Abstract
Laccase from Myceliophthora thermophila (MtL) was expressed in functional form in Saccharomyces cerevisiae. Directed evolution improved expression eightfold to the highest yet reported for a laccase in yeast (18 mg/liter). Together with a 22-fold increase in kcat, the total activity was enhanced 170-fold. Specific activities of MtL mutants toward 2,2′-azino-bis(3-ethylbenzthiazoline-6-sulfonic acid) and syringaldazine indicate that substrate specificity was not changed by the introduced mutations. The most effective mutation (10-fold increase in total activity) introduced a Kex2 protease recognition site at the C-terminal processing site of the protein, adjusting the protein sequence to the different protease specificities of the heterologous host. The C terminus is shown to be important for laccase activity, since removing it by a truncation of the gene reduces activity sixfold. Mutations accumulated during nine generations of evolution for higher activity decreased enzyme stability. Screening for improved stability in one generation produced a mutant more stable than the heterologous wild type and retaining the improved activity. The molecular mass of MtL expressed in S. cerevisiae is 30% higher than that of the same enzyme expressed in M. thermophila (110 kDa versus 85 kDa). Hyperglycosylation, corresponding to a 120-monomer glycan on one N-glycosylation site, is responsible for this increase. This S. cerevisiae expression system makes MtL available for functional tailoring by directed evolution.
Directed evolution by random mutagenesis and recombination followed by screening or selection is a valuable tool for the engineering of enzymes (2, 3, 16, 61). Functional gene expression in a suitable host is a prerequisite for directed evolution. Considering transformation efficiency, stability of plasmid DNA, and growth rate, Escherichia coli and Saccharomyces cerevisiae are best suited for these experiments. Heterologous expression in these hosts, however, is often limited by differences in the expression systems from the native organism (50). Different codon usage, missing chaperones, and posttranslational modifications such as disulfide bridges or glycosylation can all cause low expression levels and misfolded proteins that are degraded or driven into inclusion bodies (20). Finding the bottlenecks of a specific expression system requires consideration of many possibilities whose impact is hard to predict. Some incompatibilities between the expressed gene and heterologous host, such as codon usage or the recognition of signal sequences, can be overcome by changing the gene sequence. Thus, achieving functional expression is a good target for directed evolution (13, 42, 43).
Laccases, like other ligninolytic enzymes, are notoriously difficult to express in nonfungal systems. The laccase from Myceliophthora thermophila (MtL) used in this work was previously expressed only in Aspergillus oryzae (6). Expression in S. cerevisiae has been reported for other laccase genes (11, 33, 34, 60). Kojima et al. demonstrated expression of laccase from Coriolus hirsutus qualitatively with a plate assay (33). Larsson et al. reached an activity of 0.66 mU/ml after optimization of the fermentation conditions (34). Yasuchi et al. report a 5-mg/liter expression of C. hirsutus laccase (59). None of these laccases expressed in S. cerevisiae was purified or characterized. Trametes versicolor laccase was expressed in Pichia pastoris, but neither the expression level nor characterization of the recombinant protein was published (30). In a similar approach, 11 mg of T. versicolor laccase/liter was produced, but the specific activity was extremely low (0.65 U/mg for the untruncated version) (19) compared to the same enzyme expressed in fungus (40 U/mg) (57). Laccase from Pycnoporus cinnabarinus was expressed in P. pastoris to a level of 8 mg/liter. The recombinant enzyme was not isolated or further characterized (45).
Laccases belong to the family of blue copper proteins, together with ascorbate oxidase and ceruloplasmin (40). Laccases are monomeric glycoproteins (23) that catalyze the four-electron reduction of oxygen to water with one-electron oxidation of substrate without producing peroxide (27, 28). Important applications for laccase are pulp bleaching and delignification for the paper industry (4, 7, 10, 53) and biosensors for phenols or oxygen (22, 36). Degradation of polycyclic aromatic hydrocarbons (29, 39) and the cathodic reaction in biofuel cells (31) are also potential applications. Improvement and realization of these applications will require tailored catalysts. We report here the functional expression of MtL in S. cerevisiae and improvement of catalysis and expression by directed evolution.
MATERIALS AND METHODS
Materials.
All chemicals were reagent-grade purity. 2,2′-Azino-bis(3-ethylbenz-thiazoline-6-sulfonic acid) (ABTS) was purchased from Sigma (St. Louis, Mo.). Taq DNA polymerase was from GIBCO BRL (Grand Island, N.Y.). Polymerase Pfu and E. coli XL2-Blue competent cells were from Stratagene (La Jolla, Calif.). Plasmid pJRoC30 containing the wild-type gene of MtL was kindly provided by Novozymes (Davis, Calif.). Yeast cells transformed with this vector can grow on selective media (media without uracil). The shuttle vector also includes the E. coli ampicillin resistance gene. S. cerevisiae strain BJ5465 was obtained from the Yeast Genetic Stock Center (University of California, Berkeley). MtL expressed in A. oryzae was provided by Novozymes. The Gietz Lab yeast transformation kit was purchased from Tetra-Link (Amherst, N.Y.), and the yeast plasmid miniprep kit was from Zymo Research (Orange, Calif.). Minimal medium contained 50 ml of 20% sterile raffinose, 5 ml of 0.25% l-His, 5 ml of 0.25% l-Trp, 5 ml of 0.25% l-Leu, 10 ml of 0.25% adenine hemisulfate, 150 ml of double-distilled H2O (ddH2O), and 25 ml of 6.7% yeast nitrogen base. Expression medium contained 325 ml of YP medium, 25 ml of 1 M KPi buffer (pH 6.0), 2 μl of sterile 1 M CuSO4, 50 ml of 20% sterile galactose, 400 μl of chloramphenicol stock solution, and ddH2O to 400 ml. YP medium contained 10 g of yeast extract, 20 g of tryptone-peptone, and ddH2O to 650 ml. YPAD solution contained 10 g of yeast extract, 20 g of peptone-tryptone, 100 ml of 20% sterile glucose, 100 mg of adenine hemisulfate, and ddH2O to 1,000 ml. SC-drop-out plates contained 6.7 g of yeast nitrogen base, 50 mg of l-His, 50 mg of l-Trp, 50 mg of l-Leu, 50 mg of adenine hemisulfate, 15 g of Bacto agar, 100 ml of 20% sterile glucose, 1 ml of chloramphenicol stock solution, and ddH2O to 1,000 ml. Britton and Robinson (B&R) buffer contained 0.1 M boric acid, 0.1 M acetic acid, and 0.1 M phosphoric acid and was adjusted to the desired pH with 0.5 M NaOH.
Production of MtL in S. cerevisiae.
S. cerevisiae clones carrying wild-type or mutant laccase plasmids were grown in 50 ml of preculture in minimal medium. After 36 h at 30°C, 270 rpm, 450 ml of expression medium was inoculated with the preculture in a 2.8-liter baffled flask. After further incubation for 24 h at 30°C, 270 rpm, the optical density reached 30 and the laccase activity was maximal. The cells were separated by centrifugation at 1,000 × g, 4°C, and the supernatant was filtered (0.45-μm pore size) and concentrated to 10 ml in an ultrafiltration cell (Amicon; Millipore, Bedford, Mass.) equipped with a 30-kDa-cutoff membrane. A 350-ml aliquot of buffer A (Tris-HCl, 20 mM, pH 7.8) was added and the sample was concentrated to 10 ml.
