Abstract
In order to prepare for whole-genome expression analysis in Sinorhizobium meliloti, pilot DNA macroarrays were designed for 34 genes of known regulation. The experimental parameters assessed were the length of the PCR products, the influence of a tag at the 5′ end of the primers, and the method of RNA labeling. Variance and principal-component analysis showed that the most important nonbiological parameter was the labeling method. The sizes of PCR products were also found to be important, whereas the influence of 5′ tags was minimal. The variability between replicated spots on a membrane was found to be low. These experimental procedures were validated by analyzing the effects of microaerobic conditions on gene expression.
Rhizobia are soil bacteria capable of establishing an N2-fixing symbiosis with plants of the family Leguminosae. During this symbiosis, new specialized organs are formed: the nitrogen-fixing root nodules, which provide the proper physiological conditions for the bacteria to survive in the absence of competing microflora and to reduce atmospheric dinitrogen to ammonia, which is then assimilated by the host plant (15). In addition to this specific symbiotic lifestyle, the physiology of rhizobia has been investigated under a wide variety of environmental conditions, such as nutrient deprivation or resistance to environmental stress, which are relevant to their survival in soils. In recent years, Sinorhizobium meliloti—the symbiont of alfalfa—has become one of the primary model organisms for the study of microbial persistence, competition, and stress response in soil, as well as plant-microbe interactions (16). Moreover, one of its hosts, Medicago truncatula, has become a model organism for study of the plant partner of symbiotic nitrogen fixation, as well as comparative legume genetics in general (5).
The recent availability of the entire annotated genome sequence of S. meliloti (12) has opened new perspectives in the study of its biology through the use of DNA arrays. DNA arrays have already been printed and used to study gene expression in several model bacteria, such as Escherichia coli, Bacillussubtilis, and several pathogens (2, 24), but so far have not been used for model plant-associated bacteria. In addition, we are interested in evaluating the reproducibility of DNA nylon arrays, as well as identifying the main sources of experimental errors in order to make adequate technological choices. To address these issues, we have adapted the methods developed for other organisms. We have designed and tested pilot macroarrays of S. meliloti genes and have used them to evaluate the methods for studying gene expression in S. meliloti.
MATERIALS AND METHODS
Bacterial growth conditions and RNA extraction.
S. meliloti strain 1021 was grown in Vincent minimal medium (26) with succinate as a carbon source in 1-liter bioreactors (Setric, Toulouse, France) at 30°C and pH 6.8. Cultures were inoculated at an optical density at 600 nm (OD600) of 0.05 with inocula from the same medium. Oxic cultures were conducted with an oxygen pressure (pO2) always >60% of saturation by air until the cells were collected (OD600 = 0.5). In microoxic cultures, the pO2 was initially kept high (>60%, as in the aerated cultures) until an OD600 of 0.2 was reached. The reactors were then rapidly flushed with N2, and the pO2 was maintained below the detection limit of the electrodes (apparent pO2 = 0, i.e., <1 μM dissolved oxygen), although a constant input of oxygen was maintained to ensure bacterial growth and energy metabolism. This was achieved through reduced agitation (ca. 200 rpm) and airflow (3 air volumes per medium volume per min). At an OD600 of 0.5, 25 ml of culture was sampled, and the bacterial cells were rapidly separated by filtration through a 0.2-μm-pore-size filter and immediately frozen in liquid nitrogen.
RNA was extracted from free-living cultures as described by Cabanes et al. (3). The bacterial pellet from a 25-ml culture (OD600 = 0.5) was resuspended and incubated for 10 min at 65°C in 2 ml of a lysis solution (1.4% sodium dodecyl sulfate (SDS), 4 mM EDTA, 75 μg of proteinase K). Proteins were precipitated by adding 1 ml of NaCl (5 M) at 4°C. Nucleic acids were precipitated by adding 1 volume of isopropanol, and the pellet was resuspended in nuclease-free water. DNA was eliminated by the addition of 7.5 U of RNase-free DNase I (Amersham-Pharmacia Biotech). RNAs were further extracted by a phenol-chloroform-isoamyl alcohol mixture and precipitated with 90% ethanol. The absence of detectable DNA in the RNA preparation was verified by PCR analysis using S. meliloti-specific primers. To limit degradation, the RNAs were stored at −80°C.
Primer design.
Primers, whose melting temperatures (Tm) ranged from 58 to 62°C, were designed with the help of Primer 3 software (23). The primers were designed to amplify either entire open reading frames yielding PCR products ranging from 335 to 2,888 bp or internal fragments of 238 to 387 bp (Table 1). For internal fragments, both untagged and 5′-end-tagged primers were produced, whereas only tagged primers were synthesized for the full-length PCR fragments. The lengths of the primers were 20-mer on average for untagged primers and 39-mer for tagged primers. The tag sequence was a 19-mer sequence (5′-GCCCGGTTAACAATATTCC-3′; Tm = 58°C) not found in the S. meliloti genome. The tag used alone did not generate any amplification product when S. meliloti genomic DNA was used as a template (amplification consisted of 35 cycles of 30 s at 94°C, 30 s at 45°C, and 1 min at 72°C) (data not shown).
