacteria.
Eleven of the 13 sequences that fell into the α-Proteobacteria group clustered together (Fig. 2A and 3) and were found in libraries generated from the 2-, 17.5-, and 23-m samples. The nearly full-length sequence (1,391 bp) of one of these clones (ML602J-43) was 97 to 98% similar to Rhodobaca bogoriensis (46), Roseonatronobacter thiooxidans (64), and Natronohydrobacter thiooxidans. It was also 97% similar to a bacterium (ML-168a) previously isolated from Mono Lake sediment (GenBank description). R. bogoriensis is an anoxygenic, phototrophic, alkaliphilic purple nonsulfur bacterium isolated from soda lakes in the African Rift Valley (GenBank description). It is capable of both phototrophic growth and chemoautotrophic growth on a wide variety of organic compounds (46). R. thiooxidans is a strictly aerobic and obligately heterotrophic alkaliphile isolated from Russian soda lakes (64). An identical sequence was found in a DGGE band in a previous study of Mono Lake (27). The only α-Proteobacteria sequence obtained from the monimolimnion was related (96% similarity over 1,385 bp) to a Roseobacter sp.
FIG. 3.
Neighbor-joining tree of nearly full-length sequences (∼1,300 bp) of representative examples from each of the major clades from Fig. 2 showing phylogenetic relationships to 16S rRNA gene sequences from GenBank. The scale bar indicates Jukes-Cantor distances. Bootstrap values of >50% (for 100 iterations) are shown. The trees are unrooted, with Halobacterium salinarum as the outgroup. GenBank accession numbers are given in parentheses. pos, positive.
(iii) β-Proteobacteria.
Two sequences related (94% similarity) to Alcaligenes spp. of the β-Proteobacteria were retrieved from the mixolimnion and oxycline libraries (Fig. 2A).
(iv) γ-Proteobacteria.
Twenty-one sequences (only 1 from the monimolimnion) clustered with the γ-Proteobacteria (Fig. 2A and 3). Thirteen of these sequences (9 from the mixolimnion) clustered together (cluster 1). The nearly full-length sequence (1,462 bp) of one of these clones (ML602J-47) was distantly related (93 to 94% similarity) to Alcalilimnicola halodurans, Alkalispirilum mobile, and Nitrococcus mobilis. It was also 92% similar to a bacterium (MLHE-1) (49) isolated from Mono Lake. A. halodurans is an alkaliphilic, extremely halotolerant bacterium isolated from the sediment of soda-depositing Lake Natron, Tanzania (72). A. mobile is a moderately halophilic, alkaliphilic, gram-negative, obligate aerobe (58). Previous studies of Mono Lake reported sequences that were 99% similar to these sequences in oxycline samples (27, 68). Two sequences (one from the chemocline and one from the monimolimnion) fell into cluster 2 and were related (96% similarity) to an obligately autotrophic, sulfur-oxidizing, alkaliphilic bacterium (Arh1) of the genus Thioalkalivibrio. This organism was reported to use thiocyanate as a sole source of energy under highly alkaline conditions (63). A sequence found only in the clone library from the chemocline sample was related (98% similarity) (Fig. 3) to Thioalcalivibrio jannaschii, a haloalkaliphilic, obligately chemolithoautotrophic, sulfur-oxidizing bacterium recently isolated from Mono Lake (61). Three additional sequences related most closely to chemoautotrophic sulfur oxidizers (Thiomicrospira thyasirae, 98% similarity; and Thioalcalomicrobium sibericum, 96% similarity) (Fig. 2A) were recovered from the oxycline sample. T. sibericum is a member of a newly described genus isolated from soda lakes in Siberia and Kenya (62).
(v) δ-Proteobacteria.
One sequence from the chemocline clone library was distantly related (90% similarity) to a clone designated GOUTA17 (Fig. 2A) and obtained from groundwater samples contaminated with chlorobenzene (GenBank description) as well as to sulfate-reducing bacteria (<90% similarity) obtained from salt marsh sediments and deep-sea environments. A sequence from the monimolimnion library was related (95% similarity) (Fig. 2A) to Desulfonatronovibrio hydrogenovorans, an alkaliphilic, sulfate-reducing bacterium (75).
(vi) High-G+C-content gram-positive bacteria (Actinobacteria).
A total of 53 sequences clustered with the high-G+C-content gram-positive bacteria (Fig. 2B and 3). These sequences were associated with the oxygenated surface water of Mono Lake, where they were the dominant sequences in clone libraries: only three of these sequences were obtained from the chemocline library, and none were obtained from the monimolimnion library. Twenty-three sequences formed a separate lineage (cluster 1) characterized by only one previously described sequence, from an isolate identified as Chibacore 1500 (GenBank accession no. AB038407; 94% similarity). Chibacore 1500 is an alkaliphilic bacterium isolated from a depth of 1,500 m and showing only sparse growth on any culture medium; it was subsequently lost (GenBank description; H. Kaneko, personal communication). Another 27 sequences (26 from the mixolimnion and oxycline libraries) fell into a second cluster and were 94 to 96% similar (Fig. 3) to Rathayibacter tritici and Microbacterium arborescens and to an environmental isolate, FukuN101, cloned from a sample collected from an acidic forest lake in Germany (23). Three sequences that fell into a third cluster were distantly related (93% similarity) to a clone designated HTG5 and obtained from microbial populations associated with Metallogenium morphotypes and amorphous particles in a freshwater reservoir (GenBank description).
