Abstract
Direct evidence that marine cyanobacteria take up organic nitrogen compounds in situ at high rates is reported. About 33% of the total bacterioplankton turnover of amino acids, determined with a representative [35S]methionine precursor and flow sorting, can be assigned to Prochlorococcus spp. and 3% can be assigned to Synechococcus spp. in the oligotrophic and mesotrophic parts of the Arabian Sea, respectively. This finding may provide a mechanism for Prochlorococcus' competitive dominance over both strictly autotrophic algae and other bacteria in oligotrophic regions sustained by nutrient remineralization via a microbial loop.
Oxygenic phototrophic cyanobacteria (6, 11, 28) have been shown to dominate the tropical and subtropical regions of the world's oceans, thereby changing our conception of oceanic ecosystems. The cyanobacterial genus Prochlorococcus dominates phytoplankton in the central oceanic gyres, while Synechococcus spp. can become very abundant in nutrient-rich tropical regions (20, 24). Why these organisms dominate such waters remains to be understood. To explain the ecological success of cyanobacteria in these waters, we propose the hypothesis that in these oceanic ecosystems, where nutrient regeneration within a microbial loop (1, 2) predominates over nitrate production, cyanobacteria can use not only the inorganic nutrients NH4+, NO3−, and NO2− (16, 21) but also organic compounds containing reduced nitrogen.
Although cyanobacteria are unable to incorporate some organic compounds, e.g., thymidine (10), Synechococcus axenic cultures utilize urea (7, 21) and amino acids (5, 19, 23) at a low rate and even show aminopeptidase activity (18). However, Synechococcus photoheterotrophy has always been considered ecologically unimportant. Current knowledge about Prochloroccocus nutrient assimilation (24) is limited, partly because of the existence of only one axenic Prochloroccocus culture (26). Interestingly, this strain (PCC9511) was isolated in the presence of an amino acid, methionine. It was reported that Prochlorococcus cultures take up NH4+ but that they cannot utilize NO3− or NO2−; they can also use urea but not other sources of organic nitrogen (21, 26), although amino acids were not tested as a sole nitrogen source.
Genomic data from the draft annotations of the MIT9313 and MED4 strains of Prochlorococcus spp. show that they possess several transporter systems for amino acids, as shown on the Department of Energy Joint Genome Institute website (http://www.jgi.doe.gov); however, a further analysis is necessary for the prediction of possible utilization pathways. The genome of the MED4 strain lacks the nitrate and nitrite reductase genes, a further proof that this strain needs to rely on reduced nitrogen compounds, e.g., NH4+, amino acids, etc.
In working with laboratory cultures of cyanobacteria, it is very challenging to simulate oceanic oligotrophic conditions and the results of laboratory nutrient addition experiments can be extrapolated only with considerable caution. On the other hand, studies done with natural microbial communities may reveal roles of dominant groups, including cyanobacteria, more directly and unambiguously. Therefore, we chose a field study in which the 35S-labeled methionine tracer was used as a proxy for labile nitrogen-containing compounds. The present study is probably the first that demonstrates the role of Prochlorococcus in the consumption of organic, nitrogen-containing compounds in the oligotrophic oceanic ecosystem.
Sampling site.
The hypothesis was tested aboard the Royal Research Ship Charles Darwin (cruise 132) in September 2001 in a field study of bacterioplankton in the Arabian Sea by using a transect of seven stations from the equator to 24°N along the 67°E meridian. Seawater was collected with a rosette of 24 30-liter Niskin bottles mounted on a conductivity temperature density profiler with additional fluorescence and oxygen detectors. Up to 24 depths were sampled in the top 300 m of the water column, with the highest sampling resolution being in the surface mixed layer and around the deep chlorophyll maximum. The bacterial abundance was determined at all depths sampled. The rates of the total microbial consumption of amino acids were measured in water samples collected from four to eight depths. Thirty samples, used for group flow sorting of bacteria labeled with [35S]methionine], were collected from two to five depths in the surface mixed layer, the deep chlorophyll maximum (down to 65 m), and deeper waters.
