Abstract
Suppression subtractive hybridization was used to rapidly identify 18 gene differences between a citrus variegated chlorosis (CVC) strain and a Pierce's disease of grape (PD) strain of Xylella fastidiosa. The results were validated as being highly representative of actual differences by comparison of the completely sequenced genome of a CVC strain with that of a PD strain.
Identification of the relatively few genes in a pathogenic microbe that determine virulence and host range can lead to an understanding of how pathogens cause disease and to novel disease control methods. With plant pathogens, identification of such genes has been done primarily by using time-consuming functional techniques such as chemical or transposon mutagenesis followed by complementation analyses or by virulence enhancement approaches using random DNA libraries. With the advent of genomic sequencing and improved annotation, comprehensive approaches are now also available. In fact, whole-genome sequence comparisons of different strains of two plant pathogenic species, Xylella fastidiosa and Xanthomonas campestris, have been performed (11, 12, 14).
Rapid techniques for identifying gene differences between microbial strains include subtractive hybridization (4, 8, 13) and DNA microarrays (2). However, both techniques are technically demanding and DNA microarrays of bacterial genes are of limited availability. Recently, a PCR-based subtraction method, called suppression subtractive hybridization (SSH), was developed and applied for rapid identification of differences among pathogenic Helicobacter pylori strains (1). The method has also been employed to compare the Escherichia coli and Salmonella enterica serovar Typhimurium genomes (3), to compare virulent and avirulent strains of the opportunistic aquatic pathogen Aeromonas hydrophila (15), and to identify genomic differences between the uropathogenic E. coli strain 536 and the nonpathogenic E. coli strain MG1655 (7). In none of these cases were the SSH results validated by direct genomic DNA sequence comparisons. In the present study, SSH was used to identify genetic differences between two strains of X. fastidiosa that differ in pathogenicity and host range and the results were validated by using the available complete genomic DNA sequences.
X. fastidiosa.
DNA from Pierce's disease (PD) strain PD1 of X. fastidiosa (6) was used as the driver and DNA from citrus variegated chlorosis (CVC) strain CVC-SB of X. fastidiosa (isolated in São Paulo, Brazil, by R. Harakava) was used as the tester to make a CVC strain-enriched library. Also, DNA from CVC-SB was used as the driver and DNA from PD1 was used as the tester to make a PD strain-enriched library. SSH was performed using a PCR-Select kit according to the manufacturer's protocol (Clontech, Palo Alto, Calif.). Basically, genomic DNAs from the two strains were isolated and separately digested with the same restriction endonuclease to small size and two lots of tester DNA were ligated with a different adaptor, each of which was designed to self anneal. A large excess of driver DNA was then hybridized to each adaptor-ligated tester lot, resulting mainly in hybridized double-stranded DNA but also some single-stranded DNA enriched for tester DNA sequences from each lot. The two hybridized lots were then mixed together without denaturing, allowing hybridization of tester DNAs with different adaptors on each end. DNA polymerase was added to fill in the ends. The resulting mixture was then subjected to PCR, using adaptor-specific primers, to amplify the tester-specific sequences. Driver DNA cannot be amplified, and DNA with only one primer sequence amplifies only once. DNA with the same primer sequence at both ends is efficiently suppressed, since each primer sequence is strongly self annealing and forms a hairpin-like secondary structure. Only DNA with two different adaptors at each end can be amplified exponentially, and these are always tester-specific sequences.
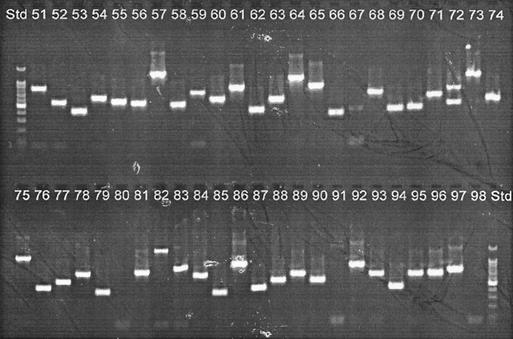
The insert sizes of 98 clones from each of the libraries were estimated by PCR amplification of cloned inserts by using vector-based primers T7 and SP6. Figure 1 depicts the screening of clones 51 through 98 of the CVC strain-enriched library. In both libraries, insert sizes varied from ∼200 to ∼1,200 bp (the PCR products contain an extra 175 bp from the pGEM-TE vector).
