Abstract
A novel enzyme with a specific phenylalanine aminopeptidase activity (ApsC) from Aspergillus niger (CBS 120.49) has been characterized. The derived amino acid sequence is not similar to any previously characterized aminopeptidase sequence but does share similarity with some mammalian acyl-peptide hydrolase sequences. ApsC was found to be most active towards phenylalanine β-naphthylamide (F-βNA) and phenylalanine para-nitroanilide (F-pNA), but it also displayed activity towards other amino acids with aromatic side chains coupled to βNA; other amino acids with nonaromatic side chains coupled to either pNA or βNA were not hydrolyzed or were poorly hydrolyzed. ApsC was not able to hydrolyze N-acetylalanine-pNA, a substrate for acyl-peptide hydrolases.
Many food products contain flavors obtained by the hydrolysis of proteins. These peptides and amino acids can taste sweet, sour, or bitter. Mixtures of endoproteases are often deliberately used in conjunction with exoproteases to improve these food flavors. Exopeptidases can reduce the amount of peptides with undesirable tastes through the removal of a single hydrophobic amino acid, or pairs of them, from the terminal ends. For example, phenylalanine-containing peptides taste >100-fold more bitter than does free phenylalanine (13, 14). Control and termination of the hydrolytic reaction are therefore crucial for obtaining hydrolysates with the desired organoleptic properties.
Enzymes from Aspergillus niger have been used in food production for several decades, and five different endoproteases (PepA to PepE [see reference 26 and references therein]), two carboxypeptidases (CpdI and PepF/CpdII [9, 24, 25]), and one aminopeptidase (ApsA [4]) have been cloned and characterized. Experiments have shown that a particular enzyme preparation of A. niger that contains a specific aminopeptidase activity can liberate phenylalanine from proteins that are present in dough and (semi)hard cheeses (8), thereby improving the flavor and aroma of these products. So far, only one phenylalanine-specific aminopeptidase has been characterized, namely, APF1 from the basidiomycetous fungus Schizophyllum commune (6). APF1 is an intracellular zinc metallo-aminopeptidase that can also hydrolyze an N-terminal tyrosine.
In this report, we describe a new aminopeptidase. The gene was cloned from A. niger, and purification and characterization of the gene product show that the gene encodes an aminopeptidase that specifically hydrolyzes amino-terminal phenylalanine and other amino acids with aromatic side groups.
MATERIALS AND METHODS
Strains and DNA and RNA techniques.
Escherichia coli DH5α and LE392 were used as hosts for recombinant plasmids and λ DNA, respectively. A. niger strains N402 (cspA1) and NW171 (cspA1 fwnA6 pyrA6 nicA1 pepA::argBnidΔA pepB::argBnidΔB pepE::argBnidΔE) (26) are derived from A. niger N400 (CBS 120.49). DNA and RNA manipulations were done as described previously (23). For Southern and Northern analyses of the transformants, the strains were grown in complete medium (minimal medium [MM] [20] supplemented with 2% [wt/vol] glucose, 0.2% [wt/vol] meat peptone, 0.1% [wt/vol] yeast extract, 0.1% [wt/vol] peptone 140, 0.03% [wt/vol] yeast RNAs, and either 0.15% [wt/vol] KH2PO4 or 1.5% [wt/vol] KH2PO4). To study the possible regulation of apsC messenger levels by the carbon and nitrogen source, strain N402 was grown on MM supplemented with either 1% glucose, 1% fructose, and 0.6% NH4Cl or 0.4% NaNO3 or on MM supplemented with 1% bovine serum albumin, 1% elastin, or 1% collagen. RNA was isolated from these cultures and subjected to Northern analysis.
Purification of extracellular ApsC from A. niger NRRL 3112 and peptide sequencing.