Purification of MtL.
MtL was purified as described elsewhere (57) with minor modifications. The concentrated and washed culture broth was loaded onto a Q-Sepharose HiTrap (5 ml; Amersham Pharmacia, Piscataway, N.J.) preequilibrated with buffer A. The column was washed with buffer A, and laccase activity was eluted in a linear gradient of 0 to 400 mM NaCl. Active fractions were pooled, desalted, and concentrated in an ultrafiltration cell. The concentrate in buffer A was loaded onto a MonoQ column (1 ml; Amersham Pharmacia) preequilibrated with buffer A. The column was washed with buffer A and laccase activity was eluted in a linear gradient of 0 to 1 M NaCl. Active fractions were pooled, desalted, and concentrated in an ultrafiltration cell.
Analysis of glycosylation, molecular mass, and protein concentration.
Purified protein samples were deglycosylated using peptide-N-glycosidase F (PNGaseF) (New England Biolabs). The protein was denatured using 5% sodium dodecyl sulfate (SDS) and 10% β-mercaptoethanol at 100°C for 10 min and incubated with PNGaseF (30 U/μl) for 3 h. The reaction product was analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). SDS-PAGE was done on a commercial apparatus (Novex precast gels; Invitrogen, Carlsbad, Calif.). Proteins were stained with Coomassie brilliant blue. Protein concentrations of purified enzymes were measured using their absorption at 280 nm. The molar absorption coefficient was calculated from the primary sequence (46).
DNA sequencing.
To locate mutations on the MtL gene, plasmids containing mutant genes were sequenced at the Caltech DNA Sequencing Core Facility, using Perkin-Elmer Applied Biosystems 373XL and 377XL automated fluorescent DNA sequencers (Perkin-Elmer Biosystems, Foster City, Calif.). The primers used were the following: sense primers RMLN (5′-CCTCTATACTTTAACGTCAAGG-3′; binds at bp 5′-160 to 180-3′ of pJROC30) and mtlsq2 (5′-GAAGGGCACCAACCTGC-3′; binds at bp 5′-643 to 659-3′ of pJROC30); and antisense primers mtlsq3 (5′-GCACGTAAAAGTCGTGG-3′; binds at bp 5′-1657 to 1673-3′ of pJROC30) and RMLC (5′-GGGAGGGCGTGAATGTAAGC-3′; binds at bp 5′-2139 to 2158-3′ of pJROC30).
Error-prone PCR.
Libraries of MtL mutants were generated by error-prone PCR (35) with a 90 nM concentration of each primer (RMLC and RMLN), 1 ng of template/μl, 0.2 mM ATP, 0.2 mM GTP, 0.6 mM CTP, 0.6 mM TTP, 3% dimethyl sulfoxide (DMSO), 0.75 mM MgCl2, 0 to 0.2 mM MnCl2, and 50 U of Taq polymerase/ml. Thermal cycling parameters were 95°C for 2 min (1 cycle), 94°C for 0.45 min, 53°C for 0.45 min, 74°C for 3 min (28 cycles), and 74°C for 10 min (1 cycle). The PCR products were purified using the QIAquick PCR purification kit (QIAGEN, Valencia, Calif.). For transformation of DNA fragments containing the MtL gene, it is necessary to linearize the plasmid and remove the MtL gene. This was done by cutting plasmid pJROC30 with restriction enzymes XhoI and BamHI and purifying the products on a preparative agarose gel followed by an agarose gel extraction (QIAEX II; QIAGEN). PCR products were mixed with linearized vector and transformed into competent yeast cells using the Gietz Lab transformation kit (Tetra Link). The error rate was checked by functional screening. In generation 9, Mutazyme (Stratagene) was used instead of Taq polymerase. The PCR conditions were adapted from the GeneMorph instruction manual, with the exception that 3% DMSO was included. Two target concentrations were used to achieve different mutation frequencies. For the low mutation rate library, we used 4 ng of template/μl, and for the high mutation rate library we used 4 pg of template/μl.
C-terminal truncation.
The gene of the best mutant of generation 7, 37A7, was mutagenized to incorporate a stop codon at the position of the C-terminal protease cleavage site. For the four-primer method the following primers were used: 4prcod (5′-CCAAGTCCGACTCGGGCCTCTAGCGCCGCTGGG-3′) and 4prgen (5′-CCACTCGCCCTCCTCGACCCAGCGGCGCTAGAGGCCCGAG-3′). (Bases that mismatch with the template are in boldface.) PCR was done with RMLN/4prgen and 4prcod/RMLC. PCR conditions were as described for site-directed recombination (see below). PCR products were gel purified, and 1 μl of each was used in the second PCR with RMLN and RMLC without template under the previous PCR conditions, but with a 3-min elongation time. The product of the second PCR was gel purified, mixed with open vector, and transformed into S. cerevisiae.
Homologous recombination in yeast.
For the first in vivo shuffling experiment (generation 2), mutant MtL genes were cut out of the vector with AseI. These fragments of the four best mutants of the first error-prone PCR were mixed equimolarly and transformed into yeast together with the linearized vector. The fragments contained 188 bp of homology at the 5′ end and 282 bp of homology on the 3′ end. In the sixth, seventh, and eighth generations, primers RMLC and RMLN were used to produce PCR products with 35-bp (5′) and 66-bp (3′) overhangs with the linearized vector.
Site-directed recombination.
In the first recombination the template was 6D6, and adjacent sense and antisense primers for the mutation sites were synthesized that were 50% mutated at the mutation sites. Together with the primers RMLN and RMLC, which bind outside of the gene, three PCR products were generated with homologous sequences on each end. Those were assembled by transformation into yeast with the open vector, yielding a library of all combinations of the mutations. The primers were the following: 6D6sense1 (5′-GCTGGTTAACAATRGTATARTCGGACCAACAATCTTTGC-3′), 6D6antisense1 (5′-GCAAAGATTGTTGGTCCGAYTATACYATTGTTAACCAGC-3′), 6D6sense2 (5′-CGTCCTCACGCAGARCACCAGCTTCCCACCCGGGYACAACATTGTCG-3′), and 6D6antisense2 (5′-CGACAATGTTGTRCCCGGGTGGGAAGCTGGTGYTCTGCGTGAGGACG-3′). PCRs were done with 6D6sense1 and -antisense2, sense2 and RMLC, and RMLN and antisense1. For a 50-μl PCR mixture, we used a 90 nM concentration of each primer, 250 pg of template/μl, 0.3 mM deoxynucleoside triphosphates, 3% DMSO, and 40 U of Pfu-turbo/ml. Thermal cycling parameters were 95°C for 2 min (1 cycle), 94°C for 0.45 min, 55°C for 0.45 min, 74°C for 2 min (28 cycles), and 74°C for 10 min (1 cycle). In the second recombination, 8H9 was the parent and the primers were the following: n20sense (5′-CATTCTCACCCCTAGCGTTCCTGCTGCCCCTCCATCC-3′), n20antisense (5′-GATGGAGGGGCAGCAGGAACGCTAGGGGTGAGAATG-3′), 65sense (5′-GCTGGTTAACAATAGTATAATCGGACCAACAATCTTTGC-3′), 65antisense (5′-GCAAAGATTGTTGGTCCGATTATACTATTGTTAACCAGC-3′), 303sense (5′-CTGCGGCGGCTCCAGGAGTCCCTACCCGGCCGCCATC-3′), 303antisense (5′-GATGGCGGCCGGGTAGGGACTCCTGGAGCCGCCGCAGAGC-3′), 396sense (5′-CGTCCTCACGCAGAACACCAGCTTCCCACCCGGGCACAACATTG-3′), 396antisense (5′-GGTGGGAAGCTGGTGTTCTGCGTGAGGACG-3′), 552sense (5′-GCTACTGGCCTACCARCCCCWACCCCAAGTCCGACTCGGGCCTC-3′), 552antisense(5′-CGAGTCGGACTTGGGGTWGGGGYTGGTAGGCCAGTAGCGGCGCCAG-3′), RMLN −382 (5′-CTGAAACGCAGATGTGCCTCG-3′), and RMLC +314 (5′-CGTTGGCCGATTCATTAATGC-3′). PCR mixtures were made with RMLN −382 and n20antisense, n20sense and 65antisense, 65sense and 303antisense, 303sense and 396antisense, 396sense and 552antisense, and 552sense and RMLC +314. The PCR conditions were the same as in recombination 1.
