Abstract
We describe the effects of modulating the activities of glucokinase, phosphofructokinase, and phosphoglucomutase on the branching point between sugar degradation and the biosynthesis of sugar nucleotides involved in the production of exopolysaccharide biosynthesis by Lactococcus lactis. This was realized by using a described isogenic L. lactis mutant with reduced enzyme activities or by controlled expression of the well-characterized genes for phosphoglucomutase or glucokinase from Escherichia coli or Bacillus subtilis, respectively. The role of decreased metabolic flux was studied in L. lactis strains with decreased phosphofructokinase activities. The concomitant reduction of the activities of phosphofructokinase and other enzymes encoded by the las operon (lactate dehydrogenase and pyruvate kinase) resulted in significant changes in the concentrations of sugar-phosphates. In contrast, a >25-fold overproduction of glucokinase resulted in 7-fold-increased fructose-6-phosphate levels and 2-fold-reduced glucose-1-phosphate and glucose-6-phosphate levels. However, these increased sugar-phosphate concentrations did not affect the levels of sugar nucleotides. Finally, an ∼100-fold overproduction of phosphoglucomutase resulted in 5-fold-increased levels of both UDP-glucose and UDP-galactose. While the increased concentrations of sugar-phosphates or sugar nucleotides did not significantly affect the production of exopolysaccharides, they demonstrate the metabolic flexibility of L. lactis.
Exopolysaccharides (EPS) include a range of diverse polymers that play vital roles in a variety of biological processes. In addition, EPS have a variety of industrial applications, including their use as biothickeners in foods. Notably, EPS produced by lactic acid bacteria (LAB) contribute significantly to the structure and viscosity of fermented milk products. Furthermore, several reports indicate that they can confer health benefits on consumers arising from their immunogenic and cholesterol-lowering properties. Although the production of EPS by LAB in milk is relatively low (<1 mg liter−1) in comparison with the milk sugar concentration (>40 g liter−1), some EPS appear to be effective thickeners (19), especially since they interact with milk proteins (40).
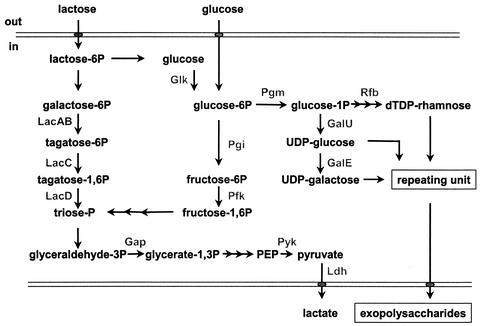
To study EPS biosynthesis by LAB, we have focused on Lactococcus lactis NIZO B40, since it produces a phosphopolysaccharide with known structure (for a recent review, see reference 2). Moreover, the NIZO B40 EPS-producing capacity is encoded by a 42,180-bp plasmid, pNZ4000, that can be transferred to genetically well-studied model strains of L. lactis (38, 39). The EPS plasmid pNZ4000 contains 14 eps genes involved in the formation of a repeating unit by sequential addition of sugars to a membrane-anchored lipid carrier and in export and polymerization of these repeating units. Upon its transfer to model strains, the NIZO B40 phosphopolysaccharide, which contains glucose, galactose, and rhamnose moieties, is produced. Therefore, their production requires the sugar nucleotides UDP-glucose, UDP-galactose, and dTDP-rhamnose, which are formed by enzymes encoded by genes on the chromosome (Fig. 1). The central intermediate, glucose-1-phosphate (glucose-1P), is converted to dTDP-rhamnose by the sequential activities of the rfbACBD gene products (2). GalU activity catalyzes the conversion of glucose-1P into UDP-glucose, which is subsequently converted into UDP-galactose by GalE activity (3). All the genes that encode the enzymes involved in the biosynthesis of these sugar nucleotides from glucose-1P (galU, galE, and rfbACBD [Fig. 1]) have been cloned from L. lactis MG1363, and their roles in controlling sugar nucleotide levels have been investigated (2, 3).
FIG. 1.
Schematic representation of pathways involved in sugar fermentation via glycolysis to lactate and/or other acids and biosynthesis of EPS in L. lactis. Glucose and lactose are transported via phosphotranseferase systems. The following enzymes are involved: Glk, Pgi, Pfk, Gap (glyceraldehyde 3-phosphate dehydrogenase), LacABCD (tagatose-6-phosphate pathway), Pyk (pyruvate kinase), Ldh (lactate dehydrogenase), Pgm, GalU (UDP-glucose pyrophosphorylase), GalE (UDP-galactose epimerase), Rfb (dTDP-rhamnose biosynthetic system consisting of RfbA and glucose-1P thymidylyltransferase), RfbB (dTDP-glucose-4,6 dehydratase), RfbC (dTDP-4-keto-6-deoxy-D-glucose-3,5 epimerase), and RfbD (dTDP-4-keto-L-rhamnose reductase).