(vii) CFB group.
Forty sequences (7 from the mixolimnion, 6 from the oxycline, 10 from the chemocline, and 17 from the monimolimnion libraries) fell into the CFB clade (Fig. 2C and 3). Fifteen monimolimnion and 5 chemocline sequences formed a separate lineage (cluster 1) and were not affiliated with any previously documented sequences. Two other sequences from the monimolimnion library were distantly related (92%) to the partial sequence (∼900 bp) of a clone designated BURTON-42 and obtained from the anoxic sediment of a meromictic lake in eastern Antarctica (5). Three deeply branching sequences from the chemocline sample (ML623J-25, -29, and -58) were distantly related (89% similarity over positions 514 to 1236) to the partial sequence of clone kpc103f (GenBank accession no. AF195431) (not shown in Fig. 2C and 3), found to co-occur with the brown-tide phytoplankter Aureococcus anophagefferens (GenBank description). Eight sequences from the mixolimnion and oxycline libraries that clustered together were related (95% similarity) to the partial sequence (∼1,000 bp) of a clone, designated MPD-32, obtained from a selenium-contaminated, hypersaline evaporation pond (12). They were 90% similar to the partial sequence (∼900 bp) of a clone, designated MT70, obtained from hydrogen sulfide-rich, black mud from a coastal marine environment (GenBank description). No other previously documented sequences fell into this cluster. Three sequences from the mixolimnion library were distantly related (86 to 88% similarity) to the sequence of a clone designated WCHB1-53 and obtained from a hydrocarbon- and chlorinated-solvent-contaminated aquifer (13); to a clone designated MERTZ_21CM_238 and obtained from continental shelf sediments off the coast of Antarctica (GenBank description); and to the 16S rRNA gene sequence of bacterium JTB132 (GenBank accession no. AB015260) (not shown in Fig. 2C and 3), isolated from a cold seep in the Japan Trench (40). A previous study (27) of Mono Lake reported the presence of a similar sequence in mixolimnion samples.
(viii) Low-G+C-content gram-positive bacteria (Bacillus and Clostridium spp.).
Low-G+C-content gram-positive bacterial sequences dominated libraries from the anoxic water of Mono Lake. Forty sequences (2 from the mixolimnion, 9 from the chemocline, and 29 from the monimolimnion) fell into the low-G+C-content gram-positive clade (Fig. 2D and 3). None of these sequences was closely related to any 16S rRNA gene sequence in the GenBank database. Twenty-seven sequences (19 from the monimolimnion and 8 from the chemocline) formed a separate lineage (cluster 1). Two sequences were distantly related (86%) to Anaerobranca bogoriae, a recently described thermoalkaliphile (55). This organism is an obligate anaerobe capable of growth on a variety of carbohydrates and does not use sulfate as an electron acceptor. Two sequences were distantly related (85%) to Natronoanaerobium salstagnum, an obligately anaerobic alkaliphile isolated from salterns of alkaline, hypersaline Lake Magadi, Kenya (32). The only two sequences obtained from oxic water (mixolimnion library) and related to the low-G+C-content gram-positive bacteria were distantly related (83%) to Enterococcus spp., facultative anaerobes that are part of the normal gut microflora, and thus might have originated from fecal material produced by the numerous birds that feed on the brine shrimp population of Mono Lake during the summer.
(ix) Spirochaetales.
Sequences of three clones obtained from the chemocline library (Fig. 2D) were related to moderately halophilic, alkaliphilic Spirochaeta spp. isolated from various soda lakes (74), including Mono Lake (GenBank description).
(x) Others.
We also retrieved eight sequences from the chemocline and monimolimnion libraries that clustered with the Verrucomicrobiales (Fig. 2D). Three sequences clustered with candidate divisions (Fig. 2D). A short sequence obtained from the monimolimnion library was related (97% similarity) to the marine cyanobacteria Synechococcus and Prochlorococcus (data not shown).
Distribution of bacterial groups.
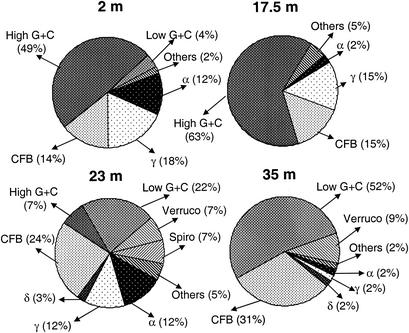
The relative abundances of different phylogenetic groups in the libraries (excluding chloroplast sequences) were calculated for all four samples (Fig. 4). The phylogenetic compositions of our clone libraries changed completely with depth. A total of 49 and 63% of the sequences retrieved from the mixolimnion (2-m) and oxycline (17.5-m) libraries, respectively, were affiliated with high-G+C-content gram-positive bacteria, whereas 52% of the sequences retrieved from the monimolimnion (35-m) library were affiliated with the low-G+C-content gram-positive group. A total of 22% of the sequences retrieved from the chemocline (23-m) library were affiliated with low-G+C-content gram-positive bacteria, and 7% were affiliated with high-G+C-content gram-positive bacteria. Sequences affiliated with the CFB group were less abundant in the mixolimnion library than in the chemocline and monimolimnion libraries (14% versus 24 and 31%, respectively) and, as mentioned above, the mixolimnion and monimolimnion libraries were dominated by different CFB.