TABLE 1.
Genes selected, characteristics of PCR products spotted on membranes, and ratios of normalized signals obtained under micro-oxic and oxic conditionsa
| Gene | Protein description | Zone amplifiedb | Length of PCR product (bp)
|
Expression ratio (−O2/+O2)c | ||
|---|---|---|---|---|---|---|
| Constant size
|
Full length with tag | |||||
| Without tag | With tag | |||||
| bacA | Putative transport protein | 119-460 | 342 | 380 | 1,298 | 1.0 |
| dctA | C-4-dicarboxylate transport protein | 401-700 | 300 | 338 | 1,358 | 3.3 |
| dnaJ | Chaperone | 736-1,015 | 280 | 318 | 1,178 | 1.3 |
| dnaK | Heat shock protein 70 (HSP70) chaperone | 314-655 | 342 | 380 | 1,958 | 0.9 |
| exoX | Posttranscriptional regulator; repressor | 1-282 | 282 | 320 | 335 | 1.7 |
| exoY | Galactosyltransferase | 131-430 | 300 | 338 | 720 | 1.4 |
| fixJ | FixJ; transcriptional activator | 60-311 | 251 | 289 | 654 | 2.0 |
| fixK1 | FixK1; transcriptional activator | 266-565 | 300 | 338 | 675 | 10.9 |
| fixL | FixL; oxygen-regulated histidine kinase | 181-480 | 300 | 338 | 1,538 | 1.8 |
| fixN1 | FixN1; heme b/copper cytochrome c oxidase subunit | 587-882 | 296 | 334 | 1,638 | 11.0 |
| fixP1 | FixP1; Diheme cytochrome c | 350-649 | 300 | 338 | 909 | 9.3 |
| fixT1 | FixT1; inhibitor of FixL autophosphorylation | 25-326 | 302 | 340 | 387 | 2.8 |
| ftsK | Cell division protein FtsK-like protein | 300-598 | 299 | 337 | 2,678 | 1.4 |
| ftsZ1 | Cell division protein | 244-592 | 349 | 387 | 1,808 | 1.2 |
| fusA | Elongation factor G | 234-577 | 344 | 382 | 2,138 | 0.4 |
| glnB | Nitrogen regulatory protein PII | 77-335 | 259 | 297 | 378 | 2.2 |
| glnD | Protein-PII uridylyltransferase | 559-866 | 308 | 346 | 2,888 | 1.2 |
| glnII | Glutamine synthetase II | 524-855 | 331 | 369 | 1,068 | 1.8 |
| groEL1 | 60-kDa chaperonin A | 231-531 | 301 | 339 | 1,688 | 0.3 |
| groES1 | 10-kDa chaperonin A | 45-284 | 240 | 278 | 335 | 0.3 |
| hemA | 5-Aminolevulinic acid synthase | 39-276 | 238 | 276 | 1,258 | 2.7 |
| ndiA | Nutrient deprivation-induced protein | 220-492 | 272 | 310 | 458 | 1.4 |
| ndiB | Probable nutrient deprivation-induced protein | 489-765 | 276 | 314 | 1,014 | 2.7 |
| nifA | NifA; transcriptional activator | 1248-1553 | 306 | 344 | 1,608 | 3.1 |
| nifH | NifH; nitrogenase Fe protein | 402-699 | 298 | 336 | 932 | 2.2 |
| nodA | NodA; N-acyltransferase | 179-482 | 304 | 342 | 629 | 1.3 |
| nodC | NodC; N-acetylglucosaminyltransferase | 956-1256 | 301 | 339 | 1,318 | 1.2 |
| nodD1 | NodD1; transcription regulator | 373-662 | 290 | 328 | 966 | 1.3 |
| nodE | NodE; beta-ketoacyl ACP synthase | 570-868 | 299 | 337 | 1,248 | 1.4 |
| ntrB | Nitrogen regulation protein | 143-477 | 335 | 373 | 1,188 | 1.3 |
| ntrC | Nitrogen assimilation regulatory protein | 429-765 | 337 | 375 | 1,488 | 0.9 |
| rpsA | 30S ribosomal protein S1 | 101-418 | 318 | 356 | 1,738 | 0.3 |
| trpE | Anthranilate synthase | 365-663 | 299 | 337 | 2,228 | 0.7 |
| tspO | Tryptophan-rich sensory protein homologue | 24-356 | 333 | 371 | 578 | 2.8 |
See Materials and Methods.
Position from start codon.
−O2, micro-oxic; +O2, oxic.
PCR amplification.