Insight into the biosynthesis of EPS is crucial for the exploitation of microorganisms for the production of EPS of industrial or medical importance. For the design of metabolic-engineering strategies that aim at increased fluxes to EPS production, it is relevant to include control factors in sugar degradation and EPS formation pathways (13). In L. lactis used in dairy fermentation, sugar degradation starts with lactose uptake via a phosphotransferase system, which yields, after hydrolysis, galactose-6P and glucose moieties. Subsequently, the galactose-6P moiety can be catabolized completely via the tagatose pathway and glycolysis for the generation of biomass and energy, while the glucose moiety can be used for EPS production (13). The uncoupling of lactose-derived glucose and galactose metabolism has been established in an L. lactis mutant impaired in glucokinase (Glk; EC 2.7.1.2) and glucose phosphotransferase system activity that accumulated glucose (35). Assuming that the linkage between glycolysis and EPS formation occurs at the branching point starting from the glycolysis intermediate glucose-6P, it is tempting to speculate that it might be possible to engineer EPS overproduction by increasing the pool of this sugar-phosphate.
The availability of glucose-6P can be affected by the modulation of glycolytic activity (1). It has been reported that the lactococcal glycolytic flux can be affected by the activities of phosphofructokinase (Pfk) (1) and the global catabolite control protein CcpA (28). The latter protein acts as an activator of transcription of the las operon (24) encoding the glycolytic enzymes Pfk and pyruvate kinase and the lactate-forming enzyme lactate dehydrogenase. Furthermore, modulation of glucose-6P could potentially be achieved by engineering the enzyme activities that are involved in the formation of this intermediate, such as Glk (Fig. 1).
Another possible key step in sugar nucleotide biosynthesis is the interconversion of the glycolysis intermediate glucose-6P and glucose-1P, which can be regarded as the central precursor in sugar nucleotide biosynthesis (34), performed by phosphoglucomutase (Pgm; E.C. 2.7.5.1) (Fig. 1). L. lactis contains two distinct forms of Pgm, one specific for β-glucose-6P (i.e., β-Pgm) and the other specific for α-glucose-6P (i.e., α-Pgm) (30). Since the phosphotransferase sugar uptake or the Glk-mediated phosphorylation of glucose yields α-glucose-6P (32), α-Pgm could be a key enzyme in sugar nucleotide biosynthesis. However, only the gene encoding β-Pgm has been identified in L. lactis (31).
We studied the effects of the modulation of enzyme activities at the branching point between sugar degradation and sugar nucleotide and/or EPS biosynthesis. By influencing the expression of the corresponding genes, we evaluated the roles of CcpA (28) and controlled overproduction of heterologous Pgm and Glk in the biosynthesis of NIZO B40 EPS in the plasmid-free model strain L. lactis MG1363. It could be established that the overproduction of Bacillus subtilis Glk and Escherichia coli Pgm significantly increased the levels of sugar-phosphates and sugar nucleotides, respectively. However, this did not affect the NIZO B40 EPS production level. Furthermore, although the ccpA mutant strain did not show any changes in the levels of the sugar nucleotides, the mutant produced significantly less EPS than the wild-type strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The lactococcal strains and plasmids used in this study are listed in Table 1. E. coli MC1061 (6), which was used as a host in cloning experiments, was grown with aeration in tryptone-yeast extract broth at 37°C. L. lactis was grown without aeration at 30°C in a chemically defined medium (CDM) (25) or in M17 broth (Merck, Darmstadt, Germany) supplemented with 0.5% (wt vol−1) glucose or lactose. When appropriate, the media contained chloramphenicol (10 μg ml−1), erythromycin (10 μg ml−1), tetracycline (2 μg ml−1), or ampicillin (100 μg ml−1). To analyze the effect of gene overexpression, the nisin-controlled expression (NICE) system was used (10, 20). For enzyme activity analysis, L. lactis cells were grown to an optical density at 600 nm (OD600) of ∼0.5, and for EPS analysis the cells were grown to an OD600 of ∼0.1. Subsequently, the culture was split into two cultures. Nisin (1 ng ml−1) was added to one of the cultures, and both cultures were grown for an additional 2 to 24 h.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Strains | ||
| L. lactis | ||
| MG1363 | 16 | |
| MG5267 | 41 | |
| NZ3800 | MG5267 pepN::nisRK | This work |
| NZ9870 | MG1363 ccpA::ery | 28 |
| NZ9000 | MG1363 pepN::nisRK | 20 |
| E. coli MC1061 | 6 | |
| B. subtilis ATCC 6633 | ATCCb | |
| Plasmids | ||
| pNZ4123 | Cmr; pNZ8048 derivative containing a functional E. coli pgmU gene | This work |
| pNZ4124 | Cmr; pNZ8048 derivative containing a functional B. subtilis glk gene | This work |
| pNZ4030 | Eryr Eps+ | 38 |
| pNZ4130 | Tetr Eps+ | 3 |
| pNZ9573 | Eryr; nonreplicative lactococcal plasmid; pepN::nisRK | 10 |
| pNZ8048 | Cmr; inducible expression vector carrying the nisA promoter | 20 |
EPS+, EPS-producing phenotype; Cmr, chloramphenicol resistant; Eryr, erythromycin resistant; Tetr, tetracycline resistant.
ATCC, American Type Culture Collection.
DNA manipulations and DNA sequence analysis.