Consumption of labeled amino acids by total bacterioplankton.
The water samples used for bulk rate measurements were initially collected into acid-washed 1-liter thermos flasks by using acid-soaked silicone tubing and processed within 1 h after the sampling. Two differently labeled amino acids, [3H]leucine, [14C]leucine, and [35S]methionine (Amersham Biosciences, Little Chalfont, United Kingdom), were used as precursors. The [35S]methionine and [3H]leucine were added at ∼1 and 5 nM final concentrations, respectively, to determine the approximate in situ rates of amino acid turnover. The [14C]leucine was added at 20 nM final concentrations to determine the rates of uptake at saturated precursor concentrations.
The samples (1.6 ml) were inoculated with l-[4,5-3H]leucine (specific activity, 171 Ci/mmol) at a 5 nM final concentration or l-[35S]methionine (specific activity, >1,000 Ci/mmol) at an ∼1 nM final concentration and incubated in the dark at the in situ temperatures. The subsamples (0.5 ml) were fixed with 1% paraformaldehyde (PFA) at 0.5, 1, and 1.5 h. The samples (33 ml) were inoculated with l-[U-14C]leucine (295 mCi/mmol) at a 20 nM final concentration. The subsamples (10 ml) were fixed at 0.5, 1.0, and 1.5 h by being mixed with an equal volume of 10% trichloroacetic acid (TCA). The sample particulate materials were harvested onto 0.2-μm-pore-size nylon filters (Pall Corporation, Ann Arbor, Mich.). Radioactivity retained on the filters was measured as disintegrations per minute with a RackBeta 1409 liquid scintillation counter (LKB-Wallac, Turku, Finland). The rate of precursor incorporation was calculated as the slope of the linear regression of radioactivity against the incubation time (r2 > 0.96, P < 0.0001) (Fig. 1a). The time series experiments showed that the bacterioplankton had low nonspecific absorption of precursors. Uptake or incorporation rate constants, i.e., fractions of the added precursors consumed by the bacterioplankton per day, were used for rate comparison.
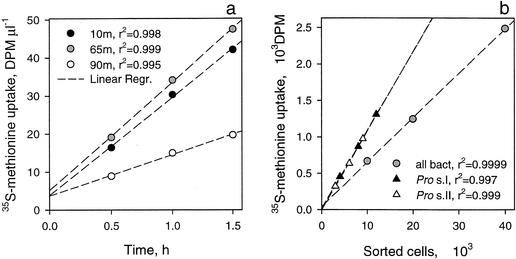
FIG. 1.
[35S]methionine uptake by total bacterioplankton collected at different depths and by flow cytometrically sorted general bacterial cells (all bact) and Prochlorococcus (Pro). Prochlorococcus was sorted from the replicated fixed samples on two successive days (s.I and s.II). (a) Time series; (b) sorted cell number series. Dashed lines show linear regressions (Regr.), and corresponding regression coefficients (r2) are presented in the keys. DPM, disintegrations per minute.
Fixation of the bacterioplankton samples with PFA, which precluded specific protein precipitation, was necessary for flow cytometric sorting to avoid cell loss. However, the different fixation procedures used for protein precipitation (5% TCA) and bacterioplankton sorting (1% PFA) should have had little effect on the determined rates (32, 34).
Flow cytometry and flow sorting of bacterioplankton.
Cyanobacteria were enumerated in freshly collected unstained samples by using their specific chlorophyll and phycoerythrin autofluorescence (6, 28). The abundance of bacterioplankton was determined with a FACSort flow cytometer (Becton Dickinson, Oxford, United Kingdom) after they were stained with SYBR Green I DNA dye (17) in unfrozen samples on board the ship and in frozen samples on the return. For flow cytometric analyses, replicated 2-ml samples were fixed with 1% PFA and stored at 2°C for 24 h to enhance cell fixation. Subsequently, the samples were kept at −20°C. Yellow-green beads with a 0.5-μm diameter (Fluoresbrite Microparticles; Polysciences, Warrington, Pa.) were used in all analyses of fixed samples as an internal standard.