Preparation of libraries and screening (9).
Subsequent to error-prone PCR, in vivo shuffling or staggered extension process (StEP) recombination, the mutated and recombined libraries were used to transform S. cerevisiae BJ5465. Yeast transformations were carried out with a modified LiAc method as described previously (24, 25). The cells were plated on SC-drop-out plates by using glass beads (6 mm; Sigma) and incubated at 30°C for 3 to 4 days to recover transformants. Single colonies were transferred into 96-well plates containing 25 μl of minimal medium per well by using a picking robot (Qpix; Genetix). Plates were incubated at 30°C, 270 rpm for 24 h in a humidity shaker (ISF-1-W; Kuhner). One row per plate was inoculated with wild type or parent; one well was not inoculated (control). After 24 h, 80 μl of expression medium was added to each well and the plates were incubated at 30°C, 270 rpm for 24 h in the humidity shaker. The plates were then centrifuged for 5 min at 1,500 × g, 4°C. Twenty microliters of supernatant was transferred to a new 96-well plate, using a 96-channel pipetting station (Multimek; Beckman, Fullerton, Calif.). A 180-μl aliquot of assay solution (final concentrations in the well, 3 mM ABTS, 5% polyethylene glycol 6000 [PEG 6000], 50 mM B&R buffer; pH 6) was added to each well. Initial activity was calculated from the measurement of the absorption change at 418 nm at 25°C on a plate reader (Spectra max Plus 384; Molecular Devices, Sunnyvale, Calif.). As a first rescreen, aliquots of the best clones (approximately 50) were used to inoculate 20 μl of minimal medium in 96-well plates. Five wells on the microtiter plates were inoculated with the same clone. Five wells on each plate were used for the standard. The screening procedure was the same as above. For the second rescreen, plasmids were extracted from the most active clones (Zymoprep yeast plasmid miniprep; Zymo Research, Orange, Calif.). The extracted DNA was very dilute and impure. Therefore, the plasmids were transformed into competent E. coli cells. Plasmids were extracted (QIAprep spin miniprep kit; QIAGEN), and S. cerevisiae was transformed and screened as described above.
Laccase activity assays (1).
Syringaldazine (5.5 mM in dimethylformamide) oxidation was performed in B&R buffer (50 mM, pH 6.0) at 25°C, monitoring the change of absorption at 530 nm with an extinction coefficient of 65 mM−1 cm−1 (5) to calculate the rate of oxidation. ABTS oxidation was done with 3 mM ABTS in B&R buffer (50 mM; pH 6) including PEG 6000 (5% wt/vol) by monitoring the change in absorption at 418 nm with an extinction coefficient of 36 mM−1 cm−1 (12). For samples with low laccase activity, an endpoint assay was used. One unit is defined as the amount of enzyme that oxidizes 1 μmol of substrate per min under the assay conditions. For pH activity measurements, 0.5 to 1.5 mU of MtL mutants and wild types were assayed for ABTS oxidation activity in B&R buffer adjusted to pH 2, 3, 4, 5, 6, 7, 8, and 9. Stability at pH 3 was assessed by incubating 0.12 to 0.36 U of MtL/ml in 10 mM B&R buffer. After 10, 15, and 30 min, 4-μl samples were diluted fivefold in a 96-well plate and ABTS oxidation activity was measured. For thermostability measurements, 0.13 to 0.39 U of MtL/ml in B&R buffer (10 mM, pH 6) was incubated at various temperatures between 23 and 80°C. After 0, 1, 7, and 24 h, 4-μl samples were diluted fivefold and ABTS activity was measured at 23°C.
RESULTS
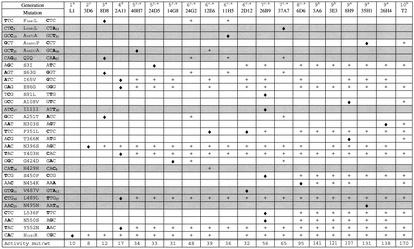
We expressed the gene of MtL in a protease-deficient strain of S. cerevisiae (BJ5465) (48). Transformants secreted a low but significant laccase activity of 0.6 U/liter. A high-throughput screen for laccase activity was optimized (9), and a coefficient of variation of less than 10% was achieved at the higher activity levels of later generations. For directed evolution of laccase activity, different mutagenesis and recombination methods were explored. A summary of mutations and the methods used in each generation is given in Fig. 1. Error-prone PCR (35) with Taq polymerase or Mutazyme was carried out at different mutagenic rates. Based on our previous experience, a rate corresponding to one to two amino acid changes per gene per generation was preferred in order to avoid accumulating neutral mutations, which might affect properties of the enzyme other than those leading to improved activity. Both in vitro StEP (60) and in vivo shuffling (49) were used for random recombination; in vivo shuffling was the easier and faster method (8). We found the highest increases in total activity when we recombined error-prone PCR products to introduce new mutations simultaneously with recombination. To eliminate neutral mutations, the best mutant of the first generation was included during recombination for backcrossing. The length of the homologous overhangs was optimized, and the best compromise between recombination rate and transformation efficiency was to use 30 to 70 bp of homology. These conditions created the best mutant of generation 6 (24G2), which showed 1.4 times higher activity than the parent 40H7, effectively recombining beneficial mutations from 8D8 and 14G8 (Fig. 1).
FIG. 1.
Mutations in the selected MtL mutants during the evolution of laccase total activity in S. cerevisiae. +, mutation; ⧫, new mutation. Grey indicates synonymous mutations; subscript numbers on codons indicate codon usage. Footnote symbols: a, error-prone PCR; b, in vivo gap repair; c, in vivo recombination; d, StEP.