Small-scale isolation of E. coli plasmid and chromosomal DNAs and standard recombinant-DNA techniques were performed as described by Sambrook et al. (33). Large-scale isolation of E. coli plasmid DNA for nucleotide sequence analysis was performed with JetStar columns (Genomed GmbH, Bad Oberhausen, Germany) by following the instructions of the manufacturer. Isolation and transformation of L. lactis DNA were performed as previously described (12). Isolation of chromosomal DNA of B. subtilis was performed as described previously (5).
Automatic double-stranded DNA sequence analysis was performed on both strains with an ALFred DNA sequencer (Pharmacia Biotech, Roosendaal, The Netherlands). Sequence reactions were performed with an Autoread kit, were initiated by using Cy5-labeled universal and reverse primers, and were continued with synthetic primers purchased from Pharmacia Biotech in combination with fluorescein-15-dATP by following the instructions of the manufacturer. Sequence data were assembled and analyzed by using the PC/GENE program version 6.70 (Intelli-Genetics).
Construction of strains and plasmids.
To make the L. lactis MG1363 derivative MG5267, a suitable host for the use of the NICE system, the lactococcal nisRK genes were introduced into the pepN locus as described by De Ruyter et al. (11). The resulting strain, designated NZ3800, contains the nisRK genes under the control of their own promoter integrated in the pepN locus. The expected genetic configuration of the pepN::nisRK locus was confirmed by PCR analysis.
The EPS-producing capacity was introduced into the L. lactis CcpA mutant strain NZ9870 (28) by transformation of the plasmid pNZ4130 (3), a tetracycline-resistant derivative of pNZ4000.
The E. coli α-Pgm gene (pgmU) was amplified by PCR using Tth polymerase and chromosomal DNA of E. coli MC1061 (6) as template DNA with the primers 5′-CATGCCATGGCAATCCACAATCGTGCAG-3′ and 5′-CTAGTCTAGATTACGCGTTTTTCAGAACTTCGC-3′. The 1.64-kb PCR product generated was cloned into pNZ8048 (20) using the NcoI and XbaI restriction sites that were introduced by the primers used (underlined), yielding the pgm overexpression plasmid pNZ4123.
The B. subtilis Glk gene (glk) was amplified by PCR using Tth polymerase and chromosomal DNA of B. subtilis ATCC 6633 as template DNA with the primers 5′-CATGCCATGGACGAGATATGGTTTGCG-3′ and 5′-CTAGTCTAGATTAACAATTTTGATGTTTCA-3′. The 0.98-kb PCR product generated was cloned into pNZ8048 (20) using the NcoI and XbaI restriction sites that were introduced by the primers used (underlined), yielding the glk overexpression plasmid pNZ4124. The plasmid pNZ4123 or pNZ4124 was introduced into L. lactis strain NZ3800 by transformation. Subsequently, the EPS-producing capacity was introduced into L. lactis NZ3800 harboring pNZ4123 or pNZ4124 by electroporation of plasmid pNZ4030 (38).
Preparation of CEs and protein analysis.
Lactococcal cells (50 ml) were harvested by centrifugation (3,500 × g; 10 min; 4°C), and the cell pellets were resuspended in 1 ml of 20 mM sodium-phosphate buffer (pH 6.5) containing 50 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol. These suspensions were mechanically disrupted by bead beating in the presence of zirconium beads (37), and cell debris was removed by centrifugation (3,500 × g; 10 min; 4°C), resulting in the cell extract (CE). The protein content of the CE was determined by the method of Bradford (4) using bovine serum albumin as the standard.
For protein analysis, lactococcal CE was mixed with an equal amount of twofold-concentrated Laemmli buffer and, after the mixture was boiled, 10 μg of each sample was analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (21).
Enzyme assays.
Enzyme reactions were performed at 30°C in a total volume of 1 ml containing various amounts of freshly prepared CE. The formation of NADH or NADPH was determined by measuring the change in absorbency at 340 nm. The values given are the means of at least two independent measurements. The control contained the reaction buffer, cofactors, and the substrate but lacked the CE.
The Glk reaction assay mixture contained 100 mM triethanolamine hydrochloride buffer (pH 7.8), 5 mM MgCl2, 1 mM NADP+, 4 U of glucose-6P dehydrogenase, 2 mM ATP, and CE. The reaction was initiated by the addition of 10 mM glucose (29).
The α-Pgm reaction assay mixture contained 50 mM triethanolamine buffer (pH 7.2), 5 mM MgCl2, 0.4 mM NADP+, 50 μM glucose-1,6P, 4 U of glucose-6P dehydrogenase, and CE. The reaction was initiated by the addition of 1.4 mM α-glucose-1P (30).
Estimation of intracellular metabolites.
The intracellular metabolites glucose-6P, glucose-1P, and fructose-6P were fixed by rapid inactivation of cell metabolism. Therefore, 4 ml of cell culture in the logarithmic growth phase was mixed with an equal amount of cold (−80°C) methanol. The methanol was removed by evaporation in a heating block, and the cell metabolites were measured by coupling appropriate enzyme assays with fluorimetric determination of NADPH as described by Garrigues et al. (15).
Sugar nucleotide and EPS analyses.