A. niger NRRL 3112 was grown in a medium containing 1.5% (wt/vol) potato flour, 2% (wt/vol) Bacto Peptone, 0.7% (wt/vol) yeast extract, 30 mM KH2PO4, 4.2 mM MgSO4, 4.5 mM CaCl2, and 3.7 mM ZnCl2 at pH 4.8 for 120 h at 30°C. Twenty milliliters of culture broth was harvested by filtration and transferred to 10 mM Tris (pH 7.5) over PD10 columns (Pharmacia, Wiesbaden, Germany). The desalted culture broth was fractionated by anion-exchange chromatography on a 1-ml Mono Q column. Bound protein was eluted by using a 25-ml linear gradient of 0 to 1 M NaCl in 10 mM Tris-HCl (pH 7.5). The fraction size was 1 ml. The fractions containing phenylalanine aminopeptidase (F-AP) activity were pooled and concentrated by ultrafiltration, desalted over a PD10 column, transferred to 10 mM Tris (pH 7.5), concentrated fivefold by ultrafiltration, and then applied to a preparative polyacrylamide gel (a 15% polyacrylamide gel was used for separation, and a 5% polyacrylamide gel was used for stacking). Two lanes were used for a zymogram to locate F-AP activity with an overlay of 0.9 mM phenylalanine in 7.5 mM HCl. The protein band showing F-AP activity was marked, and from the remaining untreated lanes, a protein band located at the same position was cut from the gel and subjected to standard (internal) amino acid sequencing procedures, which were performed at Eurosequence (Groningen, The Netherlands) as described previously (21).
Cloning of apsC.
Olignucleotide sequences of aspC from A. niger NRRL 3112 were obtained by reverse translation of the peptide sequences indicated by underlining. For peptide sequence VSWIQWN, forward primer Sap-1 (SNTGGATHCARTGGAAY) and reverse primer Sap-2 (RTTCCAYTGDATCCA) were used. For peptide sequenceWGPDGTLFFVSDR, forward primer Sap-3 (TGGGGNCCNGAYGGNAC) and reverse primer Sap-4 (GTNCCRTCNGGNCCCCA) were used. For peptide sequence AEPQTAPFGTWDSPIT, forward primer Sap-5 (GARCCICARACNGCICCNTT) was used. A standard PCR was performed with genomic DNA from A. niger NRRL 3112 by using equal amounts of forward and reverse primers and an annealing temperature of 54°C. The amplified product obtained with primers Sap-5 and Sap-4 was cloned in pGEM-T (Promega, Madison Wis.), followed by sequence analysis. The PCR product was used as a probe in the screening of a λ EMBL4 genomic library of A. niger N400 by standard methods to obtain the apsC gene (23). Nine phages were isolated, and from one positive phage a 5.5-kb EcoRI fragment and a partially overlapping 1.8-kb XhoI fragment were subcloned in pUC19 and sequenced over both strands. cDNA of apsC was generated by reverse transcription (RT)-PCR, by using the enhanced avian RT-PCR kit from Sigma (St. Louis, Mo.).
Protein and nucleotide sequence analyses were done with the program DNAstar (Lasergene, Madison, Wis.). The inferred ApsC protein sequence was used in a search of the public nonredundant protein and DNA databases with the BLASTP and TBLASTN algorithms, respectively, and a Blosum 62 substitution matrix (2). Sequences with expected values higher than 10 were ignored. Multiple alignments were made with Clustal X (15).
Plasmid construction, overexpression, and purification of aminopeptidase C.
The 5.5-kb EcoRI fragment and the partially overlapping 1.8-kb XhoI fragment were merged, resulting in pIM4103 (Fig. 1). Plasmid pIM4103 and plasmid pGW635 (12), which contains the A. niger pyrA gene, were used to cotransform A. niger NW171 according to the method described previously (17). PyrA+ transformants were screened for enhanced F-AP activity in cell extracts in 100 mM sodium-potassium phosphate buffer (PB) at pH 7.2. Strains with enhanced F-AP activity were subjected to Southern and Northern analyses to determine the copy numbers and expression levels.
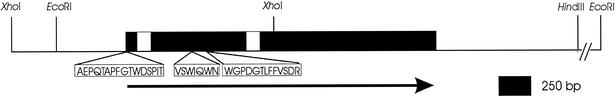
FIG. 1.
Architecture of the A. niger apsC gene. The positions of the restriction enzymes used in the cloning strategy and construction of pIM4103 are indicated. The ORF is represented by filled boxes, and an arrow indicates the direction of transcription. The positions of the two introns encountered in the ORF are indicated with open boxes. The positions in the ORF corresponding to the amino-terminally sequenced peptides are indicated.