A minimum distance between mutations is required in order for them to be recombined independently in random recombination experiments. PCR and in vivo gap repair (38, 44) were used to recombine neighboring mutations in a site-directed fashion. As with in vivo shuffling, this method takes advantage of the high level of homologous recombination in yeast, which makes in vivo approaches fast, efficient, and nonmutagenic. In generation 9, we separated two double-mutation sites, (65, 86) and (396, 403), in this way, eliminating two deleterious mutations (65 and 396) to generate 3A6 with 1.4-fold increased activity (Fig. 1). Site-directed recombination was also used in generation 10 to combine the stability of 8H9 with the increased activity of the activity mutants from generation 9. This experiment removed the mutations at 65 and 396, and the double mutation site at (550, 552) was analyzed by using primers with 50% wild-type sequence at these positions. Mutation 552 was eliminated in the best mutant T2 (Fig. 1), which retained the stability of parent 8H9 and further increased activity to 170-fold compared to that of the wild type. The specific activity of T2 (35 U/mg with syringaldazine) is comparable to the specific activity of wild-type MtL produced in A. oryzae (36 U/mg).
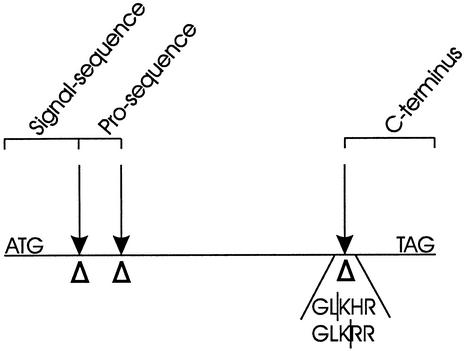
The sequence targeted for directed evolution included signal- and prosequences as well as the sequence of the C-terminal tail of the gene (Fig. 2), which code for parts of the protein that are cleaved during maturation. The signal sequence targets the gene for secretion, while the function of the other two sequences is not known (6). The single most beneficial mutation was His(c2)Arg, increasing total activity 10-fold in the first generation. Amino acid c2 is located in the C-terminal tail (Fig. 2). The exchange of the native His by an Arg introduces a cleavage site for the Kex2 protease (18, 32, 51). Since Kex2 is present in S. cerevisiae, this mutation likely adjusts the sequence to the different protease specificity in the heterologous host.
FIG. 2.
Positions of the mutations in the most active laccase (T2), relative to the processing sites of the MtL protein. The mutations in T2 are A(n20)P, S3I, and H(c2)R. The open triangles mark the positions of the mutations. The arrows mark the protease cleavage sites. For mutant H(c2)R and the wild type, sequences are given. The | indicates the cleavage position.
Comparison of the kinetics of the wild type and the mutant (L1) revealed that a 4.5-fold increase in specific activity was mainly responsible for the improved total activity, demonstrating the importance of processing in the MtL C terminus for enzyme activity. Since the function of the C terminus might be restricted to expression of the enzyme in M. thermophila, we found it worthwhile to express a C-truncated laccase. A stop codon was introduced at the proposed processing site by site-directed mutagenesis. The catalytic efficiency (kcat/Km) of the resulting enzyme (4P) was reduced 10-fold, almost to the value measured for wild type. 4P retained the high secretion level of its parent, 37A7 (Table 1). The C terminus as well as its processing are apparently important for optimizing the activity of MtL.
TABLE 1.
Comparison of kinetic parameters of mutants and wild-type MtL expressed in S. cerevisiae and wild type expressed in A. oryzae
| Clone | Substrate | Km (mM) | kcat (min−1) | kcat/Km (min−1 mM−1) | Total activitya | Improvement
|
|
|---|---|---|---|---|---|---|---|
| kcat | Expressionb | ||||||
| wt (fungus) | ABTS | 290 +/− 30 | 790 +/− 30 | 2.7 | |||
| Syringaldazine | 1.6 +/− 1 | 2,120 +/− 150 | 1,300 | ||||
| wt (yeast) | ABTS | 270 +/− 30 | 80 +/− 2.5 | 0.3 | 1 | 1 | 1 |
| Syringaldazine | 1.5 +/− 0.3 | 80 +/− 3.5 | 52 | 1 | 1 | ||
| L1, generation 1 | ABTS | 230 +/− 16 | 360 +/− 8 | 1.5 | 10 | 4.6 | 2.2 |
| Syringaldazine | 1.3 +/− 0.3 | 355 +/− 19 | 273 | 10 | 4.5 | ||
| 37A7, generation 7 | ABTS | 200 +/− 35 | 800 +/− 39 | 4 | 65 | 10 | 6.5 |
| Syringaldazine | 1 +/− 0.2 | 690 +/− 35 | 690 | 56 | 8.6 | ||
| 4P, generation 7 | ABTS | 300 +/− 30 | 125 +/− 5 | 0.4 | 11 | 1.6 | 6.9 |
| Syringaldazine | 1.6 +/− 0.5 | 135 +/− 11 | 84 | 12 | 1.7 | ||
| 8H9, generation 9 | ABTS | 190 +/− 30 | 910 +/− 38 | 4.8 | 107 | 12 | 9.3 |
| Syringaldazine | 2 +/− 0.3 | 1,220 +/− 64 | 610 | 141 | 15 | ||
| 3A6, generation 9 | ABTS | 190 +/− 10 | 1,880 +/− 37 | 9.9 | 141 | 24 | 5.9 |
| Syringaldazine | 1.9 +/− 0.9 | 2,040 +/− 130 | 1,070 | 150 | 25 | ||
| 3E3, generation 9 | ABTS | 170 +/− 20 | 1,220 +/− 36 | 7.2 | 121 | 16 | 7.8 |
| Syringaldazine | 2 +/− 0.3 | 1,420 +/− 73 | 710 | 138 | 18 | ||
| T2, generation 10 | ABTS | 290 +/− 15 | 1,740 +/− 34 | 6 | 170 | 22 | 7.7 |
| Syringaldazine | 4 +/− 1.1 | 2,210 +/− 200 | 550 | 212 | 27 | ||
Total activity for ABTS was measured in supernatants of cultures grown in 96-well plates. Numbers are averages of quintuplet measurements. Total activity for syringaldazine as substrate was calculated as the product from the increase in expression and the increase in kcat for syringaldazine.
The improvement in expression is the difference calculated between total activity increase and increase in kcat for ABTS as substrate.
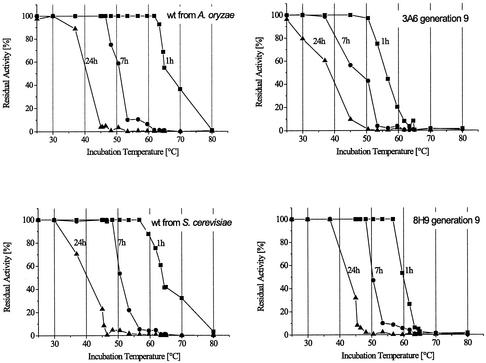
The stability of wild-type MtL expressed in S. cerevisiae is similar to the same enzyme expressed in A. oryzae (Fig. 3). After nine generations of activity evolution, the stability of the mutants was significantly reduced, as shown for 3A6. The highest temperature at which mutant 3A6 retains full activity was reduced by 10°C for all incubation times, compared to that for the wild type expressed in yeast. To recover this important property, a stability screen was introduced in generation 9. The residual activity of the mutants was measured after incubation at 37°C for 30 h. Mutant 8H9 showed about the same total activity as the parent 6D6, but its residual activity after incubation was 2.3-fold higher than the residual activity measured for 6D6.