Sugar nucleotides were separated from CEs, and individual sugar nucleotide contents were determined by high-performance liquid chromatography as previously described by Looijesteijn et al. (26). The values reported are the averages of at least two independent determinations. EPS was isolated, quantified, and characterized as described by Looijesteijn and Hugenholtz (25).
RESULTS
Glycolytic-intermediate modulation.
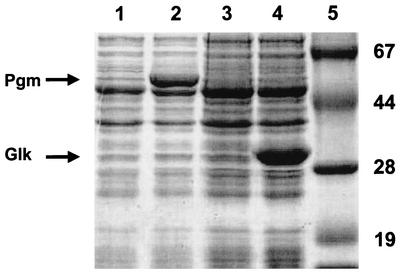
By modulation of the pools of glycolytic intermediates, we investigated the possibilities for increasing the pool of glucose-6P, the levels of sugar nucleotides, and subsequently the level of EPS production. We evaluated the effect of reduced glycolytic flux on EPS biosynthesis by analyzing NIZO B40 EPS biosynthesis in the L. lactis ccpA disruption strain NZ9870. In this strain, the transcription levels of the las operon genes (pfk, pyk, and ldh) are reduced fourfold compared to those in the parental strain (28). Moreover, the activities of other enzymes may be affected as well, since CcpA is a global control protein. The ccpA disruption strain NZ9870 showed a reduced growth rate, reduced glycolytic flux, and a switch from homolactic to mixed-acid fermentation (28). In this strain, the levels of the first two glycolytic intermediates, glucose-6P and fructose-6P, appeared to be two- and sevenfold increased relative to the levels measured in the wild-type strain (Table 2). However, neither the level of glucose-1P (Table 2) nor the levels of the sugar nucleotides UDP-glucose, UDP-galactose, and dTDP-rhamnose (data not shown) were influenced by the changed level of glucose-6P. Consequently, no effect of the ccpA mutation on the EPS production level was expected. However, EPS production in this strain appeared to be reduced twofold relative to that of the wild-type strain (Table 3), although this could suggest that CcpA exerts control on EPS production at the level of the EPS machinery itself. To evaluate the effect of stimulated phosphorylated-sugar intermediates in glycolysis on EPS biosynthesis in L. lactis, we studied the effect of controlled glk overexpression, using the NICE system (10, 20). Since the sequence of the L. lactis glk gene was not available, we used a heterologous approach as a proof of principle. Therefore, we cloned the B. subtilis ATCC 6633 glk gene under the control of the lactococcal nisA promoter, and the resulting plasmid (pNZ4124) was introduced into strain NZ3800. Strain NZ3800 harboring pNZ4124 was grown under inducing and noninducing conditions, and CEs of the cultures were prepared and analyzed by SDS-PAGE (Fig. 2). Growth in the presence of nisin resulted in the appearance of an additional protein band with an apparent molecular mass of ∼34-kDa, which is the expected size of Glk. Moreover, >25-fold-increased Glk specific activity was obtained with CEs of the induced cultures compared to that in the control cultures (Table 2). Functional overexpression of glk resulted in an almost sevenfold-increased fructose-6P level compared to that of the wild-type strain (Table 2). Remarkably, both the level of glucose-6P and the level of glucose-1P were reduced, suggesting that substrate activation may take the place of phosphoglucoisomerase (Pgi) activity. Pgm apparently maintains a constant ratio between glucose-6P and glucose-1P which is independent of the glucose-6P concentration. The increased Glk activity did not affect the sugar nucleotide levels (data not shown) or the NIZO B40 EPS production level (Table 3), suggesting that Glk does not play an important role in the control of sugar nucleotide and EPS biosynthesis in L. lactis.
TABLE 2.
Enzyme activities and sugar nucleotide and sugar-phosphate concentrations in Lactococcus lactis subsp. cremoris MG1363 derivatives grown in M17 medium with Glc or Lac as sole carbon source
| Strain (plasmid) | Sugar source | Nisin (ng ml−1) | Relevant enzyme activity (mU of protein−1a) | Sugar-phosphates (μM mg of protein−1)
|
||
|---|---|---|---|---|---|---|
| Glucose-6P | Fructose-6P | Glucose-1P | ||||
| NZ3800 | Glc | 0 | ND | 23 | 9.7 | 39 |
| NZ3800 | Lac | 0 | ND | 31 | 44 | 37 |
| NZ9870 | Glc | 0 | ND | 47 | 68 | 46 |
| NZ3800(pNZ4123) | Lac | 0 | 222 ± 42b | 30 | 25 | 35 |
| Lac | 1 | 24,767 ± 506b | 30 | 33 | 33 | |
| NZ3800(pNZ4124) | Lac | 0 | 352 ± 51c | 5.2 | 74 | 10 |
| Lac | 1 | 9,482 ± 93c | 13 | 290 | 10 | |
Values (± standard deviations) are based on at least two independent experiments. ND, not determined.
Pgm-specific activity.
Glk-specific activity.
TABLE 3.