Throughout the purification procedure, the fractions with F-AP activity were determined by using standard conditions and phenylalanine para-nitroanilide (F-pNA) as the substrate (see below). Multicopy strain Tr10 (transformant 10) was grown for 17 h in 400 ml of complete medium at 30°C. Mycelium was collected by filtration over nylon gauze, frozen in liquid nitrogen, and then ground in a Braun II dismembrator (B. Braun A.G., Melsungen, Germany). The ground mycelium was suspended in PB (pH 7.2) and stirred for 15 min at 4°C. Cellular debris was removed by centrifugation. This cell extract of Tr10 in PB was applied to a 15.5-ml Source 30Q column (Pharmacia Biotech) and equilibrated in 20 mM triethanolamine (TEA), pH 7.0, and bound protein was eluted by using a 124-ml linear gradient of 0 to 0.4 M NaCl in 20 mM TEA, pH 7.0. The fraction size was 5 ml. Active fractions 13 and 14 were diluted 10-fold in 20 mM Bis-Tris (pH 6.5) and separately loaded onto a 1-ml Resource Q column (Pharmacia Biotech) that was preequilibrated with 20 mM Bis-Tris (pH 6.5). The bound protein was then eluted by using a 20-ml linear gradient of 0 to 0.4 M NaCl in 20 mM Bis-Tris (pH 6.5). The fraction size was 1 ml.
Biochemical characterization of aminopeptidase C.
Protein concentrations were determined by the bicinchoninic acid method as described by the supplier (Sigma).
(i) Artificial substrates.
Aminopeptidase activity was determined as described previously (3), with different amino acids coupled to pNA as the substrate. Standard conditions were 1 min of incubation at 30°C with 1 mM amino acid-pNA substrate in 20 mM Na citrate (pH 5.2) at a final volume of 1 ml. Aminopeptidase activity was also determined as described previously (10), with amino acids coupled to β-naphthylamide (βNA) as the substrate. Standard conditions were then 1 min of incubation at 30°C with 1 mM βNA substrate in 20 mM Na citrate (pH 5.2) at a final volume of 1 ml. One unit of enzyme activity was defined as the amount of enzyme that produces 1 μmol of pNA or βNA min−1. L-pNA, P-pNA, R-pNA, F-pNA, A-pNA, M-pNA, K-pNA, W-βNA, H-βNA, N-βNA, S-βNA, L-βNA, and F-βNA were obtained from Sigma, and V-pNA, G-pNA, I-pNA, E-pNA, T-βNA, and Y-βNA were obtained from Bachem (Bubendorf, Switzerland).
(ii) Peptide substrates.
The liberated amino acids were detected by a method described previously (22). Standard conditions were 3 mM substrate in 20 mM Na citrate (pH 5.2). Phenylalanine and leucine concentrations were calculated from a standard curve. The peptides F-G-G, L-G-G, F-G-G-F, and F-L-E-E-I were obtained from Sigma. One unit of enzyme activity was defined as the amount of enzyme that produces 1 μmol of phenylalanine or leucine min−1. The optimal pH for enzymatic activity was determined by using McIlvaine buffers at pH values ranging from 4 to 8. The pH stability of aminopeptidase C was tested by preincubation of the purified enzyme in McIlvaine buffer with pH values ranging from 4 to 8 at 30°C for 90 min, followed by the standard enzyme reaction. The temperature stability of aminopeptidase C was tested by preincubation of the purified enzyme at 0, 30, 40, 50, and 60°C for 60 min in 100 mM PB (pH 7.2), which was followed by the standard enzyme reaction. The sample preincubated at 0°C was used as a reference to calculate the residual activity.
Nucleotide sequence accession number.
The nucleotide sequence of the apsC gene from A. niger and the encoded amino acid sequence have been deposited in the EMBL nucleotide database under accession number AJ316576.
RESULTS AND DISCUSSION
Cloning and analysis of A. niger apsC.
Since A. niger strain NRRL 3112 was used as a source for the F-AP hydrolyzing activity in an enzyme preparation described previously (8), culture broth of this strain was used to purify a protein with high activity towards F-pNA. The N-terminal sequences were determined for the N-terminal end of the mature protein and for two peptides of a tryptic digest thereof. A combination of forward primer Sap-5, based on the N-terminal peptide sequence AEPQTAPFGTWDSPIT, and reverse primer Sap-4, based on the peptide sequence WGPDGTLFFVSDR, was used to amplify a 751-bp genomic DNA fragment of A. niger NRRL 3112. The third peptide sequence, VSWIQWN, exactly matched an internal region of the inferred amino acid sequence translated from the amplified fragment.