The amplification and reamplification conditions for untagged internal PCR fragments were identical for the 34 genes tested: amplification consisted of 30 cycles of 30 s at 94°C, 1 min at 55°C, and 1 min at 72°C. For the tagged internal PCR fragments, the amplification conditions consisted of 5 cycles of 30 s at 94°C, 30 s at 52°C, and 30 s at 72°C, followed by 25 cycles of 30 s at 94°C and 1 min at 72°C. These conditions were used for the first amplification. The reamplification conditions using the tag as a primer consisted of 30 cycles of 30 s at 94°C, 30 s at 62°C, and 30 s at 72°C. For the full-length tagged PCR fragments, the amplification and reamplification conditions used were similar to the conditions used for the tagged internal fragments, with adjustments of the hybridization temperature and elongation time (except for ftsK, for which it was not possible to find acceptable conditions). All amplifications were carried out with Taq DNA polymerase (Gibco BRL). For some full-length PCR fragments (dctA, dnaJ, ftsZ1, glnD, hemA, ndiB, ntrB, and trpE), the Expand Long Template Taq polymerase (Roche) was preferred. For Corynebacterium striatum control genes, conditions for both the first and the second amplifications consisted of 30 cycles of 45 s at 94°C, 45 s at 45°C, and 30 s at 72°C, using Taq DNA polymerase from Gibco BRL and universal T3 and T7 primers.
All PCR products were analyzed by agarose gel electrophoresis. The products were then precipitated with isopropanol and resuspended in 50% dimethyl sulfoxide. The final concentrations were 0.2 to 0.5 μg/μl.
In vitro transcription.
In order to prepare control RNAs, the pTFCS6, pTFCS15, pTFCS19, pTFCS20, and pTFCS37 plasmids containing C. striatum genes were linearized downstream of the insert by cleavage with SacI, and runoff transcripts were produced using the RiboMAX T7 large-scale RNA production system (Promega). The C. striatum genes cloned in the pTFCS6, pTFCS15, pTFCS19, pTFCS20, and pTFCS37 plasmids are the ABC transporter tetA gene, the tnp5564 gene, the ytpC gene, the gcrR gene, and the aphaA1-IAB gene, respectively. The DNA was eliminated by RQ1 RNase-free DNase treatment, and the transcripts were purified using the Cleanup RNeasy protocol (Qiagen).
Macroarray production.
The spotting of DNA probes was carried out at the Toulouse Génopôle (Institut National des Sciences Appliquées), using a Eurogridder robot (Eurogentec). The concentrated PCR products, at concentrations of 0.2 to 0.5 μg/μl, were spotted onto Immobilon-NY+ (Millipore) membranes (100 nl of PCR product per spot). Preliminary tests revealed good spot homogeneity, good reproducibility, and a good response to repeated rehybridization. For direct labeling of total RNA with digoxigenin (DIG), the PCR products were spotted on positively charged nylon membranes (Roche). All PCR products were spotted in triplicate.
Probe labeling, hybridization conditions, and signal detection.
Radioactively labeled cDNAs were generated by reverse transcription of 5 μg of total RNA using Superscript II (Gibco BRL), a mixture of random primers [d(N)6 and d(N)9 (Biolabs)], and [α-33P]dCTP (New England Nuclear) for labeling (adapted from reference 21). The RNA template was degraded by alkaline treatment at high temperature, and the probe was purified using MicroSpin S-200 columns (Amersham Pharmacia Biotech). A classical hybridization procedure was used (21). It was performed overnight at 65°C in hybridization buffer (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 5× Denhardt solution, 100 μg of sheared salmon sperm DNA/ml, 0.5% SDS), with a freshly denatured (heated) probe, after a 2-h prehybridization in the same buffer at 65°C. Stringent washes were performed with 0.2× SSC-0.5% SDS at 65°C. Hybridization signals were visualized using a low-energy screen (Kodak-Molecular Dynamics) and a Storm 840 PhosphorImager (Molecular Dynamics).
Direct chemical labeling was performed using 1 μg of total RNA in 20 μl with 2 μl of DIG-Chem-Link (Roche) for 30 min at 85°C. The reaction was stopped by the addition of 5 μl of stop solution. The hybridization and washing conditions were the same as for 33P-labeled cDNA, except for the hybridization buffer composition (5× SSC, 1% [wt/vol] blocking reagent [Roche], 0.02% N-lauroylsarcosine sodium salt, 0.02% SDS). Hybridization signals were revealed using alkaline phosphatase-coupled antisera directed against DIG and analyzed using a Storm 840 PhosphorImager after a 30-min incubation in the presence of the ECF chemiluminescent substrate (Amersham).
Quality control.
For PCR reamplified with the tag, the amounts of PCR products spotted on membranes were checked by hybridization of radioactively labeled tag primer. Primer (500 pmol) was 5′ end labeled using 15 U of polynucleotide kinase (Gibco BRL) and [γ-33P]ATP (New England Nuclear) for 1 h at 37°C. The primer was purified using MicroSpin G-25 columns (Amersham Pharmacia Biotech). Both membrane hybridization and washing were performed at 38°C using the same protocol as for 33P-labeled cDNAs.