EPS production by L. lactis subsp. cremoris NZ9000 derivatives harboring pNZ4030 grown in CDM with Glc or Lac as sole carbon source
| Strain (plasmid) | Nisin (ng ml−1) | EPSa (mg liter−1 · OD600−1)
|
|
|---|---|---|---|
| Glc | Lac | ||
| MG1363(pNZ4030) | 0 | 36 ± 1 | ND |
| NZ9870(pNZ4130) | 0 | 16 ± 2 | ND |
| NZ3800(pNZ4030, pN8048) | 0 | 46 ± 1 | 39 ± 1 |
| NZ3800(pNZ4030, pNZ4123) | 0 | 42 ± 1 | 38 ± 1 |
| 1 | 49 ± 1 | 39 ± 1 | |
| NZ3800(pNZ4030, pNZ4124) | 0 | 46 ± 1 | 37 ± 3 |
| 1 | 50 ± 1 | 39 ± 1 | |
Values (± standard deviations) are averages based on at least two independent experiments. ND, not determined.
FIG. 2.
Coomassie blue-stained gel after SDS-PAGE of CE of L. lactis NZ3800 harboring pNZ4123 (lanes 1 and 2) or pNZ4124 (lanes 3 and 4) grown in the absence (lanes 1 and 3) and in the presence (lanes 2 and 4) of nisin. Lane 5 contained a set of protein standards, whose molecular masses (in kilodaltons) are indicated on the right. Additional bands resulting from nisin induction and representing the Pgm protein (lane 2) and Glk protein (lane 4) are indicated.
Branching-point modulation.
To evaluate the effect of increased Pgm activity on UDP-sugar levels and EPS production in L. lactis, we studied the effect of controlled pgm overexpression by using the NICE system (10, 20). Similar to our study of glk overexpression, we used a heterologous approach and cloned the E. coli K-12 pgmU gene under the control of the lactococcal nisA promoter (pNZ4123). Strain NZ3800 harboring pNZ4123 was grown under inducing and noninducing conditions, and CEs of the cultures were analyzed by SDS-PAGE (Fig. 2). Nisin induction resulted in the appearance of a protein band with an apparent molecular mass of ∼58-kDa, corresponding to the expected size of PgmU. An ∼100-fold-increased Pgm specific activity was determined in the CEs of the induced cultures (Table 2). These results demonstrate the functional overexpression of the pgmU gene. Despite this high-level Pgm production, no differences in growth rates between nisin-induced NZ3800 harboring pNZ4123 and the uninduced strain were observed (data not shown).
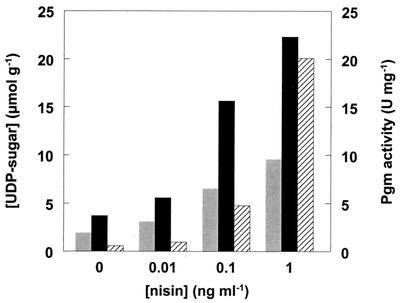
To study the effect of pgmU overexpression on sugar nucleotide levels, the concentrations of UDP-glucose and UDP-galactose were determined in NZ3800 harboring pNZ4123 grown in the presence of nisin. Overexpression of pgmU resulted in a fivefold increase in the levels of both UDP-glucose and UDP-galactose (Fig. 3), while dTDP-rhamnose levels remained the same (data not shown). These results indicate that the activity level of Pgm to some extent affects UDP-glucose and UDP-galactose levels in L. lactis. In contrast, the increased level of PgmU activity did not have a significant effect on the level of EPS production when cells were grown in CDM with glucose or lactose as a sole carbon source (Table 3). These results indicate that the increased UDP-glucose and UDP-galactose levels in NZ3800 harboring pNZ4123 have no effect on EPS production efficiency in this strain.
FIG. 3.
UDP-glucose (solid bars) and UDP-galactose (shaded bars) levels and Pgm activity (hatched bar) of L. lactis NZ9000 harboring pNZ4123 and grown in the presence of different levels of nisin.
DISCUSSION
The biosynthesis of EPS involves a large number of housekeeping enzymes required for the production of EPS precursors and EPS-specific enzymes that are encoded in eps gene clusters. In this study, we focused on the regulation of EPS production by three household enzymes, Glk, Pgm, and Pfk, all acting at the branching point between glycolysis and EPS precursor biosynthesis (Fig. 1), which is a potential bottleneck in sugar nucleotide and subsequent EPS biosyntheses. In this respect, increasing the intracellular glucose-6P pool, which is the central branching-point intermediate, might push metabolism toward sugar nucleotide biosynthesis and EPS production. It has been reported that glucose-6P could be toxic at enhanced levels (1, 14), although accumulation of lactose-phosphate was tolerated by L. lactis deficient in LacG activity (8). Therefore, it is feasible that concomitant enhancement of sugar anabolism, leading to the increased production of sugar nucleotides and subsequently EPS, might relieve glucose-6P toxic effects by decreasing the pool of glucose-6P in the cell and could lead to enhanced EPS production.