FIG. 4.
Frequencies of phylotypes affiliated with major phylogenetic groups in libraries from 2-, 17.5-, 23-, and 35-m samples. Chloroplast-like sequences were omitted from the data set used to calculate percentages. α, γ, and δ, Proteobacteria; Verruco, Verrucomicrobiales; Spiro, Spirochaetales.
Coverage, sequence diversity, and similarity of libraries.
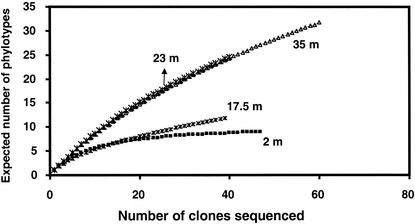
The coverage values that we obtained were 98, 83, 59, and 73% for the mixolimnion, oxycline, chemocline, and monimolimnion samples, respectively (Table 1), indicating that the sequence population from the chemocline sample was the most diverse. Coverage curves (Fig. 5) indicated different patterns of diversity in clone libraries from different samples. Coverage analysis indicated that the chemocline and monimolimnion libraries were similarly diverse and more diverse than the mixolimnion and oxycline libraries, which had similar diversities. These conclusions are supported by an analysis of diversity (Table 1). The diversity index ranged from 2.4 in the oxycline sample to 10.3 in the chemocline sample. The total number of phylotypes expected per sample ranged from 195 in the oxycline sample to 4,719 in the chemocline sample. The similarities of the phylotype populations in these libraries ranged from 0 to 67% (Table 2). The monimolimnion library had no phylotypes in common with the mixolimnion and oxycline libraries. LIBSHUFF analysis (Table 2) indicated that libraries from aerobic and anaerobic samples were highly significantly different and that libraries from aerobic samples were not significantly different from each other, nor were libraries from anoxic samples. LIBSHUFF analysis also indicated that the chemocline and monimolimnion libraries were characterized by deeper divergence than the mixolimnion and oxycline libraries.
TABLE 1.
Properties of the distributions of phylotypes in clone libraries from Mono Lake samplesa
| Sample source (depth, m) | Nt (109) | % Coverage | Nt/Nmax | St |
|---|---|---|---|---|
| Mixolimnion (2) | 4.4 | 98 | 4.0 | 846 |
| Oxycline (17.5) | 4.5 | 83 | 2.4 | 195 |
| Chemocline (23) | 2.6 | 59 | 10.3 | 4,719 |
| Monimolimnion (35) | 4.0 | 73 | 6.1 | 2,102 |
Nt, total number of cells sampled; coverage, library coverage calculated as described in the text; Nt/Nmax, diversity index calculated as the total number of cells in each sample divided by the number of cells of the most abundant phylotype in each sample; St, estimated total number of phylotypes in the sample. See Curtis et al. (10) for details.
FIG. 5.
Rarefaction curves generated for 16S rRNA genes in clone libraries from samples collected at 2, 17.5, 23, and 35 m.
DISCUSSION
A conclusive result of our study is the complete shift in the composition of the bacterial assemblage between the mixolimnion and the monimolimnion. None of the phylotypes found in the mixolimnion or oxycline libraries were the same as those found in the monimolimnion library (Table 2). The difference between surface water and deep water samples was primarily due to the distributions of two major clades. The clone libraries from the mixolimnion and oxycline samples were dominated (49 and 63%, respectively) by sequences affiliated with high-G+C-content gram-positive bacteria, while the monimolimnion library was dominated (52%) by sequences related to low-G+C-content gram-positive bacteria. It is unlikely that the shift in the composition that we observed was simply due to PCR bias, cloning bias, or a similar analytical error, as these changes reflect shifts in banding patterns detected by DGGE (27) (which uses a different primer set) and are consistent with what is known about the physiology and ecology of these groups. Examination of these samples by epifluorescence microscopy also indicated differences in the populations of bacteria from these two habitats. In general, more chain-forming colonies and more large spirochetes were present in the monimolimnion. This finding is consistent with previously reported findings for other soda lakes (15, 73). Casamayor et al. (9), in a study of meromictic Lake Vilar, Spain, found that shifts in the distributions of closely related sequences reflected the distributions and ecological properties of the organisms possessing the sequences.
Even more striking was the dramatic difference in the pattern of diversity in oxic versus anoxic samples (Fig. 2 and 3). Relative to what has been found for other environments that have been characterized (7, 21, 36, 42, 48), the diversity of Bacteria in Mono Lake is low, as indicated by the coverage, rarefaction, and diversity analyses that we performed. However, within the lake, diversity was higher, with more deeply diverging taxa in anoxic deep water than in oxic surface water. The mixolimnion and oxycline libraries contained 9 and12 phylotypes in four and five clades that accounted for 78 and 85%, respectively, of all of the sequences that we retrieved from these libraries. In contrast, the chemocline and monimolimnion libraries contained 25 and 27 phylotypes in five and six major clades that accounted for only 44 and 48%, respectively, of all of the sequences retrieved from these libraries. This difference is also reflected in the coverage estimates for these libraries, the estimated total number of phylotypes in the samples, and their diversity indices (Table 1) and by their rarefaction curves (Fig. 5).