For all experiments, the quality of probe labeling was checked using internal standards. The PCR products, corresponding to five C. striatum genes (gifts from A. Becker, University of Bielefeld) not present in the S. meliloti genome and with a GC percentage varying from 47 to 67%, were spotted on all membranes. Known amounts of in vitro-synthesized transcripts corresponding to these genes were added to S. meliloti total RNA preparations before they were labeled. In all experiments, no significant variations were observed.
The signals from replicated spots are highly reproducible, and it is not necessary to increase the number of these replicates.
Quantification and statistical analysis.
Hybridization results were analyzed using variance analysis as described by Sekowska et al. (24). The factors studied were the genes (34 genes), the oxygen status (two levels), the RNA labeling (two levels), the type of PCR product spotted (three levels), the repetitions (two levels), and the replicates on a given membrane (three levels). Images obtained from the PhosphorImager were quantified (the median for each spot) using ImaGene software (Biodiscovery). The data to be analyzed were grouped in 72 data sets containing one value for each of the 34 genes. In order to compare the data sets by variance analysis, the data should exhibit similar distributions (8). Therefore, each raw data set was log10 transformed, centered (mean = 0), and reduced (σ = 1) to yield normal distributions. As recommended (24), the background was not subtracted in the data presented, but similar conclusions could be drawn when the background was subtracted (data not shown). The transformed data were analyzed by performing both analysis of variance (ANOVA) and principal component analysis (PCA) using GeneAnova software (8). Unidentified factors are represented by the residual in the statistical model. As the measured residual here was found to be low (Table 2), other factors appear to play only a minor role in these experiments. In all analyses presented here, the background was not subtracted, except for the calculation of ratios of hybridization signals between full-length and constant-size products. In the latter case, the subtracted background was the mean signal from six spots of two nontarget C. striatum genes. The variations in gene expression presented in Table 1 were determined as follows. Ratios were calculated from the data obtained from radioactive hybridizations with tagged PCR products. Raw data with the background subtracted were normalized with the amounts of PCR products spotted and the total signal of each membrane as proposed by Nguyen et al. (17). Ratios between normalized signals obtained under both conditions (with and without oxygen) were then calculated.
TABLE 2.
Global ANOVA of gene expression based on all datasetsa
| Factor(s) | Sum of squares | DOFb | Variance | Fc | P value |
|---|---|---|---|---|---|
| Genes | 1,112.81 | 33 | 33.72 | 214.35 | <10−6 |
| Genes × oxygen | 547.64 | 33 | 16.6 | 105.49 | <10−6 |
| Genes × labeling | 153.95 | 33 | 4.67 | 29.66 | <10−6 |
| Genes × repetitions | 69.2 | 33 | 2.1 | 13.33 | <10−6 |
| Genes × PCR type | 133.48 | 66 | 2.02 | 12.86 | <10−6 |
| Genes × triplicates | 15.5 | 66 | 0.23 | 1.49 | 0.00677 |
| Residual | 343.42 | 2,183 | 0.16 | ||
| Total | 2,376 | 2,447 | 0.97 |
Degree of interaction = 2.
DOF, degrees of freedom.
F, Fischer coefficient.
RESULTS AND DISCUSSION
To investigate global relative variations in mRNA abundance by using DNA arrays and to carry out a pilot experiment before investigating the whole genome, we decided to validate macroarray technology on a small set of 34 S. meliloti genes selected because of their known regulation (Table 1 and Fig. 1). We analyzed the results using ANOVA, a classical statistical approach which was recently applied to transcriptome analysis (2, 24). Our experimental design was intended to include four experimental parameters (RNA labeling, type of PCR spotted, repetition of experiments, and replication of spots) using RNA generated under two biological conditions (aerobic and microaerobic growth) and to examine their influence on the expression of a limited number of genes. Normalized results of all experiments (see Materials and Methods) were analyzed by performing both ANOVA (Table 2) and PCA (Fig. 2) in order to identify the main sources of experimental variability and to sort out the optimal experimental conditions.
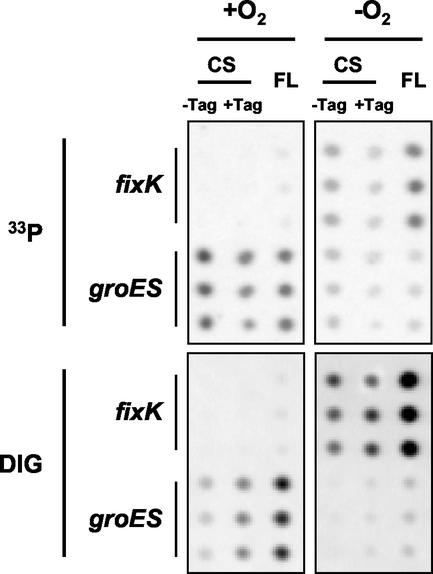
FIG. 1.