A 5.5-kb EcoRI fragment and a partially overlapping 1.8-kb XhoI fragment of A. niger N400 which hybridized to the amplified fragment were subcloned and sequenced on both strands up to the HindIII site 3′ of the gene (Fig. 1). Sequence analysis showed that together these fragments harbor the complete putative aminopeptidase C gene (apsC) and 5′- and 3′-flanking regions. RT-PCR techniques were used to generate apsC cDNA, which was identical to the genomic DNA except for two intervening sequences in the open reading frame (ORF), one 111 bp and one 917 bp downstream from the start codon (Fig. 1). The previously determined N-terminal sequence of the protein starts at amino acid 5 of the inferred protein, indicating that no cleavable amino-terminal signal sequence is present in this protein. The loss of the first four amino acids is probably due to proteolysis. The NRRL 3112-derived N-terminal sequence differs by one amino acid (a Pro-to-Ala change at position 7) from the N400-derived amino acid sequence, which is probably due to strain differences.
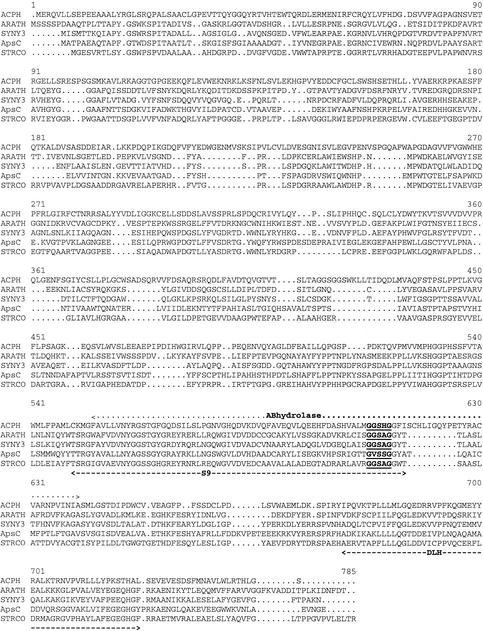
Public DNA and protein databases were searched for similarly characterized aminopeptidases, and none were found. The highest overall homology was found with some putative acylaminoacyl aminopeptidases, also referred to as acyl-peptide hydrolases (ACPHs), from Synechocystis sp. strain slr0825 (41% identity), Streptomyces coelicolor (37% identity), Caenorhabditis elegans (36% identity), and Arabidopsis thaliana (35% identity). None of these proteins, however, have been biochemically characterized. Lower scores were obtained with some genuine ACPHs from pigs, rats, and humans (all 15% identities) that catalyze the hydrolysis of an N-terminal acetylated peptide to release an N-acetylated amino acid. A comparison at the protein domain level shows that at the C-terminal end these mammalian ACPHs at least have a putative α/β-hydrolase fold domain, a putative S9-prolyloligopeptidase domain, and a putative dienelactone hydrolase domain (5). ApsC has a putative S9-prolyloligopeptidase domain and a putative dienelactone hydrolase domain at the C-terminal end (Fig. 2). In the multiple alignment, there are multiple insertions in the ACPH sequence, six of which are at least four amino acids long, that are not present in ApsC and the putative proteins of S. coelicolor, C. elegans, and A. thaliana. One of these insertions, starting at coordinate 616, is part of the α/β-hydrolase domain. In addition, there is one insertion of four amino acids, starting at position 514, which is present in ApsC and in the putative proteins but not in ACPH (Fig. 2). The putative active-site serine that is part of the conserved motif GXSXG (16, 18, 19) is also indicated in Fig. 2. As we show in the next section, ApsC is not an ACPH but is a novel aminopeptidase. Therefore, the putative protein sequences that cluster with ApsC more likely belong to a new subfamily of aminopeptidases.
FIG. 2.
Multiple alignment of A. niger ApsC and putative proteins from S. coelicolor (STRCO), a Synechocystis sp. (SYNY3), A. thaliana (ARATH), and human ACPH. Only the human ACPH is shown, since rat and pig ACPHs are more than 90% identical to human ACPH. The conserved S9-prolyloligopeptidase (S9) and dienelactone hydrolase (DLH) domains are indicated with dashed lines. The ACPH-specific α/β-hydrolase fold (ABhydrolase) domain is indicated with a dotted line. The putative active-site motif (GXSXG) is underlined and boldfaced.