FIG. 1.
Screening of the CVC strain-specific minilibrary through colony PCR. Clone inserts were amplified by PCR using vector-based primers and were run in a 1.5% agarose gel. Shown are inserts from clones 51 through 98. Std, 100-bp ladder (Promega).
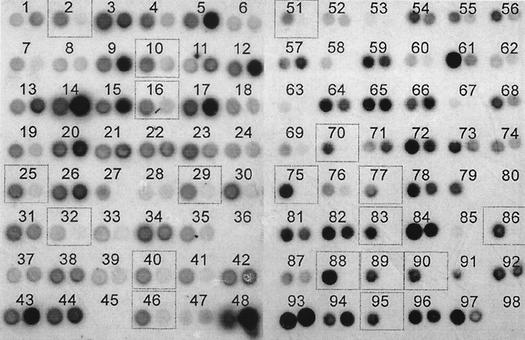
Dot blot hybridizations were performed using digoxigenin-labeled total DNA from each of the strains as probes to identify strain-specific clones among those of the strain-enriched minilibraries. Inserts from each clone of the minilibraries were PCR amplified and spotted on positively charged nylon membranes. To facilitate comparison of the dot blot signals, the X-ray films resulting from hybridization of the CVC strain-enriched minilibrary against both CVC strain and PD strain probes were superimposed (Fig. 2). Among 94 clones screened from the CVC strain-enriched library, 36 appeared to be CVC strain specific. The same procedure was performed for 47 clones of the PD strain-enriched library, but no PD strain-specific clones were found (data not shown).
FIG. 2.
Dot blot hybridization of PCR products from the CVC strain-specific minilibrary against digoxigenin-labeled CVC strain DNA (left spot of each pair) and PD strain DNA (right spot of each pair). Dotted boxes indicate clones that were selected for sequencing. Dots at position 48 correspond to 100 ng of genomic DNA from the CVC (left) and PD (right) strains.
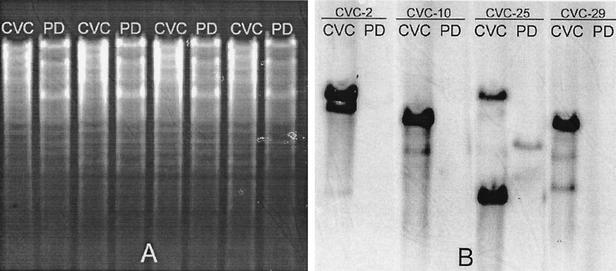
Southern blot analyses were performed on four of the CVC strain-specific clones and confirmed the results of the dot blot analyses by showing strongly hybridizing CVC strain but not PD strain genomic DNA bands (Fig. 3). Eighteen CVC strain-specific clones were sequenced; all were unique to the CVC strain according to BLAST searches of the complete genomes of CVC strain 9a5c and PD strain Temecula. The location of each sequenced CVC-SB DNA fragment in the CVC 9a5c genome (GenBank accession no. AE003849) was also determined by using BLAST. Seven of these clones were from the 51-kb megaplasmid of the CVC strain; the other 11 clones were chromosomal. CVC-89 and CVC-95 carried duplicated sequences.
FIG. 3.
Southern blot analyses using four CVC strain-specific clones as probes. (A) Agarose gel stained with ethidium bromide before transfer. (B) Membrane strips after hybridization against CVC strain-specific probes. After blotting, the nylon membrane was cut into four strips and hybridized against the digoxigenin-labeled probes CVC-2, CVC-10, CVC-25, and CVC-29.