We evaluated the effect of reduced glycolytic flux on EPS biosynthesis by analyzing a ccpA mutant. Such mutants are known to have reduced glycolytic flux, and the L. lactis ccpA mutant has a fourfold-reduced transcription level of the las operon, containing the pfk, pyk, and ldh genes (28). However, it is also known that CcpA is a global regulator and has pleiotropic effects. The concentrations, as well as the ratio, of fructose-6P and glucose-6P were increased in the ccpA mutant strain. These results confirm reported data on a lactococcal strain with reduced Pfk activity (1). Andersen et al. (1) showed clearly that Pfk activity had a very high negative control of the level of upstream metabolites, and by decreasing the Pfk activity merely twofold, the sugar phosphates glucose-6P and fructose-6P increased two- to fourfold. In contrast, we determined a fivefold-higher glucose-6P/fructose-6P ratio for the wild-type strain. We suspect that this may be a consequence of the different stages of growth when the cells were harvested, since we collected the cells later in the exponential phase than Andersen et al. did (1). The results establish that overall reduction of the enzymatic activities encoded by the las operon leads to accumulation of intermediates high up in the glycolytic pathway. Although the ccpA mutation led to an increase of the pool of glucose-6P, the EPS production level was approximately twofold reduced compared to that in the wild type. Since no catabolite control elements were found in any of the sequences of genes involved in sugar nucleotide or EPS biosynthesis (data not shown), it seems unlikely that CcpA regulates the expression of any of these genes directly. The lack of success with respect to the increase of EPS flux may well be a consequence of the pleiotropic effects resulting from the ccpA mutation. Since the ccpA mutation caused a shift to mixed-acid fermentation (28), it is possible that the overall reduced metabolic rate in the ccpA mutant results in reduced energy availability, which could negatively influence EPS production. In another approach to increase EPS production, we aimed at glucose-6P accumulation as a result of increased Glk activity. Heterologous overexpression of B. subtilis glk in L. lactis resulted in a >25-fold increase in the enzyme activity level. Remarkably, the levels of glucose-6P and glucose-1P were decreased while the level of fructose-6P was increased almost sevenfold. These observations suggest that Pgi activity is subject to substrate glucose-6P activation. However, the increased Glk activity level did not result in more EPS precursors and NIZO B40 EPS. These results imply that the level of Glk enzyme activity does not control NIZO B40 EPS production levels.
It has been shown that mutations in the pgm gene affect the morphology of E. coli (27) and that they affected the polysaccharide composition of Pseudomonas aeruginosa (7, 42) and Streptococcus pneumoniae (17, 18). However, in L. lactis, α-Pgm is likely to be indispensable for sugar nucleotide formation and thus essential for the growth and viability of the species (32). To investigate the role of Pgm at the branching point between sugar catabolism and anabolism in L. lactis, we evaluated the effect of increased levels of Pgm activity. Heterologous overexpression of E. coli pgmU in L. lactis resulted in a >100-fold increase in the enzyme activity level and fivefold increases in the UDP-glucose and UDP-galactose levels. These results show that the level of PgmU enzyme activity controls the level of production of UDP-glucose and UDP-galactose in wild-type cells. However, the sugar-phosphate analysis of the Glk overproduction strain suggests that the endogenous Pgm activity maintains a stable glucose-6P/glucose-1P ratio. These results suggest that Pgm overproduction will have no effect on sugar nucleotide synthesis. Therefore, it is likely that the increased sugar nucleotide levels are the result of favorable kinetic parameters of E. coli PgmU. However, increased Pgm activity did not affect NIZO B40 EPS production (Table 3). Analogously, GalU overproduction led to the same result (3). Similar findings were reported for Sphingomomas, in which a sixfold increase of Pgm activity did not affect EPS production (36). Contrasting results were reported for Streptococcus thermophilus strain LY03. Degeest and de Vuyst (9) showed a linear relationship between Pgm activity and EPS production, while Levander and Rådström (22) reported that overexpression of pgm in the same strain did not change EPS production. It is suggested that precursors for EPS biosynthesis in a galactose-fermenting strain of S. thermophilus originate from fermentation of the galactose moiety of lactose, thereby circumventing the need for a functional Pgm in these cells (22). Interestingly, when pgm was overexpressed in combination with galU, the EPS yield was shown to increase twofold in this strain (23).
Evaluation of the EPS biosynthesis model described here allowed us to assess the roles of Glk and Pfk in L. lactis by modulating their enzyme activity levels. Although early glycolytic intermediates like glucose-6P and fructose-6P accumulated in some cases, these modulations did not affect the sugar nucleotide levels and, eventually, the EPS production level. In contrast, modulation of Pgm activity resulted in significantly increased internal pools of both UDP-glucose and UDP-galactose. However, Pgm overproduction did not result in a significant increase of NIZO B40 EPS. Remarkably, a ccpA mutant strain produced only half the amount of EPS that wild-type cells did, which might be caused by the overall reduced metabolic rate in the ccpA mutant, resulting in reduced energy availability for EPS biosynthesis. The results presented here provide insight into enzymatic control factors that determine the carbon distribution between catabolic and anabolic reactions involved in conversion of the central glycolytic intermediate glucose-6P. Such insight is essential for the design of metabolic-engineering strategies that aim at resolving bottlenecks in EPS biosynthesis in L. lactis. Moreover, the possibility of stably overproducing significant amounts of phosphorylated glycolytic intermediates in L. lactis demonstrates the flexibility of its metabolism, which may be instrumental in further optimizing its metabolic activities in dairy and other food fermentations.