Note that the oxycline sample was identified as the least diverse by the analytical approach described by Curtis et al. (10), whereas the mixolimnion sample was identified as the least diverse by coverage estimates and rarefaction curves. Curtis et al. (10) assume a lognormal distribution of species abundance. We tested this assumption with our data sets and found that it was met (P < 0.05) only by data from the monimolimnion sample. However, the data distributions were highly skewed to the right tail of the curves (toward the most abundant phylotypes), rather than being distributed over the whole range of species abundances (i.e., rare phylotypes were not enumerated), greatly reducing the power of the statistical test of lognormality. As pointed out by Curtis et al. (10), correcting this bias would involve sequencing of a much larger number of clones from each library.
Our results are in contrast to those of a previous study of Mono Lake microbial assemblages based on DGGE analysis (27). That study concluded that diversity was higher in oxic surface water samples than in anoxic deep water samples. A similar DGGE-based study of another stratified lake (50) also reported higher diversity in oxic surface water than in anoxic deep water. A likely explanation for this discrepancy, at least with regard to Mono Lake, is that most of the phylotypes found in deep water were present at a very low relative abundance and thus formed faint bands that were not readily detected by DGGE. This explanation is supported by DGGE analysis of these samples (Hollibaugh, unpublished) by using technology superior to that available to Hollibaugh et al. (27).
Although it is an anthropocentric view, the bottom water of Mono Lake might be considered to be more extreme than the surface water because of the potential toxicity of the high concentrations of sulfide and ammonia that have accumulated after 8 years of meromixis. Current ecological theory would lead to the expectation that the species (here, sequence) diversity of populations in the extreme environment of bottom water would be low relative to the diversity of populations from surface water, where less extreme conditions prevail, yet we found the opposite to be true.
We considered the possibility that heterogeneity of the 16S rRNA gene in different rrn operons of the same genome might contribute to an artifactually high apparent diversity of Mono Lake bottom water (16, 35), since our analyses indicated that these samples were dominated by low-G+C-content gram-positive bacteria related to Bacillus and Clostridium, which are known to contain multiple copies of rrn operons. We tried to minimize the effect of intragenomic heterogeneity of rrn operons on our analysis by using a low similarity value (97%) to decide whether or not two sequences were the same.
If intragenomic heterogeneity is not the cause of the high diversity in the chemocline and monimolimnion samples, then environmental factors must be contributing to the diversity that we found. At least for the monimolimnion sample, it is unlikely that this diversity can be ascribed to mixing of disparate populations because these waters have been isolated for several years. Sinking Artemia fecal pellets may also have transported surface water microorganisms into deep water. This process may account for the presence in the chemocline library (and thus partly for the greater diversity of that library) of sequences related to the cluster 1 low-G+C-content gram-positive bacteria that were dominant in the mixolimnion and oxycline libraries (Fig. 2A). However, these sequences were not found in the monimolimnion library. Similarly, resuspension of bacterial assemblages from sediments seems unlikely because the sediments in this portion of the lake are isolated from wind-driven surface currents (41), and there is no bioturbation.
Although the strong physicochemical gradients in Mono Lake no doubt contribute to the overall diversity of the bacterioplankton in the lake by supporting a range of redox environments (niches), it is unlikely that this element of habitat diversity is responsible for the elevated diversity in the monimolimnion, as these gradients are weak there relative to other locations in the water column (the oxycline, for example). Microscope observations and transmissometer data indicated elevated particle and floc concentrations in Mono Lake bottom water, which may contribute to habitat diversity and thus elevated phylogenetic diversity. Another hypothesis to explain the elevated diversity of monimolimnion bacterial populations is that it may be possible to maintain a higher diversity of energetic pathways in anoxic environments because organisms whose metabolism is based on oxygen as an electron acceptor do not out-compete organisms that use other, less energetically efficient, forms of metabolism; such a situation would lead to the retention of higher metabolic diversity and thus ecological diversity under anaerobic conditions than under aerobic conditions. Madrid et al. (42) also found higher diversity in the deep (1,310-m) anoxic water of the Cariaco Basin than in the overlying oxic oceanic water. Regardless of the reason, these results suggest that the ecological forces that act to structure aerobic microbial communities are fundamentally different from those that act to structure anaerobic microbial communities.
The differences in the phylogenetic affinities of dominant phylotypes from mixolimnion versus monimolimnion samples are consistent with the differences in the physicochemical milieus of these two habitats and with what is known from other studies of the physiology, ecology, and distribution of representatives of these groups. Although they are not closely related to the Mono Lake high-G+C-content gram-positive bacteria, previous studies have shown that marine planktonic high-G+C-content gram-positive bacteria are generally found in the photic zone (22). The frequency of Picocystis chloroplast 16S rRNA gene sequences in our libraries is consistent with the vertical distribution of Picocystis cells in the lake. The bacterial community that we found in the monimolimnion sample is not typical of the water column but rather resembles that of sediment or soil. For example, low-G+C-content gram-positive bacteria are characteristic of deep sea sediments (40). The largest clade of sequences retrieved from sulfide-rich, anoxic sediment from a meromictic eastern Antarctica lake clustered with low-G+C-content gram-positive bacteria (5). Madrid et al. (42) also reported that the diversity and composition of 16S rRNA gene sequences from their deep anoxic water sample resembled those of a sediment or soil sample; however, low-G+C-content gram-positive bacteria were not abundant in this library, which was dominated by sequences most closely affiliated with the γ-Proteobacteria.