FIG. 3.
Thermostability of wild-type MtL expressed in the yeast S. cerevisiae and in A. oryzae, and of two mutants found in generation 9. Mutant 8H9 was discovered with the stability screen, and 3A6 was discovered with the activity screen. Tubes containing 50 μl of B&R buffer (10 mM; pH 6) containing 0.3 U of enzyme activity/ml were incubated at various temperatures and assayed for residual activity after 0, 1, 7, and 24 h at 23°C with ABTS as substrate. The activities were normalized to the initial activity at 23°C before the incubation.
Activity (kcat) of wild-type MtL with ABTS is 10-fold lower if the enzyme is expressed in S. cerevisiae instead of A. oryzae (Table 1). For the phenolic substrate syringaldazine, kcat is 25-fold lower. Evolution for higher total activity increased kcat 22 and 27 times, respectively. Improvement of the catalytic activity was very similar for both substrates and higher for syringaldazine, although we screened for activity with ABTS as substrate. Thus, the activity improvement is not substrate dependent, and it is likely that the substrate specificity of MtL and its conversion of typical laccase substrates like syringaldazine has not changed. The Km values of most mutants are similar to that of the wild type. The stability mutant 8H9 has a relatively low kcat; the improvement in total activity is due to a ninefold-higher expression level. It may be that the improved stability allows more enzyme to be secreted, as has been observed for other proteins expressed in yeast (52).
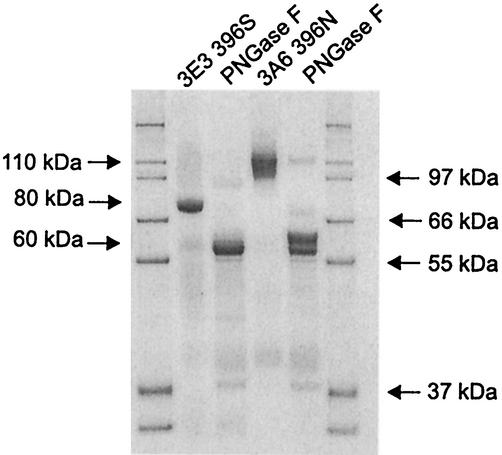
The molecular mass of MtL expressed in S. cerevisiae was 110 kDa, as determined by SDS-PAGE (Fig. 4), which is about 30% higher than the molecular mass reported for MtL expressed in M. thermophila and A. oryzae. S. cerevisiae is known to hyperglycosylate N-glycosylation sites. When the N-glycosylation site at position 396 is mutated (N396S), the mass of the mutant is reduced to 80 kDa (Fig. 4, cf. 3A6 and 3E3, which only differ in the N396S mutation). This 30-kDa decrease corresponds to loss of a 120-monomer glycan at that site. Deglycosylation of any mutant reduces the molecular mass to 55 to 60 kDa, the molecular mass reported for the deglycosylated enzyme expressed in fungus (6) (Fig. 4). The appearance of additional bands in lane 5 of the SDS-PAGE might be due to incomplete digestion by PNGaseF. The kinetic parameters show the influence of glycosylation on catalysis (Table 1). 3A6 secretes 1.2-fold more laccase activity in culture supernatants. This results from a 1.5-fold higher kcat of the glycosylated mutant, which is expressed 1.3-fold lower than 3E3.
FIG. 4.
SDS-PAGE of two mutants from generation 9. 3E3 is identical to 3A6 except for mutation N396S. The purified enzymes were deglycosylated using the N-glycosidase PNGaseF. Samples were analyzed before and after deglycosylation. The gel was stained with Coomassie brilliant blue.
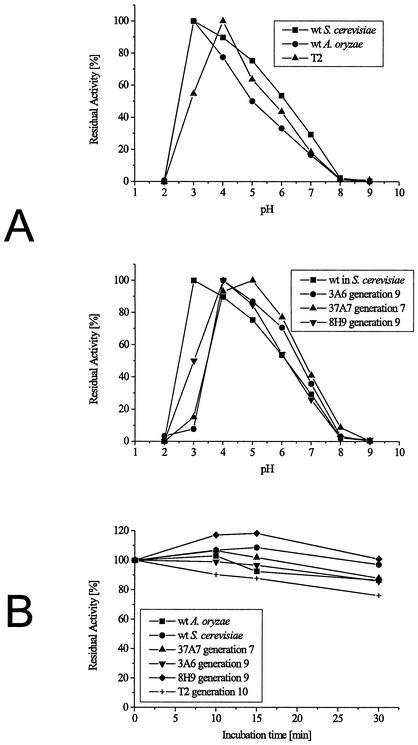
The activities of the wild type and mutants expressed in fungus and in yeast at different pHs were compared (Fig. 5A). Wild type expressed in S. cerevisiae is less sensitive to changes in pH than wild type expressed in A. oryzae. The wild-type optimum at pH 3 is shifted to pH 5 in 37A7 of generation 7. All mutants found in later generations are most active at pH 4. Activity at pH 3 is almost completely lost in the activity mutants of generations 7 and 9. The stability mutant of generation 9 and the derived T2 regain 50% of the wild-type activity at pH 3. This increase is not due to improved stability under these acidic conditions, which can be seen from the stability measurements at pH 3 (Fig. 5B). 8H9 is most stable under these conditions, but T2 is the least stable, and in general the loss in activity during the incubation time of the pH activity measurement of any of the enzymes tested is negligible. Differences in pH activity between wild type and the evolved enzymes of generations 9 and 10 are minor. The low activity at pH 3 is not significant for current applications of this enzyme (e.g., pH for laccases in pulp processing range from 4 to 6). For future applications for which pH 3 might be required, the stability of the enzyme under these conditions can be improved by directed evolution. The stability of T2 at pH 6 is not lower than the stability of 8H9 at that pH (data not shown). Only stability at pH 3 is influenced negatively by the combined introduction of five mutations in generation 10. This result underscores the fact that all properties not addressed in the screen (e.g., stability at pH 3) will drift. Since most mutations are deleterious, these properties are likely to be compromised.
FIG. 5.
(A) pH-activity profiles of wild-type (wt) MtL expressed in S. cerevisiae, wt expressed in A. oryzae, and of mutants 37A7 (generation 7), 8H9 (stability mutant, generation 9), 3A6 (activity mutant, generation 9), and T2 (generation 10). Activities were measured in B&R buffer at different pHs with ABTS as substrate. Laccase activity is normalized to the optimum activity value of the enzymes. (B) Stability of MtL wild type and mutants at pH 3. Enzyme samples (0.3 U/ml) were incubated in B&R buffer, pH 3, and residual activity at pH 6 was measured with ABTS as substrate. The activities were normalized to the initial activity at pH 6 before the incubation.