Acknowledgments
We thank Jan van Riel for determining sugar nucleotide and EPS contents and Arno Wegkamp for assistance with the sugar-phosphate analysis.
Part of this work was supported by EC research grant BIOT-CT96-0498.
REFERENCES
- 1.Andersen, H. W., C. Solem, K. Hammer, and P. R. Jensen. 2001. Twofold reduction of phosphofructokinase activity in Lactococcus lactis results in strong decrease in growth rate and in glycolytic flux. J. Bacteriol. 183:3458-3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boels, I. C., R. van Kranenburg, J. Hugenholtz, M. Kleerebezem, and W. M. de Vos. 2001. Sugar catabolism and its impact on the biosynthesis and engineering of exopolysaccharide production in lactic acid bacteria. Int. Dairy J. 11:721-730. [Google Scholar]
- 3.Boels, I. C., A. Ramos, M. Kleerebezem, and W. M. de Vos. 2001. Functional analysis of the Lactococcus lactis galU and galE genes and their impact on sugar nucleotide and exopolysaccharide biosynthesis. Appl. Environ. Microbiol. 67:3033-3040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Bron, S. 1990. Plasmids, p. 75-174. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. J. Wiley and Sons, Chichester, United Kingdom.
- 6.Casadaban, M. J., and S. N. Cohen. 1980. Analysis of gene control signals by DNA fusion and cloning in E. coli. J. Mol. Biol. 138:179-207. [DOI] [PubMed] [Google Scholar]
- 7.Coyne, J. R., M. J. Coyne, Jr., K. S. Russell, C. L. Coyle, and J. B. Goldberg. 1994. The Pseudomonas aeruginosa algC gene encodes phosphoglucomutase, required for the synthesis of a complete lipopolysaccharide core. J. Bacteriol. 176:3500-3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crow, V. L., and T. D. Thomas. 1984. Properties of a Streptococcus lactis strain that ferments lactose slowly. J. Bacteriol. 157:28-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Degeest, B., and L. de Vuyst. 2000. Correlation of activities of the enzymes α-phosphoglucomutase, UDP-galactose-epimerase and UDP-glucose pyrophosphorylase with exopolysaccharide biosynthesis by Streptococcus thermophilus LY03. Appl. Environ. Microbiol. 66:3519-3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Ruyter, P. G. G. A., O. P. Kuipers, and W. M. de Vos. 1996. Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl. Eviron. Microbiol. 62:3662-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Ruyter, P. G. G. A., O. P. Kuipers, M. M. Beerthuyzen, I. J. van Alen-Boerrigter, and W. M. de Vos. 1996. Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J. Bacteriol. 178:3434-3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Vos, W. M., P. Vos, H. de Haard, and I. Boerrigter. 1989. Cloning and expression of the Lactococcus lactis ssp. cremoris SK11 gene encoding an extracellular serine protease. Gene 85:169-176. [DOI] [PubMed] [Google Scholar]
- 13.De Vos, W. M. 1996. Metabolic engineering of sugar catabolism in lactic acid bacteria. Antonie Leeuwenhoek 70:223-242. [DOI] [PubMed] [Google Scholar]
- 14.Fraenkel, D. G. 1968. The accumulation of glucose-6-phosphate from glucose and its effect in an Escherichia coli mutant lacking phosphoglucose isomerase and glucose 6-phosphate dehydrogenase. J. Biol. Chem. 243:6451-6457. [PubMed] [Google Scholar]
- 15.Garrigues, C., P. Loubiere, N. D. Lindley, and M. Cocaign-Bousquet. 1997. Control of the shift from homolactic acid to mixed fermentation in Lactococcus lactis: predominant role of the NADH/NAD+ ratio. J. Bacteriol. 179:5282-5287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gasson, M. J. 1983. Plasmid complements of NCDO 712 and other lactic streptococci after protoplast-induced curing. J. Bacteriol. 154:1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardy, G. G., M. J. Caimano, and J. Yother. 2000. Capsule biosynthesis and basic metabolism in Streptococcus pneumoniae are linked through the cellular phosphoglucomutase. J. Bacteriol. 182:1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy, G. G., A. D. Magee, C. L. Ventura, M. J. Caimano, and J. Yother. 2001. Essential role for cellular phosphoglucomutase in virulence of type 3 Streptococcus pneumoniae. Infect. Immun. 69:2309-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hess, S. J., R. F. Roberts, and G. R. Ziegler. 1997. Rheological properties of nonfat yoghurt stabilized using Lactobacillus delbrueckii ssp. bulgaricus producing exopolysaccharide or using commercial stabilizer systems. J. Dairy Sci. 80:252-263. [Google Scholar]
- 20.Kuipers, O. P., P. G. G. A. de Ruyter, M. Kleerebezem, and W. M. de Vos. 1998. Quorum sensing-controlled gene expression in lactic acid bacteria. J. Biotechnol. 64:15-21. [Google Scholar]