The metabolic activities of bacterioplankton groups, including cyanobacteria, were determined by using the consumption rate of [35S]methionine (34). Methionine was added at an ∼1 nM concentration to determine approximate in situ rates of amino acid turnover. Four replicated samples were incubated at in situ temperature in the dark, fixed with 1% PFA after 3 h of incubation, and kept frozen. Thawed samples were stained with SYBR Green I (see above), and visualized bacterioplankton groups and whole communities were flow sorted by using FACSort and FACSCalibur flow cytometers (Becton Dickinson), both of which were set to single-cell sort mode at a sorting rate of 10 to 150 particles s−1. Deionized water (18 MΩ) was used as a sheath fluid to enhance the washing of sorted PFA-fixed cells. Cyanobacteria were identified in the stained samples by using their red autofluorescence. Sorted cells were collected onto 0.2-μm-pore-size nylon filters, washed with deionized water, and radioassayed (see above). Three proportional numbers of the cells (e.g., 1 × 103, 2 × 103, and 3 × 103 to 10 × 103, 20 × 103, and 30 × 103 cells, respectively) were sorted, and the cellular metabolic activities were determined as the slopes of the linear regressions of radioactivity against the numbers of sorted bacteria (Fig. 1b). The reproducibility of gate sorting was checked with six samples, which were sorted in duplicate on different days or by using different instruments (Fig. 1b). The determined [35S]methionine cellular uptake correlated with a 1:1 relationship (r2 = 0.99, P < 0.0001, n = 6).
To confirm cyanobacterial identification, several thousand SYBR Green I-stained cells from cyanobacterial flow cytometric clusters in two pooled samples representative of the surface mixed layer of the two main regions studied were sorted for molecular analysis with a FACStar Plus flow cytometer (Becton Dickinson, San Jose, Calif.) (9). The 16S rRNA genes of the cyanobacterial cell samples were amplified by using PCR with a pair of general bacterial primers, 8F and 1492R (22). A 16S rDNA clone library was generated by using standard molecular techniques (33). Sequence data were analyzed by using the ARB software package (http://www.arb.home.de).
Methodological aspects.
To test the suitability of methionine for quantifying bacterioplankton amino acid turnover in the Arabian Sea, we compared the microbial turnover rate constants of [35S]methionine (∼1 nM) and [3H]leucine (5 nM) and the rates of [14C]leucine (20 nM) incorporation into TCA-insoluble material, i.e., proteins. The correlation between the turnover rate constants of [35S]methionine and [3H]leucine was very strong (slope, 1.01 ± 0.06; n = 11; r2 = 0.96; P < 0.0001). By adding 20 nM [14C]leucine, we deliberately elevated the ambient amino acid concentration significantly; nevertheless, the rates of [35S]methionine turnover strongly correlated with the corresponding estimates of leucine incorporation (slope, 0.46 ± 0.03; n = 25; r2 = 0.92; P = <0.0001). The turnover rate constant of methionine was about twice that for leucine at 20 nM and similar to that for leucine at 5 nM. Therefore, both amino acids were most likely taken up similarly, and [35S]methionine and [3H]leucine were added, if not at tracer concentrations, then at least at subsaturating concentrations. The strong correlation between methionine and leucine turnover rate constants by bacterioplankton agrees well with the previous observations of marine (34) and limnic (12) bacterioplankton. Therefore, we could conclude that [35S]methionine was a representative amino acid for estimating rates of bacterioplankton amino acid turnover.