Sequences from mixolimnion and oxycline libraries that were related to the CFB group were affiliated with the order Cytophaga-Flavobacteria, which is dominated by aerobic bacteria. CFB sequences from the monimolimnion and some of the chemocline CFB sequences fell within the order Bacteroidales, which is dominated by anaerobes that are commonly members of gut microflora.
Although the compositions of the mixolimnion and oxycline libraries were similar, two mixolimnion phylotypes were not found in the oxycline samples and four oxycline phylotypes were not found in the mixolimnion samples. One of the unique oxycline phylotypes was closely related (98% similarity) to Thiomicrospira spp., chemolithoautotrophic bacteria that use reduced sulfur compounds, such as sulfide, thiosulfate, and sulfur, as an energy source. The oxycline is a logical place in the physicochemical gradient of the lake to find sulfide-oxidizing bacteria like Thiomicrospira spp. It was shown that Thiomicrospira spp. increased at an interface between the oxidized surface and the anaerobic layer of black mud, assimilating the reduced product from the lower anoxic zone (6). Another unique oxycline phylotype was distantly related (88% similarity) to Hahella chejuensis, a facultative anaerobe capable of using several different carbohydrates as a carbon source (38). The two other unique oxycline phylotypes were related to Cytophaga spp. The planktonic alga P. salinarium was abundant in the oxycline samples, coinciding with peaks in fluorescence, chlorophyll, and DOC, suggesting that algal exudates provided the major source of organic carbon in this environment. Members of the CFB have been identified as being associated with the degradation of complex organic substrates (53, 57). Yager et al. (71) found that Cytophaga spp. dominated during the peak of an algal bloom. The CFB phylotypes that appeared to be unique to the oxycline samples may have been involved in degrading organic matter produced by Picocystis, which has an unusual composition (39).
If we accept the criterion that two organisms with 16S rRNA gene sequences that are 93% similar are related at the level of genus (48), 75% of our cloned sequences, representing 50 phylotypes, belonged to taxonomic groups that are distinct at the genus level or higher and that have not been previously described. None of the sequences that we obtained and that were related to CFB, low-G+C-content gram-positive bacteria, and candidate division groups were similar (>97% sequence similarity) to 16S rRNA gene sequences of cultivated organisms or to environmental sequences recovered from other sites. The sequences found in this study are also different from sequences previously retrieved from other alkaline environments. Most previous studies of alkaline environments have been based on classical isolation techniques (15),which may not completely sample the diversity present in a habitat (32). It is estimated that >99% of microorganisms observable in nature typically are not cultivated by standard techniques (2, 18). Thus, the data presented in this report significantly advance the understanding of the microbial ecology of alkaline, hypersaline environments.
Acknowledgments
This work was supported by National Science Foundation grant MCB 99-77886. Field laboratory facilities, limnological data processing, and other survey and logistical support were provided by R. Jellison, S. Roll, and the Sierra Nevada Aquatic Research Laboratory with support from the National Science Foundation (MCB-9977901).
We are grateful to G. Lecleir for help with sample collection and for providing cell count and DOC data. S. B. Joye and S. Carini supplied sulfide and ammonia data, and A. Burd helped with diversity analyses. E. Biers helped with suggestions and excellent cakes.
REFERENCES
- 1.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed]
- 2.Amann, R., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Riel, and F. Thingstad. 1983. Ecological role of water column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 4.Azam, F., and R. A. Long. 2001. Oceanography—sea snow microcosms. Nature 414:495.. [DOI] [PubMed] [Google Scholar]
- 5.Bowman, J. P., S. M. Rea, S. A. McCammon, and A. McMeekin. 2000. Diversity and community structure within anoxic sediment from marine salinity meromictic lakes and a coastal meromictic marine basin, Vestfold Hills, Eastern Antarctica. Environ. Microbiol. 2:227-237. [DOI] [PubMed] [Google Scholar]
- 6.Brinkhoff, T., and G. Muyzer. 1997. Increased species diversity and extended habitat range of sulfur-oxidizing Thiomicrospira spp. Appl. Environ. Microbiol. 63:3789-3796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Britschgi, T. B., and S. J. Giovannoni. 1991. Phylogenetic analysis of a natural marine bacterioplankton population by rRNA cloning and sequencing. Appl. Environ. Microbiol. 57:1707-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brosius, J., M. L. Palmer, J. K. Poindexter, and H. F. Noller. 1978. Complete nucleotide sequence of a 16S ribosomal RNA gene from Escherichia coli. Proc. Natl. Acad. Sci. USA 75:4801-4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Casamayor, E. O., C. Pedros-Alio, G. Muyzer, and R. Amann. 2002. Microheterogeneity in 16S ribosomal DNA-defined bacterial populations from a stratified planktonic environment is related to temporal changes and to ecological adaptations. Appl. Environ. Microbiol. 68:1706-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeLong, E. E., and N. R. Pace. 2001. Environmental diversity of Bacteria and Archaea. Syst. Biol. 50:470-478. [PubMed] [Google Scholar]
- 12.de Souza, M. P., A. Amini, M. A. Dojka, I. J. Pickering, S. C. Dawson, N. R. Pace, and N. Terry. 2001. Identification and characterization of bacteria in a selenium-contaminated hypersaline evaporation pond. Appl. Environ. Microbiol. 67:3785-3794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dojka, M. A., P. Hugenholtz, S. K. Haack, and N. R. Pace. 1998. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl. Environ. Microbiol. 64:3869-3877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Domagalski, J. L., W. H. Orem, and H. P. Eugster. 1989. Organic geochemistry and brine composition in Great Salt, Mono, and Walker Lakes. Geochim. Cosmochim. Acta 53:2857-2872. [Google Scholar]
- 15.Duckworth, A. W., W. D. Grant, B. E. Jones, and R. van Steenbergen. 1996. Phylogenetic diversity of soda lake alkaliphiles. FEMS Microbiol. Ecol. 19:181-191. [Google Scholar]
- 16.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein, J. 1993. PHYLIP (Phylogeny Inference Package), v. 3.5 ed. University of Washington, Seattle.