Examples of macroarray portions. The focus was set on the spots corresponding to the fixK and groES genes. Membranes were hybridized with RNA extracted from micro-oxic (−O2) or oxic (+O2) cultures. DIG, RNA was directly chemically labeled with DIG-Chem-Link; 33P, radioactive labeling of cDNA; CS − Tag, PCR products of constant size without tag; CS + Tag, PCR products of constant size with tag; FL, full-length PCR products with tag. Each PCR product is spotted three times (vertical replicates).
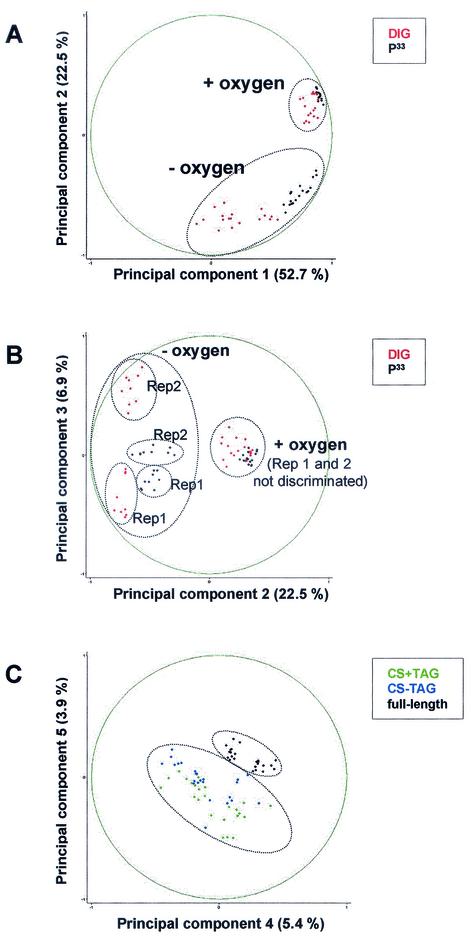
FIG. 2.
PCA of expression results. Each point represents a complete data set of the expression of the 34 test genes. The data are plotted using the axes representing the highest variability. (A and B) Black points represent data obtained after 33P labeling; red points represent data obtained after DIG labeling. (C) Green points correspond to the results obtained with the constant-size PCR products obtained with tagged primers (CS + TAG); blue points correspond to the results obtained with the constant-size PCR products obtained with primers without tag (CS − TAG); black points correspond to the results obtained with full-length PCR products obtained with tagged primers (full-length). (B) Rep1 and Rep2, experiments 1 and 2 representing repetitions of the same experiment.
Probe labeling: random priming is sensitive and reproducible.
Unlike eukaryotic mRNA, prokaryotic transcripts are generally not polyadenylated and cannot be labeled by reverse transcription using poly(dT) primers. Therefore alternative strategies are required to label the complex mixture of mRNAs extracted from bacterial cells. These include the generation of labeled cDNA by reverse transcription using random primers or a mixture of specific primers (2), 5′ end labeling using polynucleotide kinase (20), and chemical modification by DIG (9, 10). The use of a mixture of specific primers was proven to yield strong biases (2), and the 33P end labeling of mRNAs with polynucleotide kinase yields signals of low intensity, as only one radiolabeled atom is incorporated into each molecule. Finally, both random priming (2) and chemical labeling (9) were successfully used to monitor bacterial gene expression in DNA arrays, but these were not previously directly compared. One main advantage of DIG labeling lies in direct labeling of the RNA and the reduced RNA requirement. The amount of bacterial RNA is generally not limiting when free-living cultures are used; however, when symbiotic or pathogenic bacteria are to be studied within their hosts, the amount of biological material becomes rapidly limiting.
ANOVA showed that the biological factors studied (gene identity and oxygen status) were responsible for most of the variance (Table 2). RNA labeling was the first nonbiological factor introducing variation in our experimental design. ANOVA yielded a high Fischer value (F = 29.66) associated with a low P value (<10−6), indicating that the type of protocol used to label RNA strongly influenced the results. PCA analysis showed that data sets obtained with DIG-labeled RNA were more scattered than when 33P was used, especially under microaerobic conditions (Fig. 2A). This is mostly due to high variation between experiments, as shown in Fig. 2B. When DIG-labeled probes were used, the background was strongly increased when membranes were rehybridized, thus preventing their multiple use. As a result, the signal/background ratio was lower with DIG-labeled than with 33P-labeled probes. Therefore, and despite the smaller amounts of RNA required, we do not recommend the chemical labeling of RNA with DIG for transcriptome studies and prefer to use bacterial RNA labeled by reverse transcription with random primers. Such variations among results obtained by different labeling methods were described earlier (22).
Effect of PCR fragment size.