Table 1 shows the position of each clone in the CVC 9a5c genome along with the reference CVC strain genome open reading frame (ORF) annotation number and indicated function, if any. Most of the ORFs were of unknown function and located either on the 51-kb megaplasmid or in chromosomal regions rich in phage-related or plasmid-related genes. For example, clones CVC-10, CVC-29, CVC-75, and CVC-89/95 were clustered within an 8-kb region of the chromosome; CVC-10 and CVC-75 show low similarities to genes of the filamentous bacteriophage Pf3 of Pseudomonas aeruginosa. Sequences from the mulberry leaf scorch and periwinkle wilt strains of X. fastidiosa with similarity to genes of the same phage have also been reported (9). CVC-40 was located within Xf-4, one of the four large prophages found in the CVC strain chromosome. CVC-16 is located in a cluster of genes related to plasmid conjugal transfer (tra and trb genes) that may correspond to a plasmid that has integrated into the chromosome.
TABLE 1.
CVC strain-specific clones corresponding to chromosomal genes
| Clone | Position(s) in the CVC strain genome (size [bp])a | ORF | Function |
|---|---|---|---|
| CVC-2 | 32536-32817 (282)b | XFa0042 | Conjugal transfer |
| 1953158-1953439 (282) | XF2051 | Conjugal transfer | |
| CVC-10 | 1781908-1782274 (367) | XF1867 | Unknown |
| XF1868 | Unknown | ||
| CVC-16 | 1958074-1958272 (199) | XF2057 | Unknown |
| XF2058 | Conjugal transfer | ||
| CVC-25 | 495465-495850 (386+) | XF0496 | Unknown |
| CVC-29 | 1781622-1781907 (286) | XF1867 | Unknown |
| CVC-32 | 50439-50636 (198)b | XFa0064 | Unknown |
| CVC-40 | 2383787-2384141 (355+) | XF2505 | Phage related |
| CVC-46 | 20910-21078 (169)b | XFa0025 | Unknown |
| CVC-51 | 43160-43554 (395+)b | XFa0054 | Unknown |
| XFa0055 | Unknown | ||
| CVC-70 | 42215-42426 (212)b | XFa0053 | Unknown |
| CVC-75 | 1782759-1783098 (340+) | XF1869 | Phage related |
| XF1870 | Phage related | ||
| CVC-77 | 4416-4729 (314)b | XFa0007 | Conjugal transfer |
| CVC-83 | 1335614-1336004 (391+) | XF1384 | Unknown |
| CVC-86 | 531864-532260 (397+) | XF0539 | Fimbrillin |
| CVC-88 | 3560-3895 (336)b | XFa0005 | Conjugal transfer |
| XFa0006 | Conjugal transfer | ||
| CVC-89/95 | 1789260-1789644 (385+) | XF1881 | Unknown |
| CVC-90 | 343764-344102 (339) | XF0328 | Unknown |
The position in the CVC 9a5c chromosome, ORF identification, and assigned function were obtained at the X. fastidiosa genome website (http://aeg.lbi.ic.unicamp.br/xf). A plus sign indicates that the clone is larger than the size presented and has not been completely sequenced.
The sequence lies in the 51-kb megaplasmid.