Northern analysis of A. niger N402 grown on several carbon and nitrogen sources showed that the apsC messenger levels were independent of the carbon or nitrogen source used (results not shown). A. niger apsA transcript levels are also independent of the C and N source used (4). Furthermore, the yeast AapI- and ApeII-encoding genes also show constitutive expression.
Overexpression, purification, and characterization of the enzyme.
ApsC was overexpressed in A. niger strain NW171 by transformation with plasmid pIM4103 (Fig. 1). Twelve transformed strains were analyzed for the occurrence of multiple integrations of the plasmid in the genome. Southern analysis showed that Tr10 has the highest copy number (approximately 75 copies) of the integrated plasmid (results not shown). Scanning of Northern blots revealed that Tr10 had at least a 70-fold-higher messenger level of the correct size than the untransformed strain did (results not shown).
Although ApsC was purified from culture broth of strain NRRL 3112, we were not able to detect ApsC in culture broth of strain N402. However, we could find ApsC activity in cell extracts of this strain. Tr10 had a 50-fold-higher activity towards F-pNA than the untransformed strain did, i.e., 3.6 and 0.07 U mg−1, respectively. ApsC was purified from cell extracts of Tr10. This resulted in an enzyme preparation with a specific activity of 189 U mg−1 when F-pNA was used as the substrate (Table 1).
TABLE 1.
Purification of aminopeptidase C from A. niger Tr10
| Step | Total activity (U)a (103) | Total protein (mg) (103) | Sp act (U mg−1) | Purifi- cation (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract | 4.6 | 1.3 | 3.6 | 1 | 100 |
| Source 30Q | 1.4 | 0.9 | 15 | 4.2 | 31 |
| Resource Q | 0.42 | 0.0022 | 189 | 53 | 9 |
One unit of enzymatic activity is defined as the amount of enzyme that produces 1 μmol of pNA min−1.
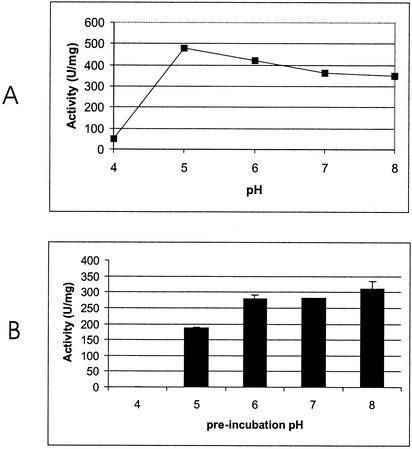
An analysis by sodium dodecyl sulfate-10% polyacrylamide gel electrophoresis of the active fractions revealed a protein band with an apparent molecular mass of 72 kDa, identical to the calculated molecular mass of the protein inferred from the amino acid sequence. The fraction with the highest activity was used for further characterization of the enzyme. The N-terminal sequence of the purified protein determined by Edman degradation exactly matched amino acids 5 to 16 of the predicted amino acid sequence. Although there is no secretion signal, ApsC is found both intra- and extracellularly in A. niger NRRL 3112. NRRL 3112 is a strain that was selected from a screen for high extracellular levels of F-AP activity. This strain probably shows more lysis than the N400 derivative strain used for overexpression under these circumstances. F-pNA was used as a substrate for the determination of the pH optimum, the pH stability, and the temperature stability of the purified enzyme. The optimal pH is 5, which is different from the pH optima reported for A. niger ApsA and other intracellular aminopeptidases, such as the yeast aminopeptidases which all have pH optima around 7.5 (1, 4, 7, 11).
The purified protein is stable between pH 5 and 8 (Fig. 3) for 90 min, with an optimal stability at pH 8. The purified protein is stable up to a temperature of 50°C for 60 min at pH 7.0. After 1 h at 60°C, 5% of the activity remained. ApsC specifically hydrolyzed not only F-pNA and F-βNA substrates but also other amino acids with aromatic side chains coupled to βNA. The activity towards amino acids with nonaromatic side chains was low (at least 16 times less) (Table 2). All ACPHs tested with the substrate N-acetylalanine-pNA could hydrolyze this substrate. ApsC, however, is not able to hydrolyze this substrate, and thus ApsC is not an ACPH. This apparent high specificity for amino acids with aromatic side chains was further demonstrated by the hydrolysis rate of peptides with phenylalanine or leucine at the N-terminal position. The peptides F-G-G, F-G-G-F, and F-L-E-E-I were hydrolyzed by ApsC at comparable rates (2.4 ± 0.049 [mean ± standard deviation], 2.8 ± 0.025, and 1.1 ± 0.028 U mg−1, respectively). In agreement with the results obtained with the artificial substrates, ApsC was not able to hydrolyze LGG, a peptide with an amino acid with a nonaromatic side chain at the N-terminal end.