S. meliloti gene size varies over a wide range, from <150 bp to >3 kb (12). In order to design DNA arrays harboring one target for each predicted gene, it is possible to use either long single oligomers (1, 19) or specific PCR products (13). The latter solution is often more economical, as large amounts of DNA can be generated with virtually no limit through PCRs. Another advantage of the use of PCR products lies in the fact that they can be used for further cloning and generation of knockout mutants or for protein production in the case of a full-length PCR product. The question then arises as to the optimal size of the PCR products to be generated for genomic arrays. In several studies, full-length genes were amplified (21), whereas other authors used gene fragments of constant size (14). In order to choose a strategy, we evaluated both options. We amplified either the genes from the predicted start codon to the stop codon, yielding PCR products from 335 to 2,888 bp (Table 1) or internal fragments of 238 to 387 bp. When the ANOVA analysis was performed with all the data generated with tagged primers, the PCR fragment size appeared to be a crucial factor influencing the results (a high Fischer value [F = 20.22] associated with a low P value [<10−6]). It was observed that the ratio between hybridization signals obtained with full-length and constant-size PCR products increased globally with the corresponding ratios of the PCR product sizes (size of full-length PCR product/size of constant-size PCR products), the slope being close to 1 (data not shown). This increase could be attributed to the higher number of labeled molecules in the probe that binds to a PCR product covering an entire gene rather than an internal fragment. Furthermore, the use of constant sizes enabled us to standardize the characteristics of the primers, as well as the Tm of the PCR products, and therefore the PCR conditions (see Materials and Methods). This was not possible when full-length genes were amplified and optimization of amplification was necessary.
Tags do not bias quantification.
Specific sequences, referred to as tags, are often included in primers used to amplify genes or gene fragments in transcriptome studies (21). These tags are most useful to enable reamplification using a single pair of primers corresponding to the tags and to generate restriction sites facilitating further cloning of PCR products. Tags also make it possible to quantify the amounts of PCR products spotted on DNA arrays, an important quality control. However, the use of tags implies that nonspecific DNA is being spotted together with the desired target sequences. Therefore, we wished to evaluate their influence on our transcriptome experiments.
The data sets obtained for gene fragments of constant size with and without tags were compared by means of ANOVA (Table 3). The results showed that the influence of the tag sequence was minimal and lower than that yielded by the repetition of a single experiment. PCA confirmed this result: none of the five most significant principal components could discriminate data sets obtained with tagged primers from those obtained with untagged primers (for example, Fig. 2C). Therefore, the presence of this 19-mer tag in all the primers did not influence the transcriptome results, and the use of tagged primers can be highly recommended.
TABLE 3.
Influence of tag sequences on expression results analyzed by global ANOVAa
| Factor(s) | Sum of squares | DOFb | Variance | Fc | P value |
|---|---|---|---|---|---|
| Genes | 682.95 | 33 | 20.7 | 116.42 | <10−6 |
| Genes × oxygen | 406.08 | 33 | 12.31 | 69.23 | <10−6 |
| Genes × labeling | 132.66 | 33 | 4.02 | 22.61 | <10−6 |
| Genes × repetitions | 67.27 | 33 | 2.04 | 11.47 | <10−6 |
| Genes × tag | 30.37 | 33 | 0.92 | 5.18 | <10−6 |
| Genes × triplicates | 15.81 | 66 | 0.24 | 1.35 | 0.0354 |
| Residual | 248.86 | 1,400 | 0.18 | ||
| Total | 1,584 | 1,631 | 0.97 |
Only data obtained with constant-size PCR products (with or without tags) were included. Degree of interaction = 2.
DOF, degrees of freedom.
F, Fischer coefficient.
Validation using oxygen-controlled genes.
The last factor considered was a biological one: gene expression levels under aerobic and microaerobic conditions were compared. A local ANOVA analysis performed as described by Didier et al. (8) enabled us to sort out those genes that were more responsive to changes in the oxygen status (Fig. 3). Genes showing both high variation in expression depending on the oxygen input and low P values testifying to the significance of the observed effect were considered to be specifically regulated by oxygen. As expected (25), the fix genes involved in micro-oxic respiration were significantly induced by oxygen limitation (induction factors ranged from 9 to 11 for the fixKNP genes [Table 1]). The induction by oxygen limitation of other genes was also detected by the macroarray experiments: fixT, nifA, tspO, and ndiB, coding for an antikinase under the control of FixK, the nitrogen fixation transcriptional activator, a tryptophan-rich sensory protein, and a nutrient deprivation-induced protein, respectively, were induced two to three times by oxygen limitation (Table 1), a result in agreement with the literature (6, 7, 11). However, nifH was not induced by oxygen limitation under free-living conditions, as previously shown by Cabanes et al. (4). Finally, we found that the dicarboxylic acid transporter dctA was induced three times by oxygen limitation. The expression of dctA was previously demonstrated to be induced by dicarboxylic acids such as succinate, as in our cultures, but the effect of oxygen had not been previously demonstrated. Finally, the hemA heme synthase gene was previously thought to be constitutive. This work, together with previous observations (F. Ampe et al., unpublished data), strongly suggests that it is in fact induced by oxygen limitation.