- 21.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 22.Levander, F., and P. Rådström. 2001. Requirement for phosphoglucomutase in exopolysaccharide biosynthesis in glucose and lactose-utilizing Streptococcus thermophilus. Appl. Environ. Microbiol. 67:2734-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levander, F., M. Svensson, and P. Rådström. 2002. Enhanced exopolysaccharide production by metabolic engineering of Streptococcus thermophilus. Appl. Environ. Microbiol. 68:784-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Llanos, R. M., C. J. Harris, A. J. Hiller, and B. E. Davidson. 1993. Identification of a novel operon in Lactococcus lactis encoding three enzymes for lactic acid synthesis: phosphofructokinase, pyruvate kinase, and lactate dehydrogenase. J. Bacteriol. 175:2541-2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Looijesteijn, P. J., and J. Hugenholtz. 1999. Uncoupling of growth and exopolysaccharide production by Lactococcus lactis subsp. cremoris NIZO B40 and optimisation of its exopolysaccharide synthesis. J. Biosci. Bioeng. 88:178-182. [DOI] [PubMed] [Google Scholar]
- 26.Looijesteijn, P. J., I. C. Boels, M. Kleerebezem, and J. Hugenholtz. 1999. Regulation of exopolysaccharide production by Lactococcus lactis subsp. cremoris by the sugar source. Appl. Eviron. Microbiol. 65:5003-5008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu, M., and N. Kleckner. 1994. Molecular cloning and characterization of the pgm gene encoding phosphoglucomutase of Escherichia coli. J. Bacteriol. 176:5847-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luesink, E. J., R. E. M. A. van Herpen, B. P. Grossiord, O. P. Kuipers, and W. M. de Vos. 1998. Transcriptional activation of the glycolytic las operon and catabolite repression of the gal operon in Lactococcus lactis are mediated by the catabolite control protein CcpA. Mol. Microbiol. 30:789-798. [DOI] [PubMed] [Google Scholar]
- 29.Petit, C., J. P. Grill, N. Maazouzi, and R. Marczak. 1991. Regulation of polysaccharide formation by Streptococcus thermophilus in batch and fed-batch cultures. Appl. Microbiol. Biotechnol. 36:216-221. [Google Scholar]
- 30.Qian, N., G. A. Stanley, B. Hahn-Hägerdal, and P. Rådström. 1994. Purification and characterization of two phosphoglucomutases from Lactococcus lactis subsp. lactis and their regulation in maltose- and glucose-utilizing cells. J. Bacteriol. 176:5304-5311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qian, N., G. A. Stanley, A. Bunte, and P. Rådström. 1997. Product formation and phosphoglucomutase activities in Lactococcus lactis: cloning and characterization of novel phosphoglucomutase gene. Microbiology 143:855-866. [DOI] [PubMed] [Google Scholar]
- 32.Ramos, A., I. C. Boels, W. M. de Vos, and H. Santos. 2001. Relationship between glycolysis and exopolysaccharide biosynthesis in Lactococcus lactis. Appl. Environ. Microbiol. 67:33-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sjöberg, A., and B. Hahn-Hägerdal. 1989. β-Glucose-1-phosphate: a possible mediator for polysaccharide formation in maltose-assimilating Lactococcus lactis. Appl. Environ. Microbiol. 55:1549-1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson, J., B. M. Chassy, and W. Egan. 1985. Lactose metabolism in Streptococcus lactis: studies with a mutant lacking glucokinase and mannose-phosphotransferase activities. J. Bacteriol. 162:217-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorne, L., M. J. Mikolajczak, R. W. Armentrout, and T. J. Pollock. 2000. Increasing the yield and viscosity of exopolysaccharides secreted by Sphingomonas by augmentation of chromosomal genes with multiple copies of cloned biosynthetic genes. J. Ind. Microbiol. Biotechnol. 25:49-57. [Google Scholar]
- 37.Van der Meer, J. R., J. Polman, M. M. Beerthuyzen, R. J. Siezen, O. P. Kuipers, and W. M. de Vos. 1993. Characterization of the Lactococcus lactis nisinA operon genes nisP, encoding a subtilisin-like protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J. Bacteriol. 175:2578-2588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Kranenburg, R., J. D. Marugg, I. I. van Swam, J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 39.Van Kranenburg, R., M. Kleerebezem, and W. M. de Vos. 2000. Nucleotide sequence analysis of the lactococcal EPS plasmid pNZ4000. Plasmid 43:130-136. [DOI] [PubMed] [Google Scholar]
- 40.Van Marle, M. E., and P. Zoon. 1995. Permeability and rheological properties of microbially and chemically acidified skim-gels. Neth. Milk Dairy J. 49:47-65. [Google Scholar]
- 41.Van Rooijen, R. J., M. J. Gasson, and W. M. de Vos. 1992. Characterization of the Lactococcus lactis lactose operon promoter: contribution of flanking sequences and lacR repressor to promoter activity. J. Bacteriol. 174:2273-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye, R. W., N. A. Zielinski, and A. M. Chakrabarty. 1994. Purification and characterization of phosphomannomutase/phosphoglucomutase from Pseudomonas aeruginosa involved in biosynthesis of both alginate and lipopolysaccharide. J. Bacteriol. 176:4851-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]