FIG. 3.
Biochemical characterization of ApsC. (A) pH optimum of ApsC determined by using McIlvaine buffer. F-pNA was used as the substrate. (B) pH stability of ApsC. McIlvaine buffer was used as the preincubation buffer.
TABLE 2.
Substrate specificity of ApsC towards (N-acetylated) amino acid-pNA or amino acid-βNA substrates
| Substratea | Relative activity (%) (mean ± SD) |
|---|---|
| F-βNA | 100 ± 1.1 |
| Y-βNA | 72 ± 2.5 |
| W-βNA | 2 ± 0.77 |
| L-βNA | 6 ± 0.46 |
| Other amino acid-βNA | <1 |
| F-pNA | 100 ± 0.40 |
| L-pNA | 6 ± 0.32 |
| M-pNA | 6 ± 0.60 |
| NacA-pNA | <1 |
| Other amino acid-pNA | <1 |
One hundred percent F-pNA activity is 189 U mg−1. NacA, N-acetylalanine, Other pNA substrates tested include A-pNA, K-pNA, R-pNA, G-pNA, I-pNA, V-pNA, and E-pNA; other βNA substrates tested include N-βNA, T-βNA, S-βNA, and H-βNA.
In this report, a novel aminopeptidase that is specific for amino acids with aromatic side chains has been described. This aminopeptidase is most active at a slightly acidic pH and therefore might be used in the controlled preparation of cheese, thereby improving flavor and aroma by reducing the content of bitter-tasting phenylalanine- and tyrosine-containing peptides. Further research will focus on the in vivo role of ApsC.
Acknowledgments
This work was financially supported by DSM Food Specialties.
REFERENCES
- 1.Achstetter, T., C. Ehmann, and D. H. Wolf. 1983. Proteolysis in eucaryotic cells: aminopeptidases and dipeptidyl aminopeptidases of yeast revisited. Arch. Biochem. Biophys. 226:292-305. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atlan, D., C. Gilbert, B. Blanc, and R. Portalier. 1994. Cloning, sequencing and characterization of the pepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397. Microbiology 140:527-535. [DOI] [PubMed] [Google Scholar]
- 4.Basten, D. E. J. W., J. Visser, and P. J. Schaap. 2001. Lysine aminopeptidase of Aspergillus niger. Microbiology 147:2045-2050. [DOI] [PubMed] [Google Scholar]
- 5.Bateman, A., E. Birney, R. Durbin, S. R. Eddy, R. D. Finn, and E. L. L. Sonnhammer. 1999. Pfam 3.1: 1313 multiple alignments and profile HMMs match the majority of proteins. Nucleic Acids Res. 27:260-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilbrey, R. E., A. R. Penheiter, A. C. Gathman, and W. W. Lilly. 1996. Characterization of a novel phenylalanine-specific aminopeptidase from Schizophyllum commune. Mycol. Res. 100:462-466. [Google Scholar]
- 7.Caprioglio, D. R., C. Padilla, and M. Werner-Washburne. 1993. Isolation and characterization of AAP1, a gene encoding an alanine/arginine aminopeptidase in yeast. J. Biol. Chem. 268:14310-14315. [PubMed] [Google Scholar]
- 8.Coenen, T. M. M., and P. Aughton. 1998. Safety evaluation of amino peptidase enzyme derived from Aspergillus niger. Food Chem. Toxicol. 36:781-789. [DOI] [PubMed] [Google Scholar]
- 9.Dal Degan, F., B. Ribadeau-Dumas, and K. Breddam. 1992. Purification and characterization of two serine carboxypeptidases from Aspergillus niger and their use in C-terminal sequencing of proteins and peptide synthesis. Appl. Environ. Microbiol. 58:2144-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Frey, J., and K.-H. Röhm. 1979. External and internal forms of yeast aminopeptidase II. Eur. J. Biochem. 97:169-173. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Alvarez, N., R. Cueva, and P. Suarez-Rendueles. 1991. Molecular cloning of soluble aminopeptidases from Saccharomyces cerevisiae. Sequence analysis of aminopeptidase yscII, a putative zinc-metallopeptidase. Eur. J. Biochem. 202:993-1002. [DOI] [PubMed] [Google Scholar]