FIG. 3.
Local ANOVA with genes and oxygen status (data with full-length open reading frames were excluded). The abscissa represents the variation due to oxygen normalized by the total variation of gene expression (i.e., for each gene, the variance due to oxygen divided by the total variance), and the ordinate represents the logarithm of the P value. Genes exhibiting the highest and most reproducible variation due to oxygen limitation are located at the bottom right.
Conversely, the expression of a few other genes was shown to decrease in oxygen-limited cultures: groESL, rspA, and fusA, coding for chaperones, a ribosomal protein, and an elongation factor, respectively. The expression levels of groES and groEL were repressed (3 and 3.7 times, respectively) (Table 1) but also highly correlated (data not shown), a result in agreement with Ogawa and Long (18), who suggested that they form an operon. The expression levels of rspA and fusA were repressed (4 and 2.5 times, respectively) when oxygen was limiting, as a probable consequence of slower growth of the bacteria (μ, <0.1 h−1) than in fully aerated cultures (μ, ∼0.3 h−1). As expected, other genes (nodACD, exoXY, glnD, ntrC, ftsZ, etc. [Table 1]) did not show significant variations with respect to the change in aeration conditions. Interestingly, with the exception of dctA, the expression of none of the genes tested here varied between minimal and rich media when growth rates were identical (Ampe et al. unpublished data), thus supporting the idea that the variations observed here are essentially due to oxygen limitation.
Conclusion.
The experiments described above were aimed at measuring the impacts of several parameters on S. meliloti transcriptome analysis. These pilot experiments, carried out using a number of genes, allowed us to validate the use of macroarrays to study changes in mRNA expression in S. meliloti. Our conclusions are that (i) internal gene fragments of constant size should be used preferentially in order to compare signal intensities within a membrane, (ii) the incorporation of a tag at the 5′ end of each primer does not significantly influence hybridization results and allows for simple reamplification and quantification of the amount of spotted DNA, (iii) labeling probes by reverse transcription is more reliable than direct incorporation of DIG into RNA, and (iv) signals from replicated spots are highly reproducible.
These pilot experiments, done on a number of limited open reading frames, allowed us to define the conditions that can be used for whole-genome S. meliloti macroarrays to study global mRNA changes.
Acknowledgments
H. Bergès, F. Ampe, and E. Lauber contributed equally to this work.
F. Ampe is supported by the Institut de Recherche pour le Développement (IRD; Montpellier, France). H. Bergès was supported by a fellowship from the Toulouse Génopôle. This work was supported by the Toulouse Génopôle, the department “Santé des Plantes et Environnement” of INRA (grant 0441-04), and the CNRS program “Puces à ADN.”
We thank V. Le Berre and A. Dagkesamanskaya for the spotting of PCR products on membranes. We are grateful to A. Becker for providing plasmids containing C. striatum genes.
REFERENCES
- 1.Aharoni, A., and O. Vorst. 2002. DNA microarrays for functional plant genomics. Plant Mol. Biol. 48:99-118. [DOI] [PubMed] [Google Scholar]
- 2.Arfin, S. M., A. D. Long, E. T. Ito, L. Tolleri, M. M. Riehle, E. S. Paegle, and G. W. Hatfield. 2000. Global gene expression profiling in Escherichia coli K12. The effects of integration host factor. J. Biol. Chem. 275:29672-29684. [DOI] [PubMed] [Google Scholar]
- 3.Cabanes, D., P. Boistard, and J. Batut. 2000. Symbiotic induction of pyruvate dehydrogenase genes from Sinorhizobium meliloti. Mol. Plant-Microbe Interact. 13:483-493. [DOI] [PubMed] [Google Scholar]
- 4.Cabanes, D., P. Boistard, and J. Batut. 2000. Identification of Sinorhizobium meliloti genes regulated during symbiosis. J. Bacteriol. 182:3632-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Catoira, R., C. Galera, F. de Billy, R. V. Penmetsa, E.-P. Journet, F. Maillet, C. Rosenberg, D. Cook, C. Gough, and J. Dénarié. 2000. Four genes of Medicago truncatula controlling components of a nod factor transduction pathway. Plant Cell 12:1647-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey, M. E., and F. J. de Bruijn. 2000. A homologue of the tryptophan-rich sensory protein TspO and FixL regulate a novel nutrient deprivation-induced Sinorhizobium meliloti locus. Appl. Environ. Microbiol. 66:5353-5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.David, M., M. L. Daveran, J. Batut, A. Dedieu, O. Domergue, J. Ghai, C. Hertig, P. Boistard, and D. Kahn. 1988. Cascade regulation of nif gene expression in Rhizobium meliloti. Cell 54:671-683. [DOI] [PubMed] [Google Scholar]