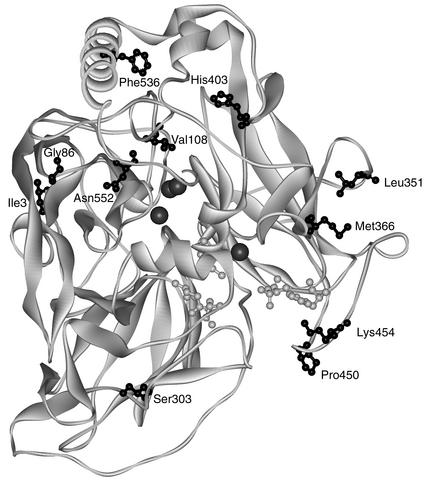
The structure of a laccase from Melanocarpus albomyces (MaL) was published recently (26). As an ascomycete laccase like MtL, its sequence identity to MtL is high (76%). We used this structure to build a model for MtL and map the locations of the mutations (Fig. 6). Most of the mutations found in the best mutant T2 are on the surface and in loop regions, where they are likely to not disrupt the three-dimensional structure (54). Apart from H403 (13 Å) and V108 (6 Å), the mutations are more than 15 Å away from the copper atoms. All mutations but one (K454; 3 Å) are further than 10 Å from the residues believed to participate in substrate binding (26).
FIG. 6.
Model of MtL mutant T2 derived from the structure of MaL. Mutations are in black ball and stick display. Residues presumably participating in substrate binding (26) are in grey ball and stick display. Copper atoms are shown as grey spheres. The model was created using DeepView and the Swiss-Model server.
DISCUSSION
We used directed evolution to increase functional expression of the laccase gene from M. thermophila in S. cerevisiae. We employed 10 generations and screened 20,000 clones to convert an activity level that was originally detectable only using a very sensitive assay system to the highest level yet reported for the expression of a laccase in yeast. The molecular masses of the purified and deglycosylated mutants are comparable to that of the wild-type enzyme produced in M. thermophila (55 to 60 kDa) (6) (Fig. 4). Thus, the maturation of the recombinant protein in S. cerevisiae seems to be similar to the process in M. thermophila, although this would have to be proven by protein sequencing. Three of the 13 amino acid substitutions found in the best mutant of the last generation are located at the three processing sites of MtL (Fig. 2). We believe these mutations adjust the MtL sequence to the different specificities of the S. cerevisiae proteases compared to those of M. thermophila. MtL processing at its C terminus is similar to that described for Neisseria crassa laccase (21); C-terminal processing was also postulated for Podospora anserina laccase (17). The C-terminal sequence of N. crassa is similar to that found in MtL. The importance of this processing for the catalytic activity is unknown. It was suggested that the highly basic C-terminal sequence functions as an inhibitor by interacting with the active center, preventing copper from binding (21). Whereas N-terminal processing and glycosylation may be required for secretion, C-terminal processing may play a role in activation of the enzyme. Supporting this hypothesis, the mutation H(c2)R resulted in an almost fivefold improvement of kcat. The dramatic loss in activity of the C-truncated mutant showed that the C terminus, or the inactivation of the enzyme caused by it, is essential for early posttranslational processing steps.
Random mutagenesis at low error rates favors the discovery of beneficial mutations at structurally tolerant sites, which are more abundant on the surface and in loop regions of a protein. Changes in the closely packed interior and within secondary structural elements are more likely to be disruptive (54). Most mutations in MtL are far away from the copper atoms and from the substrate binding residues of MtL. The effectiveness of these mutations underscores the value of directed evolution experiments, which tend to find paths for improvement that cannot be anticipated and therefore are not found by rational design.
Most of the synonymous mutations, including the one in the best mutant, T2, are changes to a more frequently used codon (Fig. 1). Low-frequency codons can cause translation pauses, depending on their position and abundance, and therefore yeast codon bias is used for optimization of yeast expression systems (15). The majority of these third base changes reduce the GC content of the MtL gene, which is 65% for the whole gene and 90% for third bases. The high GC content is typical for enzymes from thermophiles and can cause stable secondary RNA structures that interfere with translation. Formation of stable secondary structures is likely the reason why successful PCR with this gene requires addition of DMSO.
Glycosylation is believed to influence laccase secretion, sensitivity to proteases, copper retention, and thermal stability (37). Laccase from P. cinnabarinus expressed in P. pastoris had an increase in molecular mass similar to that reported here. The recombinant protein had a molecular mass of 110 kDa, which was 30% higher than expected (81 kDa), presumably due to hyperglycosylation (45). The location of the glycosylation and its effects on kinetics and expression were not determined. Complex and outer-chain carbohydrates are added in the Golgi. The size of the glycan that was introduced at position 396 in this study indicates that the residence time of the protein in this compartment is long (51) and, therefore, the transit of the protein from the Golgi to the cell membrane and its exocytosis may be bottlenecks for expression of MtL in S. cerevisiae.
Laccase is a good target for directed evolution approaches because knowledge of structure-function relations underlying their key properties is very limited. We chose to work with the laccase gene from M. thermophila because it is thermophilic, which is an advantage for applications in pulp bleaching processes (14, 41) and bioremediation. The relatively low electrochemical potential of MtL can be a starting point for the generation of mutants with increased potential because there are natural laccases that realize higher potentials with very similar copper centers (55). For most applications, laccase activity requires the presence of mediators. One example is electron transfer in biofuel cell applications. Direct electron transfer from the electrode is less efficient than transfer via a mediator (58). For the application of laccase in the cathodic compartment of a biofuel cell, the rate of direct electron transfer is insufficient (31, 47). Mediators, however, are mostly toxic, instable, or expensive. Moreover, they lead to side products and inactivate the enzyme (37). To increase the stability of laccase in the presence of mediators or to raise its activity without mediators would be suitable goals for directed evolution. The activity of laccase towards most substrates is a function of the difference in electrochemical potential between enzyme and substrate (56). Mutations that change the electrochemical potential would be of interest for the understanding of intramolecular electron transfer in copper oxidases, and a higher oxidation potential would allow the conversion of a wide range of compounds that are poor substrates for the natural laccases or are not converted at all. The functional expression system described here enables mutagenesis studies to generate new variants of this promising catalyst.
Acknowledgments
This work was supported by the U.S. Office of Naval Research. We thank the Ministerio de Educacion y Cultura of Spain (M.A.) and Deutsche Forschungsgemeinschaft (T.B., V.S.) for fellowships.