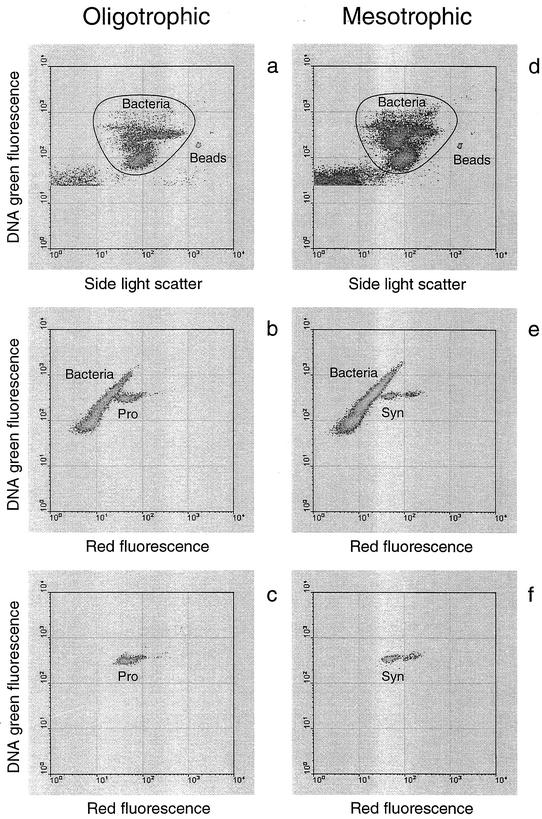
We flow-sorted cyanobacteria and other distinct cytometric groups of bacterioplankton to determine their cellular specific activities. Both Prochlorococcus and Synechococcus cyanobacteria were easily discriminated by flow cytometry from other DNA-stained bacterioplankton by using their natural red fluorescent pigments (Fig. 2). A comparison of direct counts of unstained and stained cyanobacteria showed very close agreement (slope, 0.94 ± 0.06; n = 15; r2 = 0.95; P < 0.0001). Because of the very low chlorophyll content of the cells in the surface 30 to 50 m, in six surface samples the Prochlorococcus cluster could not be clearly separated from other bacterioplankton. In such cases, we sorted only that part of the cyanobacterial cluster with clearly defined brighter-red fluorescence. To verify the accuracy of sorting, we used the determined activities and proportions of the total number of bacteria in each of the four to five groups (including the cyanobacterial group) that were sorted to determine the sum of activities of what should be an average bacterioplankton cell. These sum activities were compared with the directly measured mean activities of an average bacterioplankton cell. The latter were determined by sorting cells from the whole bacterioplankton supercluster (polygon) (Fig. 2a and d). The correlation between the mean activities and the sum activities was very strong (slope, 0.99 ± 0.03; n = 28; r2 = 0.98; P < 0.0001) and reconfirmed that group sorting was strictly targeted.
FIG. 2.
Characteristic flow cytometric signatures of natural bacterioplankton in oligotrophic (a to c) and mesotrophic (d to f) regions of the Arabian Sea. Clearly revealed bacterioplankton cells stained for DNA were gated by the polygons (a and d) for radiolabel sorting to determine [35S]methionine uptake by an average bacterioplankton cell. The bacterial cells within the polygons were plotted separately to show Prochlorococcus (Pro) (b) and Synechococcus (Syn) (e) clusters with extra red fluorescence. The clusters of Prochlorococcus (c) and Synechococcus (f) sorted for radiolabel analyses were plotted separately.
Ecological aspects.
Molecular analysis of the 16S rRNA genes of sorted bacterioplankton cells with bright-red fluorescence confirmed the flow cytometric identification of cyanobacteria. At the oligotrophic station, only sequences related to high-light-adapted Prochlorococcus clade II (29, 30) could be found, whereas at the mesotrophic sampling site, only members of Synechococcus group A were found (27).