- 8.Didier, G., P. Brézellec, E. Remy, and A. Hénaut. 2002. GeneAnova—gene expression analysis of variance. Bioinformatics 18:490-491. [DOI] [PubMed] [Google Scholar]
- 9.Even, S., N. D. Lindley, and M. Cocaign-Bousquet. 2001. Molecular physiology of sugar catabolism in Lactococcus lactis IL1403. J. Bacteriol. 183:3817-3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontaine, L., S. Even, P. Soucaille, N. D. Lindley, and M. Cocaign-Bousquet. 2001. Transcript quantification based on chemical labeling of RNA associated with fluorescent detection. Anal. Biochem. 298:246-252. [DOI] [PubMed] [Google Scholar]
- 11.Foussard, M., A.-M. Garnerone, F. Ni, E. Soupene, P. Boistard, and J. Batut. 1997. Negative autoregulation of the Rhizobium meliloti fixK gene is indirect and requires a newly identified regulator, FixT. Mol. Microbiol. 25:27-37. [DOI] [PubMed] [Google Scholar]
- 12.Galibert, F., T. M. Finan, S. R. Long, A. Pühler, P. Abola, F. Ampe, et al. 2001. The complete genome sequence of the legume symbiont Sinorhizobium meliloti. Science 293:668-672. [DOI] [PubMed] [Google Scholar]
- 13.Khodursky, A. B., B. J. Peter, N. R. Cozzarelli, D. Botstein, P. O. Brown, and C. Yanofsky. 2000. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl. Acad. Sci. USA 97:12170-12175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Loos, A., C. Glanemann, L. B. Willis, X. M. O'Brien, P. A. Lessard, R. Gerstmeir, S. Guillouet, and A. J. Sinskey. 2001. Development and validation of Corynebacterium DNA microarrays. Appl. Environ. Microbiol. 67:2310-2318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Merrick, M. J., and R. A. Edwards. 1995. Nitrogen control in bacteria. Microbiol. Rev. 59:604-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milcamps, A., D. M. Ragatz, P. Lim, K. A. Berger, and F. J. de Bruijn. 1998. Isolation of carbon- and nitrogen-deprivation-induced loci of Sinorhizobium meliloti 1021 by Tn5-luxAB mutagenesis. Microbiology 144:3205-3218. [DOI] [PubMed] [Google Scholar]
- 17.Nguyen, C., D. Rocha, S. Granjeaud, M. Baldit, K. Bernard, P. Naquet, and B. R. Jordan. 1995. Differential gene expression in the murine thymus assayed by quantitative hybridization of arrayed cDNA clones. Genomics 29:207-216. [DOI] [PubMed] [Google Scholar]
- 18.Ogawa, J., and S. R. Long. 1995. The Rhizobium meliloti groELc locus is required for regulation of early nod genes by the transcription activator NodD. Genes Dev. 9:714-729. [DOI] [PubMed] [Google Scholar]
- 19.Pease, A. C., D. Solas, E. J. Sullivan, M. T. Cronin, C. P. Holmes, and S. P. Fodor. 1994. Light-generated oligonucleotide arrays for rapid DNA sequence analysis. Proc. Natl. Acad. Sci. USA 91:5022-5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perret, X., C. Freiberg, A. Rosenthal, W. J. Broughton, and R. Fellay. 1999. High-resolution transcriptional analysis of the symbiotic plasmid of Rhizobium sp. NGR234. Mol. Microbiol. 3:415-425. [DOI] [PubMed] [Google Scholar]
- 21.Richmond, C. S., J. D. Glasner, R. Mau, H. Jin, and F. R. Blattner. 1999. Genome-wide expression profiling in Escherichia coli K-12. Nucleic Acids Res. 27:3821-3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenow, C., R. M. Saxena, M. Durst, and T. R. Gingeras. 2001. Prokaryotic RNA preparation methods useful for high density array analysis: comparison of two approaches. Nucleic Acids Res. 29:E112. [Online.] [DOI] [PMC free article] [PubMed]
- 23.Rozen, S., and H. Skaletsky. 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132:365-386. [DOI] [PubMed] [Google Scholar]
- 24.Sekowska, A., S. Robin, J. J. Daudin, A. Henaut, and A. Danchin. 2001. Extracting biological information from DNA arrays: an unexpected link between arginine and methionine metabolism in Bacillus subtilis. Genome Biol. 2:0019.1-0019.12. [DOI] [PMC free article] [PubMed]
- 25.Soupene, E., M. Foussard, P. Boistard, G. Truchet, and J. Batut. 1995. Oxygen as a key developmental regulator of Rhizobium meliloti N2-fixation gene expression within the alfalfa root nodule. Proc. Natl. Acad. Sci. USA 92:3759-3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vincent, J. M. 1970. A manual for the practical study of root nodule bacteria. IBP Handbook 15. Blackwell, Oxford, England.