- 12.Goosen, T., F. van Engelenburg, F. Debets, K. Swart, K. Bos, and H. van den Broek. 1989. Tryptophan auxotrophic mutants in Aspergillus niger: inactivation of the trpC gene by cotransformation mutagenesis. Mol. Gen. Genet. 219:282-288. [DOI] [PubMed] [Google Scholar]
- 13.Ishibashi, N., I. Ono, K. Kato, T. Shigenaga, I. Shinoda, H. Okai, and S. Fukui. 1988. Role of the hydrophobic amino acid residue in the bitterness of peptides. Agric. Biol. Chem. 52:91-94. [Google Scholar]
- 14.Ishibashi, N., K. Sadamori, O. Yamamoto, H. Kanehisa, K. Kouge, E. Kikuchi, H. Okai, and S. Fukui. 1987. Bitterness of phenylalanine- and tyrosine-containing peptides. Agric. Biol. Chem. 51:3309-3313. [Google Scholar]
- 15.Jeanmougin, F., J. D. Thompson, M. Gouy, D. G. Higgens, and T. J. Gibson. 1998. Multiple sequence alignment with Clustal X. Trends Biochem. Sci. 23:403-405. [DOI] [PubMed] [Google Scholar]
- 16.Kanatani, A., T. Yoshimoto, A. Kitazono, T. Kokubo, and S. Tsuru. 1993. Prolyl endopeptidase from Aeromonas hydrophila: cloning, sequencing and expression of the enzyme gene, and characterization of the expressed enzyme. J. Biochem. 113:790-796. [DOI] [PubMed] [Google Scholar]
- 17.Kusters-van-Someren, M. A., J. A. M. Harmsen, H. C. M. Kester, and J. Visser. 1991. Structure of the Aspergillus pelA gene and its expression in Aspergillus niger and Aspergillus nidulans. Curr. Genet. 20:293-299. [DOI] [PubMed] [Google Scholar]
- 18.Mitta, M., M. Miyagi, I. Kato, and S. Tsunasawa. 1998. Identification of the catalytic triad residues of porcine liver acylamino acid-releasing enzyme. J. Biochem. 123:924-931. [DOI] [PubMed] [Google Scholar]
- 19.Polgar, L. 1992. Structural relationship between lipases and peptidases of the prolyl oligopeptidase family. FEBS Lett. 311:281-284. [DOI] [PubMed] [Google Scholar]
- 20.Pontecorvo, G., J. A. Roper, L. J. Hemmons, K. D. MacDonald, and A. W. J. Bufton. 1953. The genetics of Aspergillus nidulans. Adv. Genet. 5:141-238. [DOI] [PubMed]
- 21.Rosenfeld, J., J. Capdevielle, J. C. Guillemot, and P. Ferrara. 1992. In-gel digestion of proteins for internal sequence analysis after one- or two-dimensional gel electrophoresis. Anal. Biochem. 203:173-179. [DOI] [PubMed] [Google Scholar]
- 22.Roth, M. 1971. Fluorescence reaction for amino acids. Anal. Chem. 43:880-882. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 24.Svendsen, I., and F. Dal Degan. 1998. The amino acid sequences of carboxypeptidases I and II from Aspergillus niger and their stability in the presence of divalent cations. Biochim. Biophys. Acta 1387:369-377. [DOI] [PubMed] [Google Scholar]
- 25.van den Hombergh, J. P., G. Jarai, F. P. Buxton, and J. Visser. 1994. Cloning, characterization, and expression of pepF, a gene encoding a serine carboxypeptidase from Aspergillus niger. Gene 151:73-79. [DOI] [PubMed] [Google Scholar]
- 26.van den Hombergh, J. P., P. J. I. van de Vondervoort, L. Fraissinet-Tachet, and J. Visser. 1997. Aspergillus as a host for heterologous protein production: the problem of proteases. Trends Biotechnol. 15:256-263. [DOI] [PubMed] [Google Scholar]