Prochlorococcus spp. were very abundant in waters from the equator to 8°N [(3.0 ± 1.6) × 108 cells liter−1] and were later replaced at the northern end of the transect by Synechococcus spp. [(1.0 ± 0.5) × 108 cells liter−1 ]. The clear switching of dominance between these two taxa coincided with a transition from the oligotrophic region, characterized by the primary production of 180 ± 16 mg of C m−2 day−1 (number of samples, 4) to the mesotrophic region with primary production of 330 ± 56 mg of C m−2 day−1 (number of samples, 4) (G. R. C. Morgan and A. P. Rees, personal communication). In the oligotrophic region, Prochlorococcus comprised 96.5% ± 3% of all cyanobacteria, while Synechococcus clearly dominated the mesotrophic region, accounting for 97% ± 5% of all cyanobacteria.
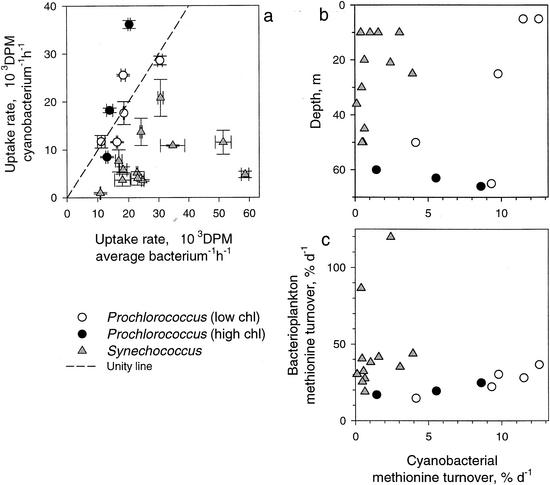
We found that both Prochlorococcus and Synechococcus cells incorporated the precursor at a rate comparable to the rates of average bacterioplankton cells (Fig. 3a). The mean cellular specific activity of a Prochlorococcus cell was 110% of the activity of an average bacterial cell in the oligotrophic region, while the mean activity of a Synechococcus cell was only 30% of the activity of an average bacterial cell in the mesotrophic region. The methionine turnover rate constants of both cyanobacteria suggested that these might decrease with depth (Fig. 3b).
FIG. 3.
Comparison of [35S]methionine uptake rates of flow-sorted Prochlorococcus and Synechococcus cells with uptake rates of average bacterioplankton cells (a) as well as vertical distributions of cyanobacterial methionine turnover (b) and its comparison with total bacterioplankton turnover (c). The uptake rates of average bacterioplankton cells were determined by flow sorting of the whole bacterioplankton community. Error bars indicate a single standard error of three measurements. Dashed line indicates a unity line. d, days. Data derived from Prochlorococcus sorted from the partially resolved clusters are shown by open circles and from the complete clusters by filled circles. DPM, disintegrations per minute.
Although both Prochlorococcus and Synechococcus cyanobacteria were able to incorporate dissolved amino acids at close to natural concentrations, the ecological significance of this differed widely between the two cyanobacterial genera (Fig. 3). In the mesotrophic region, Synechococcus consumed on average only 1.5% of the dissolved methionine pool per day, compared to an average 46% daily turnover by the other bacterioplankton; in other words, Synechococcus was responsible for only 3% of methionine turnover by all bacterioplankton (Fig. 3c). Conversely, in oligotrophic waters Prochlorococcus turned over on average 8% of the methionine pool daily, compared to 16% turned over daily by the other bacterioplankton, or 33% of the methionine turnover by all bacterioplankton, showing that it could compete directly with other bacterioplankton populations. The competitive success of Prochlorococcus was spectacular, since it was able to consume one-third of the amino acid pool and represent one-third of the abundance of the total bacterioplankton community. Considering that the contribution of Prochlorococcus to total phytoplankton production could be as high as 50% (15), Prochlorococcus is undoubtedly the key algal species in oceanic oligotrophic ecosystems.