Perhaps the most interesting genes were those which were unique to the CVC strain and neither plasmid nor phage associated. CVC-25 corresponded to ORF XF0496, which encodes a protein 25% identical to the hypothetical protein Rv2515 (GenBank accession no. Z95556) of Mycobacterium tuberculosis. CVC-83 corresponded to ORF XF1384, which encodes a protein 39% identical to protein PqaA from S. enterica serovar Typhi, which is regulated by the two-component system PhoP/PhoQ but whose function is unknown. In S. enterica serovar Typhimurium, PhoP/PhoQ regulates expression of genes involved in virulence, such as those involved in lipid A modifications that result in bacterial resistance against antimicrobial cationic peptides produced by the host cell (5). Interestingly, orthologues of ORF XF1384 were not found in the genomes of other strains of X. fastidiosa but highly similar ones were present in the genomes of the plant pathogenic bacteria Xanthomonas campestris pv. campestris (GenBank accession no. NP638290), Xanthomonas axonopodis pv. citri (GenBank accession no. NP643428), and Ralstonia solanacearum (GenBank accession no. NP521722). CVC-86 partially overlapped ORF XF0539, encoding a CVC strain-specific fimbrillin, the structural subunit of the CVC strain fimbriae. Although CVC 9a5c and PD Temecula each have five genes encoding fimbrillin proteins, the PD strain fimbrillin that is most similar to the one encoded by CVC strain ORF XF0539 shows only 46% identity. This sequence divergence of fimbrillin proteins may reflect the fact that each bacterial strain adheres to surfaces inside different plant hosts and insect vectors. Novel fimbriae are associated with virulent clonal groups of E. coli and are thought to be horizontally transferred from other pathogens (10). Clone CVC-90 corresponded to the 3′ end of ORF XF0328, which did not show similarity to any sequence in GenBank. These genes are potential candidates for functional analyses.
Besides those of CVC 9a5c and PD Temecula, nearly complete genome sequences are also available for two additional strains of X. fastidiosa (http://www.jgi.doe.gov/JGI_microbial/html/), oleander strain Ann 1 and almond strain Dixon, which have different host ranges. Comparison of the sequences of the CVC strain-specific clones with the genomes of these strains were also performed using BLAST. The results of all three comparisons are shown in Table 2. Interestingly, both the oleander and almond strains contained sequences with similarity to a few more of the CVC strain-specific clones, indicating that multiple additional strains should be included as driver DNAs to obtain absolutely strain-specific clones.
TABLE 2.
BLAST scarch results comparing the sequences of the CVC strain-specific clones against the genome sequences of PD, oleander, and almond strains of X. fastidiosaa
| Clone | PD | Oleander | Almond |
|---|---|---|---|
| CVC-10 | − | + | − |
| CVC-16 | − | − | + |
| CVC-25 | − | − | + |
| CVC-29 | − | + | − |
| CVC-40 | +/− | +/− | +/− |
| CVC-75 | +/−− | + | +/−− |
| CVC-83 | − | − | − |
| CVC-86 | +/−− | − | +/− |
| CVC-89/95 | − | +/− | − |
| CVC-90 | − | − | +/− |
+, presence of nucleotide sequences >90% identical to the clone (high similarity); +/−, presence of nucleotide sequences <90% but >70% identical to the clone (low similarity); +/−−, presence of short segments (<60 nucleotides) identical to the clone (very low similarity).
In our hands, the application of SSH in a quick survey did not result in PD strain-specific clones. These results are consistent with the fact that the 2,679,305-bp CVC strain genome is ca. 158 kb larger than the highly similar 2,521,145-bp PD strain genome. Comparison of the two sequenced genomes showed that there are 41 PD strain-specific genes and 152 CVC strain-specific genes. Only 2 of the 18 CVC strain-specific clones were redundant, so more clones could likely have been identified by using this technique before a point of diminishing returns had been reached. In order to determine virtually all differences between the two strains by using SSH, approximately 2,260 strain-specific clones with an average insert size of 350 bp would need to be identified and sequenced. Since the SSH method usually yields subtracted libraries containing at least 50% strain-specific clones (Clontech manual), screening of 4,520 of these would in theory be necessary to find all genes that are present in one strain but absent in the other. Genes that have sequence divergence of less than ∼10% between the two strains would likely not be represented.
The results of the present study confirm previous reports that SSH is a powerful tool for rapid identification of gene differences between closely related bacterial strains (1, 3, 15). Following their identification, selected genes of unknown function can then be subjected to functional pathogenicity and host range analyses, possibly reducing the overall effort needed to find previously undescribed pathogenicity determinants.