REFERENCES
- 1.Alcalde, M., and T. Bulter. Colorimetric assays for screening laccases. In F. H. Arnold and G. Georgiou (ed.), Directed evolution, vol. 2. Methods in molecular biology series. Humana Press, Totowa, N.J., in press. [DOI] [PubMed]
- 2.Arnold, F. H. 2001. Combinatorial and computational challenges for biocatalyst design. Nature 409:253-257. [DOI] [PubMed] [Google Scholar]
- 3.Arnold, F. H., and A. A. Volkov. 1999. Directed evolution of biocatalysts. Curr. Opin. Chem. Biol. 3:54-59. [DOI] [PubMed] [Google Scholar]
- 4.Bajpai, P. 1999. Application of enzymes in the pulp and paper industry. Biotechnol. Prog. 15:147-157. [DOI] [PubMed] [Google Scholar]
- 5.Bauer, R., and C. O. Rupe. 1971. Use of syringaldazine in a photometric method for estimating “free” chlorine in water. Anal. Chem. 43:421-425. [Google Scholar]
- 6.Berka, R. M., P. Schneider, E. J. Golightly, S. H. Brown, M. Madden, K. M. Brown, T. Halkier, K. Mondorf, and F. Xu. 1997. Characterization of the gene encoding an extracellular laccase of Myceliophthora thermophila and analysis of the recombinant enzyme expressed in Aspergillus oryzae. Appl. Environ. Microbiol. 63:3151-3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bourbonnais, R., M. G. Paice, B. Freiermuth, E. Bodie, and S. Borneman. 1997. Reactivities of various mediators and laccases with kraft pulp and lignin model compounds. Appl. Environ. Microbiol. 63:4627-4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bulter, T., and M. Alcalde. Preparing libraries in Saccharomyces cerevisiae. In F. H. Arnold and G. Georgiou (ed.), Directed evolution, vol. 1. Methods in molecular biology series. Humana Press, Totowa, N.J., in press. [DOI] [PubMed]
- 9.Bulter, T., M. Alcalde, and V. Sieber. Screening mutant libraries in Saccharomyces cerevisiae. In F. H. Arnold and G. Georgiou (ed.), Directed evolution, vol. 2. Methods in molecular biology series. Humana Press, Totowa, N.J., in press. [DOI] [PubMed]
- 10.Call, H. P., and I. Mucke. 1997. History, overview and applications of mediated lignolytic systems, especially laccase-mediator-systems (Lignozym® process). J. Biotechnol. 53:163-202. [Google Scholar]
- 11.Cassland, P., and L. J. Jonsson. 1999. Characterization of a gene encoding Trametes versicolor laccase A and improved heterologous expression in Saccharomyces cerevisiae by decreased cultivation temperature. Appl. Microbiol. Biotechnol. 52:393-400. [DOI] [PubMed] [Google Scholar]
- 12.Childs, R. E., and W. G. Bardsley. 1975. The steady-state kinetics of peroxidase with 2,2′-azino-di-(3-ethyl-benzthiazoline-6-sulphonic acid) as chromogen. Biochem. J. 145:93-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crameri, A., E. A. Whitehorn, E. Tate, and W. P. Stemmer. 1996. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 14:315-319. [DOI] [PubMed] [Google Scholar]
- 14.de Carvalho, M. E. A., M. C. Monteiro, and G. L. Sant'Anna. 1999. Laccase from Trametes versicolor: stability at temperature and alkaline conditions and its effect on biobleaching of hardwood kraft pulp. Appl. Biochem. Biotechnol. 77-79:723-733. [DOI] [PubMed] [Google Scholar]
- 15.Dix, D. B., and R. C. Thompson. 1989. Codon choice and gene expression: synonymous codons differ in translational accuracy. Proc. Natl. Acad. Sci. USA 86:6888-6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Farinas, E. T., T. Bulter, and F. H. Arnold. 2001. Directed enzyme evolution. Curr. Opin. Biotechnol. 12:545-551. [DOI] [PubMed] [Google Scholar]
- 17.FernandezLarrea, J., and U. Stahl. 1996. Isolation and characterization of a laccase gene from Podospora anserina. Mol. Gen. Genet. 252:539-551. [DOI] [PubMed] [Google Scholar]
- 18.Gabrielsen, O. S., S. Reppe, O. Saether, O. R. Blingsmo, K. Sletten, J. O. Gordeladze, A. Hogset, V. T. Gautvik, P. Alestrom, T. B. Oyen, and K. M. Gautvik. 1990. Efficient secretion of human parathyroid hormone by Saccharomyces cerevisiae. Gene 90:255-262. [DOI] [PubMed] [Google Scholar]
- 19.Gelo-Pujic, M., H. H. Kim, N. G. Butlin, and G. T. Palmore. 1999. Electrochemical studies of a truncated laccase produced in Pichia pastoris. Appl. Environ. Microbiol. 65:5515-5521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georgiou, G. 1996. Expression of proteins in bacteria, p. 101-126. In J. L. Cleland and C. S. Craik (ed.), Protein engineering: principles and practice. Wiley-Liss, Inc., New York, N.Y.
- 21.Germann, U. A., G. Muller, P. E. Hunziker, and K. Lerch. 1988. Characterization of 2 allelic forms of Neurospora crassa laccase amino-terminal and carboxyl-terminal processing of a precursor. J. Biol. Chem. 263:885-896. [PubMed] [Google Scholar]
- 22.Ghindilis, A. 2000. Direct electron transfer catalysed by enzymes: application for biosensor development. Biochem. Soc. Trans. 28:84-89. [DOI] [PubMed] [Google Scholar]
- 23.Gianfreda, L., F. Xu, and J.-M. Bollag. 1999. Laccases: a useful group of oxidoreductive enzymes. Bioremediation J. 3:1-25. [Google Scholar]
- 24.Gietz, R. D., and R. H. Schiestl. 1995. Transforming yeast with DNA. Methods Mol. Cell. Biol. 5:255-269. [Google Scholar]
- 25.Gietz, R. D., R. H. Schiestl, A. R. Willems, and R. A. Woods. 1995. Studies on the transformation of intact yeast cells by the LiAc/S-DNA/PEG procedure. Yeast 11:355-360. [DOI] [PubMed] [Google Scholar]
- 26.Hakulinen, N., L. L. Kiiskinen, K. Kruus, M. Saloheimo, A. Paananen, A. Koivula, and J. Rouvinen. 2002. Crystal structure of a laccase from Melanocarpus albomyces with an intact trinuclear copper site. Nat. Struct. Biol. 9:601-605. [DOI] [PubMed] [Google Scholar]
- 27.Huang, H. W., G. Zoppellaro, and T. Sakurai. 1999. Four-electron reduction of dioxygen by Rhus vernicifera laccase. J. Inorg. Biochem. 74:169.. [DOI] [PubMed] [Google Scholar]
- 28.Huang, H. W., G. Zoppellaro, and T. Sakurai. 1999. Spectroscopic and kinetic studies on the oxygen-centered radical formed during the four-electron reduction process of dioxygen by Rhus vernicifera laccase. J. Biol. Chem. 274:32718-32724. [DOI] [PubMed] [Google Scholar]
- 29.Johannes, C., A. Majcherczyk, and A. Huttermann. 1998. Oxidation of acenaphthene and acenaphthylene by laccase of Trametes versicolor in a laccase-mediator system. J. Biotechnol. 61:151-156. [Google Scholar]
- 30.Jonsson, L. J., M. Saloheimo, and M. Penttila. 1997. Laccase from the white-rot fungus Trametes versicolor: cDNA cloning of lcc1 and expression in Pichia pastoris. Curr. Genet. 32:425-430. [DOI] [PubMed] [Google Scholar]