Assuming that in the oligotrophic region Prochlorococcus contributed about 50% (15) of the total phytoplankton primary production of 180 mg of C m−2 day−1, and with a 6.6 C/N molar ratio (25), the Prochlorococcus population nitrogen demand should be about 1.1 mM N m−2 day−1. The leucine-derived bacterioplankton production was estimated to be 25 ± 9 mg of C m−2 day−1(number of samples, 4) in the photic zone of the oligotrophic region (M. V. Zubkov, unpublished data), to which Prochlorococcus contributed about 30% or 0.1 mM N m−2 day−1. Therefore, the dissolved amino acids, which are an energetically preferential nitrogen source, were estimated to satisfy about 10% of the Prochlorococcus requirements for nitrogen, while ammonium may have satisfied the remainder of their nitrogen requirements. By consuming amino acids, Prochlorococcus deprived other bacteria of an important source of organic nitrogen, forcing the latter to use inorganic nitrogen at a higher energetic cost in the organic-carbon-limited oligotrophic environment.
Such strong competition for nutrients may be the driving force behind a shift of photoheterotrophic bacteria towards photosynthesis (4, 14). It was reported that the contribution of these bacteria to total photosynthesis rises from about 1% in nutrient-rich regions to 10% in oligotrophic regions (13).
The presented results may help to explain the current controversy in balancing the carbon cycle in the oligotrophic central regions of the ocean with low primary production (31), where community respiration may exceed photosynthesis (8). The dominance of Prochlorococcus, which consumes organic as well as inorganic nitrogen pools, may generate about a 30% overestimation of bacterial secondary production and hence of computed organic matter decomposition and respiration in oligotrophic waters.
These findings suggest that the classical clear distinction between auto- and heterotrophic microorganisms in the ocean is actually rather blurred, that, at least in very oligotrophic waters, heterotrophic bacteria can use photosynthesis (3, 14), and, conversely, that photosynthetic cyanobacteria can take up key nutrients heterotrophically. When modeling oceanic ecosystems, one should be aware that we know little about some of the key players in the microbial food web and that one ought to be cautious about one's conclusions, in particular with respect to the ocean's heterotrophic components.
Nucleotide sequence accession numbers.
The sequences of five clones belonging to the Prochlorococcus clade HLII (clones A314010, A314905, A314006, A314907, and A314908) and 27 clones related to the Synechococcus group A (clones A31501 to A31504, A31506 to A31508, A315010, A315011, A315020, A315023 to A315026, A71501 to A71506, A71509, A715011, and A715812 to A715816) will appear in the EMBL nucleotide sequence database under the accession numbers AY125355 to AY125386.
Acknowledgments
We gratefully acknowledge the captain, officers, and crew aboard the Royal Research Ship Charles Darwin for their help during cruise CD132. Many thanks go to Frank-Oliver Glöckner and Marga Bauer for help with the Prochlorococcus genome data and to two anonymous reviewers for their constructive criticisms of an earlier version of the manuscript.
This work forms part of the Marine and Freshwater Microbial Biodiversity program (NER/T/S/2000/00635) and was supported by the Natural Environment Research Council (NERC), Swindon, United Kingdom, and the Max Planck Society, Bremen, Germany. The research of M.V.Z. was supported by an NERC advanced research fellowship (NER/I/S/2000/00898).