Acknowledgments
R. Harakava was the recipient of a doctoral scholarship from the Sao Paulo State Research Funding Agency (FAPESP). This research was also supported by USDA-NRI grant no. 5S143.
Footnotes
Florida Agricultural Experiment Station contribution no. R09260.
REFERENCES
- 1.Akopyants, N. S., A. Fradkov, L. Diatchenko, J. E. Hill, P. D. Siebert, S. A. Lukyanov, E. D. Sverdlov, and D. E. Berg. 1998. PCR-based subtractive hybridization and differences in gene content among strains of Helicobacter pylori. Proc. Natl. Acad. Sci. USA 95:13108-13113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 3.Bogush, M. L., T. V. Velikodvorskaya, Y. B. Lebedev, L. G. Nikolaev, S. A. Lukyanov, A. F. Fradkov, B. K. Pliyev, M. N. Boichenko, G. N. Usatova, A. A. Vorobiev, G. L. Andersen, and E. D. Sverdlov. 1999. Identification and localization of differences between Escherichia coli and Salmonella typhimurium genomes by suppressive subtractive hybridization. Mol. Gen. Genet. 262:721-729. [DOI] [PubMed] [Google Scholar]
- 4.Brown, P. K., and R. Curtiss III. 1996. Unique chromosomal regions associated with virulence of an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 93:11149-11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunn, J. S., and S. I. Miller. 1996. PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178:6857-6864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hopkins, D. L. 1985. Physiological and pathological characteristics of virulent and avirulent strains of the bacterium that causes Pierce's disease of grapevine. Phytopathology 75:713-717. [Google Scholar]
- 7.Janke, B., U. Dobrindt, J. Hacker, and G. Blum-Oehler. 2001. A subtractive hybridization analysis of genomic differences between the uropathogenic E. coli strain 536 and the E. coli K-12 strain MG1655. FEMS Microbiol. Lett. 199:61-66. [DOI] [PubMed] [Google Scholar]
- 8.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pooler, M. R., J. S. Hartung, and R. G. Fenton. 1997. Sequence analysis of a 1296-nucleotide plasmid from Xylella fastidiosa. FEMS Microbiol. Lett. 155:217-222. [DOI] [PubMed] [Google Scholar]
- 10.Pouttu, R., B. Westerlund-Wikstrom, H. Lang, K. Alsti, R. Virkola, U. Saarela, A. Siitonen, N. Kalkkinen, and T. K. Korhonen. 2001. matB, a common fimbrillin gene of Escherichia coli, expressed in a genetically conserved, virulent clonal group. J. Bacteriol. 183:4727-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, et al. 2002. Comparison of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 12.Simpson, A. J., F. C. Reinach, P. Arruda, F. A. Abreu, M. Acencio, et al. 2000. The genome sequence of the plant pathogen Xylella fastidiosa. Nature 406:151-157. [DOI] [PubMed] [Google Scholar]
- 13.Tinsley, C. R., and X. Nassif. 1996. Analysis of the genetic differences between Neisseria meningitidis and Neisseria gonorrhoeae: two closely related bacteria expressing two different pathogenicities. Proc. Natl. Acad. Sci. USA 93:11109-11114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Van Sluys, M. A., M. C. Oliveira, C. B. Monteiro-Vitorello, C. Y. Miyaki, L. R. Furlan, et al. Comparative analyses of the complete genome sequences of Pierce's disease and citrus variegated chlorosis strains of Xylella fastidiosa. J. Bacteriol., in press. [DOI] [PMC free article] [PubMed]
- 15.Zhang, Y. L., C. T. Ong, and K. Y. Leung. 2000. Molecular analysis of genetic differences between virulent and avirulent strains of Aeromonas hydrophila isolated from diseased fish. Microbiology 146:999-1009. [DOI] [PubMed] [Google Scholar]