- 31.Katz, E., B. Filanovsky, and I. Willner. 1999. A biofuel cell based on two immiscible solvents and glucose oxidase and microperoxidase-11 monolayer-functionalized electrodes. New J. Chem. 23:481-487. [Google Scholar]
- 32.Koikeda, S., K. Ando, H. Kaji, T. Inoue, S. Murao, K. Takeuchi, and T. Samejima. 1993. Molecular cloning of the gene for bilirubin oxidase from Myrothecium verrucaria and its expression in yeast. J. Biol. Chem. 268:18801-18809. [PubMed] [Google Scholar]
- 33.Kojima, Y., Y. Tsukuda, Y. Kawai, A. Tsukamoto, J. Sugiura, M. Sakaino, and Y. Kita. 1990. Cloning, sequence analysis, and expression of ligninolytic phenoloxidase genes of the white-rot basidiomycete Coriolus hirsutus. J. Biol. Chem. 265:15224-15230. [PubMed] [Google Scholar]
- 34.Larsson, S., P. Cassland, and L. J. Jonsson. 2001. Development of a Saccharomyces cerevisiae strain with enhanced resistance to phenolic fermentation inhibitors in lignocellulose hydrolysates by heterologous expression of laccase. Appl. Environ. Microbiol. 67:1163-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leung, D. W., E. Chen, and D. V. Goeddel. 1989. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique 1:11-15. [Google Scholar]
- 36.Li, J., S. Tan, and J. Oh. 1998. Silica sol-gel immobilized amperometric enzyme electrode for peroxide determination in the organic phase. J. Electroanal. Chem. 448:69-77. [Google Scholar]
- 37.Li, K. C., F. Xu, and K. E. L. Eriksson. 1999. Comparison of fungal laccases and redox mediators in oxidation of a nonphenolic lignin model compound. Appl. Environ. Microbiol. 65:2654-2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma, H., S. Kunes, P. J. Schatz, and D. Botstein. 1987. Plasmid construction by homologous recombination in yeast. Gene 58:201-216. [DOI] [PubMed] [Google Scholar]
- 39.Majcherczyk, A., C. Johannes, and A. Huttermann. 1998. Oxidation of polycyclic aromatic hydrocarbons (PAH) by laccase of Trametes versicolor. Enzyme Microb. Technol. 22:335-341. [Google Scholar]
- 40.Messerschmidt, A. 1993. Blue copper oxidases. Adv. Inorg. Chem. 40:121-185. [Google Scholar]
- 41.Monteiro, M., and M. de Carvalho. 1998. Pulp bleaching using laccase from Trametes versicolor under high temperature and alkaline conditions. Appl. Biochem. Biotechnol. 70-72:983-993. [DOI] [PubMed] [Google Scholar]
- 42.Morawski, B., Z. Lin, P. Cirino, H. Joo, G. Bandara, and F. H. Arnold. 2000. Functional expression of horseradish peroxidase in Saccharomyces cerevisiae and Pichia pastoris. Protein Eng. 13:377-384. [DOI] [PubMed] [Google Scholar]
- 43.Morawski, B., S. Quan, and F. H. Arnold. 2001. Functional expression and stabilization of horseradish peroxidase by directed evolution in Saccharomyces cerevisiae. Biotechnol. Bioeng. 76:99-107. [DOI] [PubMed] [Google Scholar]
- 44.Muhlrad, D., R. Hunter, and R. Parker. 1992. A rapid method for localized mutagenesis of yeast genes. Yeast 8:79-82. [DOI] [PubMed] [Google Scholar]
- 45.Otterbein, L., E. Record, S. Longhi, M. Asther, and S. Moukha. 2000. Molecular cloning of the cDNA encoding laccase from Pycnoporus cinnabarinus I-937 and expression in Pichia pastoris. Eur. J. Biochem. 267:1619-1625. [DOI] [PubMed] [Google Scholar]
- 46.Pace, C. N., F. Vajdos, L. Fee, G. Grimsley, and T. Gray. 1995. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 4:2411-2423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmore, G. T. R., and H. H. Kim. 1999. Electro-enzymatic reduction of dioxygen to water in the cathode compartment of a biofuel cell. J. Electroanal. Chem. 464:110-117. [Google Scholar]
- 48.Parekh, R., K. Forrester, and D. Wittrup. 1995. Multicopy overexpression of bovine pancreatic trypsin inhibitor saturates the protein folding and secretory capacity of Saccharomyces cerevisiae. Protein Expr. Purif. 6:537-545. [DOI] [PubMed] [Google Scholar]
- 49.Pompon, D., and A. Nicolas. 1989. Protein engineering by cDNA recombination in yeasts: shuffling of mammalian cytochrome P-450 functions. Gene 83:15-24. [DOI] [PubMed] [Google Scholar]
- 50.Romanos, M. A., C. A. Scorer, and J. J. Clare. 1992. Foreign gene expression in yeast: a review. Yeast 8:423-488. [DOI] [PubMed] [Google Scholar]
- 51.Schuster, J. R. 1991. Gene expression in yeast: protein secretion. Curr. Opin. Biotechnol. 2:685-690. [DOI] [PubMed] [Google Scholar]
- 52.Shusta, E. V., M. C. Kieke, E. Parke, D. M. Kranz, and K. D. Wittrup. 1999. Yeast polypeptide fusion surface display levels predict thermal stability and soluble secretion efficiency. J. Mol. Biol. 292:949-956. [DOI] [PubMed] [Google Scholar]
- 53.Srebotnik, E., and K. E. Hammel. 2000. Degradation of nonphenolic lignin by the laccase/1-hydroxybenzotriazole system. J. Biotechnol. 81:179-188. [DOI] [PubMed] [Google Scholar]
- 54.Voigt, C. A., S. L. Mayo, F. H. Arnold, and Z. G. Wang. 2001. Computational method to reduce the search space for directed protein evolution. Proc. Natl. Acad. Sci. USA 98:3778-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xu, F. 1997. Effects of redox potential and hydroxide inhibition on the pH activity profile of fungal laccases. J. Biol. Chem. 272:924-928. [DOI] [PubMed] [Google Scholar]
- 56.Xu, F. 1996. Oxidation of phenols, anilines, and benzenethiols by fungal laccases: correlation between activity and redox potentials as well as halide inhibition. Biochemistry 35:7608-7614. [DOI] [PubMed] [Google Scholar]
- 57.Xu, F., W. S. Shin, S. H. Brown, J. A. Wahleithner, U. M. Sundaram, and E. I. Solomon. 1996. A study of a series of recombinant fungal laccases and bilirubin oxidase that exhibit significant differences in redox potential, substrate specificity, and stability. Biochim. Biophys. Acta 1292:303-311. [DOI] [PubMed] [Google Scholar]
- 58.Yaropolov, A. I., O. V. Skorobogatko, S. S. Vartanov, and S. D. Varfolomeyev. 1994. Laccase: properties, catalytic mechanism, and applicability. Appl. Biochem. Biotechnol. 49:257-280. [Google Scholar]
- 59.Yasuchi, K., K. Yukio, and T. Yukiko. 1990. DNA for expression and secretion. European patent EP 0 388 166 A1.
- 60.Zhao, H., L. Giver, Z. Shao, J. A. Affholter, and F. H. Arnold. 1998. Molecular evolution by staggered extension process (StEP) in vitro recombination. Nat. Biotechnol. 16:258-261. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, H., J. C. Moore, A. A. Volkov, and F. H. Arnold. 1999. Methods for optimizing industrial enzymes by directed evolution, p. 597-604. In A. L. Demain and J. E. Davies (ed.), Manual of industrial microbiology and bio/technology, 2nd ed. ASM Press, Washington, D.C.