REFERENCES
- 1.Andersson, A., C. Lee, F. Azam, and A. Hagstrom. 1985. Release of amino-acids and inorganic nutrients by heterotrophic marine microflagellates. Mar. Ecol. Prog. Ser. 23:99-106. [Google Scholar]
- 2.Azam, F., T. Fenchel, J. G. Field, J. S. Gray, L. A. Meyer-Reil, and F. Thingstad. 1983. The ecological role of water-column microbes in the sea. Mar. Ecol. Prog. Ser. 10:257-263. [Google Scholar]
- 3.Beja, O., L. Aravind, E. V. Koonin, M. T. Suzuki, A. Hadd, L. P. Nguyen, S. Jovanovich, C. M. Gates, R. A. Feldman, J. L. Spudich, E. N. Spudich, and E. F. DeLong. 2000. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science 289:1902-1906. [DOI] [PubMed] [Google Scholar]
- 4.Beja, O., M. T. Suzuki, J. F. Heidelberg, W. C. Nelson, C. M. Preston, T. Hamada, J. A. Eisen, C. M. Fraser, and E. F. DeLong. 2002. Unsuspected diversity among marine aerobic anoxygenic phototrophs. Nature 415:630-633. [DOI] [PubMed] [Google Scholar]
- 5.Chen, T. H., T. L. Chen, L. M. Hung, and T. C. Huang. 1991. Circadian-rhythm in amino-acid-uptake by Synechococcus Rf-1. Plant Physiol. 97:55-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisholm, S. W., R. J. Olson, E. R. Zettler, R. Goericke, J. B. Waterbury, and N. A. Welschmeyer. 1988. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature 334:340-343. [Google Scholar]
- 7.Collier, J. L., B. Brahamsha, and B. Palenik. 1999. The marine cyanobacterium Synechococcus sp. WH7805 requires urease (urea amidohydrolase, EC 3.5.1.5) to utilize urea as a nitrogen source: molecular-genetic and biochemical analysis of the enzyme. Microbiology 145:447-459. [DOI] [PubMed] [Google Scholar]
- 8.Del Giorgio, P. A., J. J. Cole, and A. Cimbleris. 1997. Respiration rates in bacteria exceed phytoplankton production in unproductive aquatic systems. Nature 385:148-151. [Google Scholar]
- 9.Fuchs, B. M., M. V. Zubkov, K. Sahm, P. H. Burkill, and R. Amann. 2000. Changes in community composition during dilution cultures of marine bacterioplankton as assessed by flow cytometric and molecular biological techniques. Environ. Microbiol. 2:191-202. [DOI] [PubMed] [Google Scholar]
- 10.Fuhrman, J. A., and F. Azam. 1982. Thymidine incorporation as a measure of heterotrophic bacterioplankton production in marine surface waters: evaluation and field results. Mar. Biol. 66:109-120. [Google Scholar]
- 11.Johnson, P. W., and J. M. Sieburth. 1979. Chroococcoid cyanobacteria in the sea: a ubiquitous and diverse phototrophic biomass. Limnol. Oceanogr. 24:928-935. [Google Scholar]
- 12.Kirchman, D. L., S. Y. Newell, and R. E. Hodson. 1986. Incorporation versus biosynthesis of leucine—implications for measuring rates of protein-synthesis and biomass production by bacteria in marine systems. Mar. Ecol. Prog. Ser. 32:47-59. [Google Scholar]
- 13.Kolber, Z. S., F. G. Plumley, A. S. Lang, J. T. Beatty, R. E. Blankenship, C. L. VanDover, C. Vetriani, M. Koblizek, C. Rathgeber, and P. G. Falkowski. 2001. Contribution of aerobic photoheterotrophic bacteria to the carbon cycle in the ocean. Science 292:2492-2495. [DOI] [PubMed] [Google Scholar]
- 14.Kolber, Z. S., C. L. Van Dover, R. A. Niederman, and P. G. Falkowski. 2000. Bacterial photosynthesis in surface waters of the open ocean. Nature 407:177-179. [DOI] [PubMed] [Google Scholar]
- 15.Li, W. K. W. 1994. Primary production of prochlorophytes, cyanobacteria, and eukaryotic ultraphytoplankton: measurements from flow cytometric sorting. Limnol. Oceanogr. 39:169-175. [Google Scholar]