- 18.Ferguson, R. L., E. N. Buckley, and A. V. Palumbo. 1984. Response of marine bacteria to differential filtration and confinement. Appl. Environ. Microbiol. 47:49-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrari, V. C., and J. T. Hollibaugh. 1999. Distribution of microbial assemblages in the central Arctic Ocean basin studied by PCR/DGGE: analysis of a large data set. Hydrobiologia 401:55-68. [Google Scholar]
- 20.Garcia-Martinez, J., and F. Rodriguez-Valera. 2000. Microdiversity of uncultured marine prokaryotes: the SAR11 cluster and the marine Archaea of group I. Mol. Ecol. 9:935-948. [DOI] [PubMed] [Google Scholar]
- 21.Giovannoni, S. J., T. B. Britschgi, C. L. Moyer, and K. G. Field. 1990. Genetic diversity in Sargasso Sea bacterioplankton. Nature 345:60-63. [DOI] [PubMed] [Google Scholar]
- 22.Giovannoni, S. J., and M. Rappe. 2000. Evolution, diversity and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. Wiley-Liss, New York, N.Y.
- 23.Glöckner, F. O., E. Zaichikov, N. Belkova, L. Denissova, J. Pernthaler, A. Pernthaler, and R. Amann. 2000. Comparative 16S rRNA analysis of lake bacterioplankton reveals globally distributed phylogenetic clusters including an abundant group of actinobacteria. Appl. Environ. Microbiol. 66:5053-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grant, W. D., W. E. Mwatha, and B. E. Jones. 1990. Alkaliphiles: ecology, diversity, and applications. FEMS Microbiol. Rev. 75:255-270. [Google Scholar]
- 25.Hagstrom, A., J. Pinhassi, and U. L. Zweifel. 2000. Biogeographical diversity among marine bacterioplankton. Aquat. Microb. Ecol. 21:231-244. [Google Scholar]
- 26.Heck, K. L., Jr., G. Van Belle, and D. Simberloff. 1975. Explicit calculation of the rarefaction diversity measurement and the determination of sufficient sample size. Ecology 56:1459-1461. [Google Scholar]
- 27.Hollibaugh, J. T., P. S. Wong, N. Bano, S. K. Pak, E. M. Prager, and C. Orrego. 2001. Stratification of microbial assemblages in Mono Lake, California, and response to a mixing event. Hydrobiologia 466:45-60. [Google Scholar]
- 28.Horikoshi, K., and T. Akiba. 1982. Alkaliphilic microorganisms. Springer-Verlag KG, Berlin, Germany.
- 29.Jellison, R., R. F. Anderson, J. M. Melack, and D. Heil. 1996. Organic matter accumulation in sediments of hypersaline Mono Lake during a period of changing salinity. Limnol. Oceanogr. 41:1539-1544. [Google Scholar]
- 30.Jellison, R., and J. M. Melack. 1993. Meromixis in hypersaline Mono Lake, California. 1. Stratification and vertical mixing during the onset, persistence, and breakdown of meromixis. Limnol. Oceanogr. 38:1008-1019. [Google Scholar]
- 31.Jensen, K. M., and R. P. Cox. 1992. Effects of sulfide and low redox potential on the inhibition of nitrous-oxide reduction by acetylene in Pseudomonas nautica. FEMS Microbiol. Lett. 96:13-17. [DOI] [PubMed] [Google Scholar]
- 32.Jones, B. E., W. D. Grant, A. W. Duckworth, and G. G. Owenson. 1998. Microbial diversity of soda lakes. Extremophiles 2:191-200. [DOI] [PubMed] [Google Scholar]
- 33.Joye, S. B., T. L. Connell, L. G. Miller, R. S. Oremland, and R. S. Jellison. 1999. Oxidation of ammonia and methane in an alkaline, saline lake. Limnol. Oceanogr. 44:178-188. [Google Scholar]
- 34.Joye, S. B., and J. T. Hollibaugh. 1995. Sulfide inhibition of nitrification influences nitrogen regeneration in sediments. Science 270:623-625. [Google Scholar]
- 35.Kerkhof, L., and M. Speck. 1997. Ribosomal RNA gene dosage in marine bacteria. Mol. Mar. Biol. Biotechnol. 6:260-267. [PubMed] [Google Scholar]
- 36.Kerkhof, L. J., M. A. Voytek, R. M. Sherrell, D. Millie, and O. Schofield. 1999. Variability in bacterial community structure during upwelling in the coastal ocean. Hydrobiologia 401:139-148. [Google Scholar]
- 37.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. Academic Press, Chichester, England.