- 16.Lindell, D., and A. F. Post. 2001. Ecological aspects of ntcA gene expression and its use as an indicator of the nitrogen status of marine Synechococcus spp. Appl. Environ. Microbiol. 67:3340-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marie, D., F. Partensky, S. Jacquet, and D. Vaulot. 1997. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl. Environ. Microbiol. 63:186-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez, J., and F. Azam. 1993. Aminopeptidase activity in marine chroococcoid cyanobacteria. Appl. Environ. Microbiol. 59:3701-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montesinos, M. L., A. Herrero, and E. Flores. 1997. Amino acid transport in taxonomically diverse cyanobacteria and identification of two genes encoding elements of a neutral amino acid permease putatively involved in recapture of leaked hydrophobic amino acids. J. Bacteriol. 179:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moore, L. R., R. Goericke, and S. W. Chisholm. 1995. Comparative physiology of Synechococcus and Prochlorococcus: influence of light and temperature on growth, pigments, fluorescence, and absorptive properties. Mar. Ecol. Prog. Ser. 116:259-275. [Google Scholar]
- 21.Moore, L. R., A. F. Post, G. Rocap, and S. W. Chisholm. 2002. Utilization of different nitrogen sources by the marine cyanobacteria Prochlorococcus and Synechococcus. Limnol. Oceanogr. 47:989-996. [Google Scholar]
- 22.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gradient gel electrophoresis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 23.Paerl, H. W. 1991. Ecophysiological and trophic implications of light-stimulated amino acid utilization in marine picoplankton. Appl. Environ. Microbiol. 57:473-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partensky, F., W. R. Hess, and D. Vaulot. 1999. Prochlorococcus, a marine photosynthetic prokaryote of global significance. Microbiol. Mol. Biol. Rev. 63:106-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Redfield, A. C., B. H. Ketchum, and F. A. Richards. 1963. The influence of organisms on the composition of sea-water, p. 26-77. In M. N. Hill (ed.), The sea, vol. 2. Interscience Publishers, John Wiley & Sons, New York, N.Y.
- 26.Rippka, R., T. Coursin, W. Hess, C. Lichtle, D. J. Scanlan, K. A. Palinska, I. Iteman, F. Partensky, J. Houmard, and M. Herdman. 2000. Prochlorococcus marinus Chisholm et al. 1992 subsp. pastoris subsp. nov. strain PCC 9511, the first axenic chlorophyll a(2)/b(2)-containing cyanobacterium (Oxyphotobacteria). Int. J. Syst. Evol. Microbiol. 50:1833-1847. [DOI] [PubMed] [Google Scholar]
- 27.Urbach, E., D. J. Scanlan, D. L. Distel, J. B. Waterbury, and S. W. Chisholm. 1998. Rapid diversification of marine picophytoplankton with dissimilar light-harvesting structures inferred from sequences of Prochlorococcus and Synechococcus (Cyanobacteria). J. Mol. Evol. 46:188-201. [DOI] [PubMed] [Google Scholar]
- 28.Waterbury, J. B., S. W. Watson, R. R. L. Guillard, and L. E. Brand. 1979. Wide-spread occurrence of a unicellular, marine planktonic, cyanobacterium. Nature 277:293-294. [Google Scholar]
- 29.West, N. J., and D. J. Scanlan. 1999. Niche-partitioning of Prochlorococcus populations in a stratified water column in the eastern North Atlantic Ocean. Appl. Environ. Microbiol. 65:2585-2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.West, N. J., W. A. Schonhuber, N. J. Fuller, R. I. Amann, R. Rippka, A. F. Post, and D. J. Scanlan. 2001. Closely related Prochlorococcus genotypes show remarkably different depth distributions in two oceanic regions as revealed by in situ hybridization using 16S rRNA-targeted oligonucleotides. Microbiology 147:1731-1744. [DOI] [PubMed] [Google Scholar]
- 31.Williams, P. J. L. 1998. The balance of plankton respiration and photosynthesis in the open oceans. Nature 394:55-57. [Google Scholar]
- 32.Zubkov, M., M. Sleigh, and P. Burkill. 1998. Measurement of bacterivory by protists in open-ocean waters. FEMS Microbiol. Ecol. 27:85-102. [Google Scholar]
- 33.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
- 34.Zubkov, M. V., B. M. Fuchs, P. H. Burkill, and R. Amann. 2001. Comparison of cellular and biomass specific activities of dominant bacterioplankton groups in stratified waters of the Celtic Sea. Appl. Environ. Microbiol. 67:5210-5218. [DOI] [PMC free article] [PubMed] [Google Scholar]