- 38.Lee, H. K., J. Chun, E. Y. Moon, S. H. Ko, D. S. Lee, H. S. Lee, and K. S. Bae. 2001. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int. J. Syst. Bacteriol. 51:661-666. [DOI] [PubMed] [Google Scholar]
- 39.Lewin, R. A., L. Krienitz, R. Goericke, H. Takeda, and D. Hepperle. 2000. Picocystis salinarum gen. et sp. nov. (Chlorophyta)—a new picoplanktonic green alga. Phycologia 39:560-565. [Google Scholar]
- 40.Li, L. N., C. Kato, and K. Horikoshi. 1999. Bacterial diversity in deep-sea sediments from different depths. Biodivers. Conserv. 8:659-677. [Google Scholar]
- 41.MacIntyre, S., and R. Jellison. 2001. Nutrient fluxes from upwelling and enhanced turbulence at the top of the pycnocline in Mono Lake, California. Hydrobiologia 384:21-39. [Google Scholar]
- 42.Madrid, V., G. Taylor, M. Scranton, and A. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Magurran, A. E. 1988. Ecological diversity and its measurement. Princeton University Press, Princeton, N.J.
- 44.Melack, J. M., and R. Jellison. 1998. Limnological conditions in Mono Lake: contrasting monomixis and meromixis in the 1990s. Hydrobiologia 384:21-39. [Google Scholar]
- 45.Meyers, P. A. 1994. Preservation of elemental and isotopic source identification of sedimentary organic matter. Chem. Geol. 114:289-302. [Google Scholar]
- 46.Milford, A. D., L. A. Achenbach, D. O. Jung, and M. T. Madigan. 2000. Rhodobaca bogoriensis gen. nov. and sp. nov., an alkaliphilic purple nonsulfur bacterium from African Rift Valley soda lakes. Arch. Microbiol. 174:18-27. [DOI] [PubMed] [Google Scholar]
- 47.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 48.Mullins, T. D., T. B. Britschgi, R. L. Krest, and S. J. Giovannoni. 1995. Genetic comparisons reveal the same unknown bacterial lineages in Atlantic and Pacific bacterioplankton communities. Limnol. Oceanogr. 40:148-158. [Google Scholar]
- 49.Oremland, R. S., S. E. Hoeft, J. M. Santini, N. Bano, R. A. Hollibaugh, and J. T. Hollibaugh. 2002. Anaerobic oxidation of arsenite in Mono Lake water and by a facultative chemoautotroph, strain MLHE-1. Appl. Environ. Microbiol. 68:4795-4802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ovreas, L., L. Forney, F. L. Daae, and V. Torsvik. 1997. Distribution of bacterioplankton in meromictic Lake Saelenvannet, as determined by denaturing gradient gel electrophoresis of PCR-amplified gene fragments coding for 16S rRNA. Appl. Environ. Microbiol. 63:3367-3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paabo, S., and A. C. Wilson. 1988. Polymerase chain reaction revealing cloning artifacts. Nature 334:387-388. [DOI] [PubMed] [Google Scholar]
- 52.Paul, J. H., and B. Myers. 1982. Fluorometric determination of DNA in aquatic microorganisms by use of Hoechst 33258. Appl. Environ. Microbiol. 43:1393-1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pinhassi, J., F. Azam, J. Hempälä, R. A. Long, J. Martinez, U. L. Zweifel, and Å. Hagström. 1999. Coupling between bacterioplankton species composition, population dynamics, and organic matter degradation. Aquat. Microb. Ecol. 17:13-26. [Google Scholar]
- 54.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 55.Prowe, S. G., and G. Antranikian. 2001. Anaerobranca gottschalkii sp. nov., a novel thermoalkaliphilic bacterium that grows anaerobically at high pH and temperature. Int. J. Syst. E vol. Microbiol. 51:457-465. [DOI] [PubMed] [Google Scholar]
- 56.Reysenbach, A., L. J. Giver, G. S. Wickham, and N. Pace. 1992. Differential amplification of rRNA genes by polymerase chain reaction. Appl. Environ. Microbiol. 58:3417-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Riemann, L., G. F. Steward, and F. Azam. 2000. Dynamics of bacterial community composition and activity during a mesocosm diatom bloom. Appl. Environ. Microbiol. 66:578-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rijkenberg, M. J., R. Kort, and K. J. Hellingwerf. 2001. Alkalispirillum mobile gen. nov., sp. nov., an alkaliphilic non-phototrophic member of the Ectothiorhodospiraceae. Arch. Microbiol. 175:369-375. [DOI] [PubMed] [Google Scholar]
- 59.Sakano, Y., and L. Kerkhof. 1998. Assessment of changes in microbial community structure during operation of an ammonia biofilter with molecular tools. Appl. Environ. Microbiol. 64:4877-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorokin, D. Y., V. M. Gorlenko, T. P. Tourova, T. V. Kalganova, A. L. Tsapin, K. H. Nealson, and G. J. Kuenen. 2002. Thioalkalimicrobium cyclicum sp. nov., and Thioalkalivibrio jannaschii sp. nov., new species of haloalkaliphilic, obligately chemolithoautotrophic sulfur-oxidizng bacteria from hypersaline alkaline Mono Lake (California). Int. J. Syst. E vol. Microbiol. 52:913-920. [DOI] [PubMed] [Google Scholar]
- 62.Sorokin, D. Y., A. M. Lysenko, L. L. Mityushina, T. P. Tourova, B. E. Jones, F. A. Rainey, L. A. Robertson, and G. J. Kuenen. 2001. Thioalkalimicrobium aerophilum gen. nov., sp. nov., Thioalkalimicrobium sibericum sp. nov., Thioalkalivibrio versutus gen. nov., sp. nov., Thioalkalivibrio nitratis sp. nov., and Thioalkalivibrio denitrificans sp. nov., novel obligately alkaliphilic and obligately chemolithoautotrophic sulfur-oxidizing bacteria from soda lakes. Int. J. Syst. E vol. Microbiol. 51:565-580. [DOI] [PubMed] [Google Scholar]
- 63.Sorokin, D. Y., T. P. Tourova, A. M. Lysenko, and J. G. Kuenen. 2001. Microbial thiocyanate utilization under highly alkaline conditions. Appl. Environ. Microbiol. 67:528-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sorokin, D. Y., T. P. Turova, B. B. Kuznetsov, I. A. Briantseva, and V. M. Gorlenko. 2000. Roseinatronobacter thiooxidans gen. nov., sp. nov., a new alkaliphilic aerobic bacteriochlorophyll-alpha-containing bacteria from a soda lake. Mikrobiologiia 69:89-97. [PubMed] [Google Scholar]
- 65.Speksnijder, A. G. C. L., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stackebrandt, E., and B. M. Goebel. 1994. Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int. J. Syst. Bacteriol. 44:846-849. [Google Scholar]
- 67.Suzuki, M. T., and S. J. Giovannoni. 1996. Bias caused by template annealing in the amplification of mixtures of 16S rRNA genes by PCR. Appl. Environ. Microbiol. 62:625-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ward, B. B., D. P. Martino, M. C. Diaz, and S. B. Joye. 2000. Analysis of ammonia-oxidizing bacteria from hypersaline Mono Lake, California, on the basis of 16S rRNA sequences. Appl. Environ. Microbiol. 66:2873-2881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Weisenburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wintzingerode, F., U. B. Göbel, and E. Stackebrandt. 1997. Determination of microbial diversity in environmental samples: pitfalls of PCR-based rRNA analysis. FEMS Microbiol. Rev. 21:213-329. [DOI] [PubMed] [Google Scholar]
- 71.Yager, P. L., T. L. Connelly, B. B. Mortazavi, K. E. Wommack, N. Bano, J. Bauer, S. Opsahl, and J. T. Hollibaugh. 2001. Dynamic microbial response to an Arctic algal bloom at sub-zero temperatures. Limnol. Oceanogr. 46:790-801. [Google Scholar]
- 72.Yakimov, M. M., L. Giuliano, T. N. Chernikova, G. Gentile, W. R. Abraham, H. Lunsdorf, K. N. Timmis, and P. N. Golyshin. 2001. Alcalilimnicola halodurans gen. nov., sp. nov., an alkaliphilic, moderately halophilic and extremely halotolerant bacterium, isolated from sediments of soda-depositing Lake Natron, East Africa Rift Valley. Int. J. Syst. E vol. Microbiol. 51:2133-2143. [DOI] [PubMed] [Google Scholar]
- 73.Zehr, J. P., R. W. Harvey, R. S. Oremland, J. E. Cloern, L. H. George, and J. L. Lane. 1987. Big Soda Lake (Nevada). 1. Pelagic bacterial heterotrophy and biomass. Limnol. Oceanogr. 32:781-793. [Google Scholar]
- 74.Zhilina, T. N., G. A. Zavarzin, F. A. Rainey, V. V. Kevbrin, N. A. Kostrikina, and A. M. Lysenko. 1996. Spirochaeta alkalica sp. nov., Spirochaeta africana sp. nov., and Spirochaeta asiatica sp. nov., alkaliphilic anaerobes from the continental Soda Lakes in Central Asia and the East African Rift. Int. J. Syst. Bacteriol. 46:305-312. [DOI] [PubMed] [Google Scholar]
- 75.Zhilina, T. N., G. A. Zavarzin, F. A. Rainey, E. N. Pikuta, G. A. Osipov, and N. A. Kostrikina. 1997. Desulfonatronovibrio hydrogenovorans gen. nov., sp. nov., an alkaliphilic, sulfate-reducing bacterium. Int. J. Syst. Bacteriol. 47:144-149. [DOI] [PubMed] [Google Scholar]