Abstract
A practical enzymatic synthesis of a doubly chiral key compound, (4R,6R)-4-hydroxy-2,2,6-trimethylcyclohexanone, starting from the readily available 2,6,6-trimethyl-2-cyclohexen-1,4-dione is described. Chirality is first introduced at the C-6 position by a stereoselective enzymatic hydrogenation of the double bond using old yellow enzyme 2 of Saccharomyces cerevisiae, expressed in Escherichia coli, as a biocatalyst. Thereafter, the carbonyl group at the C-4 position is reduced selectively and stereospecifically by levodione reductase of Corynebacterium aquaticum M-13, expressed in E. coli, to the corresponding alcohol. Commercially available glucose dehydrogenase was also used for cofactor regeneration in both steps. Using this two-step enzymatic asymmetric reduction system, 9.5 mg of (4R,6R)-4-hydroxy-2,2,6-trimethylcyclohexanone/ml was produced almost stoichiometrically, with 94% enantiomeric excess in the presence of glucose, NAD+, and glucose dehydrogenase. To our knowledge, this is the first report of the application of S. cerevisiae old yellow enzyme for the production of a useful compound.
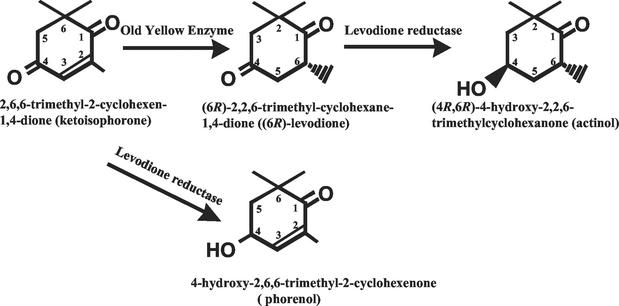
(4R,6R)-4-hydroxy-2,2,6-trimethylcyclohexanone (actinol) is an ideal precursor for the synthesis of naturally occurring, optically active hydroxylated carotenoids, such as zeaxanthin (8), cryptoxanthin, and structurally related compounds (1, 2). Two-step conversion of 2,6,6-trimethyl-2-cyclohexen-1,4-dione (ketoisophorone) to 4-hydroxy-2,2,6-trimethylcyclohexanone (Fig. 1) by bacterial cells has been reported previously (3, 11, 12, 19). However, in those cases, mixtures of isomers, (4R,6S), (4S,6R), (4R,6R), and (4S,6S), were produced and the productivity was low (∼2.5 mg/ml). The enzymes involved in these reactions have not yet been identified.
FIG. 1.
Two-step conversion of 2,6,6-trimethyl-2-cyclohexen-1,4-dione to (4R,6R)-4-hydroxy-2,2,6-trimethylcyclohexanone using OYE and LVR.
The discovery, purification, and characterization of levodione reductase (LVR), which catalyzes the regio- and stereospecific reduction of (6R)-2,2,6-trimethylcyclohexane-1,4-dione [(6R)-levodione] to actinol, has been reported (17). This LVR has also been cloned and expressed in Escherichia coli (21). Recently, it was also reported that old yellow enzyme (OYE; EC 1.6.99.1) of Candida macedoniensis catalyzes the stereospecific hydrogenation of ketoisophorone to (6R)-levodione (4). However, this enzyme has not yet been cloned, and enzyme purification from C. macedoniensis is needed for (6R)-levodione production. Thus, we tried to use OYE of Saccharomyces cerevisiae, for which the whole-genome sequence is now available. S. cerevisiae has two OYEs: OYE2, encoded by the OYE2 gene (15), and OYE3, encoded by the OYE3 gene (10). Both of them were cloned and expressed in E. coli, and the conversion efficiencies of ketoisophorone to (6R)-levodione were then compared.
Here, we report actinol production by the stereoselective hydrogenation of the double bond of ketoisophorone and by the stereoselective reduction of the carbonyl group at the C-4 position of (6R)-levodione. For the first step, hydrogenation, the OYE2 of S. cerevisiae expressed in E. coli was used, and for the second step, carbonyl reduction, the LVR of Corynebacterium aquaticum M-13 expressed in E. coli was used. For cofactor regeneration, commercially available glucose dehydrogenase (GDH) was used because it has been widely employed for enzymatic asymmetric reduction (6, 20). To our knowledge, this is the first report of the application of S. cerevisiae OYE for the production of a useful compound.
MATERIALS AND METHODS
Microorganisms and cultivation.
S. cerevisiae S288C was used as the source of chromosomal DNA. E. coli JM109 {recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 Δ(lac-proAB) F′[traD36 proAB+ lacIq lacZ ΔM15] e14− (McrA−)} was used as a host for gene cloning and for plasmid amplification. E. coli cells were grown at 37°C in Luria-Bertani (LB) medium containing 1% Bacto Tryptone (Difco Laboratories, Detroit, Mich.), 0.5% Bacto Yeast Extract (Difco Laboratories), and 1% NaCl (pH 7.0). When necessary, ampicillin (50 μg/ml) was added to the medium.
For enzyme production, E. coli JM109 was cultivated in M9 minimal medium [10% glucose, 4.5% (NH4)2SO4, 0.1% K2HPO4, 0.1% MgSO4 · 7H2O, 1% FeSO4 · 7H2O, 1% MnSO4 · 5H2O, 0.03% thiamine-HCl, and 0.01% biotin] containing 2% Casamino Acids and 50 μg of ampicillin/ml (M9CA). The pH was adjusted to 7.0 using NaOH.
Plasmid construction.
To obtain the complete coding sequence for the OYE gene without the excess flanking region, two-step PCR amplification was performed. For the first step of the amplification of OYE2, OYE2-1 (+) (5′-CGGTCCAGATATAGAATAAATCATCATATTAAG-3′) and OYE2-2 (−) (5′-GAAATGGTGCTACAAAGTACGGTTAACAC-3′) were used, and for the second step, OYE2-3 (+) (5′-TTAGAAGAATTCATGCCATTTGTTA-3′) (the EcoRI site is underlined) and OYE2-4 (−) (5′-AGATTTCTGCAGTTAATTTTTGTCC-3′) (the PstI site is underlined) were used. For the first step of the amplification of OYE3, OYE3-1 (+) (5′-GTACGTACTTGATATATACAACAACTGTAG-3′) and OYE3-2 (−) (5′-GCTGCCCTATATAAACAAAGATCGAGTC-3′) were used, and for the second step, OYE3-3 (+) (5′-TTAGAACAATTGATGCCATTTGTAA-3′) (the MfeI site is underlined) and OYE3-4 (−) (5′-AGATTTCTGCAGTCAGTTCTTGTT-3′) (the PstI site is underlined) were used. The chromosomal DNA from S. cerevisiae S288C and the first PCR product were used as the templates for the first PCR and the second PCR, respectively. The PCR-generated DNA fragment was digested with EcoRI and PstI (for OYE2) or with MfeI and PstI (for OYE3) and then ligated into the expression vector pKK223-3 (Amersham Bioscience, Piscataway, N.J.) cleaved with EcoRI and PstI. The ligated plasmids were then used to transform E. coli JM109. After selection with ampicillin, several clones were picked up, and the nucleotide sequence of the plasmid DNA was examined. The constructed plasmids were designated pKKOYE2 and pKKOYE3. The nucleotide sequences of the plasmids were confirmed using a model 377 DNA sequencer (Applied Biosystems, Foster City, Calif.) employing the dideoxy chain termination method.
Expression of LVR.
The LVR gene of C. aquaticum M-13 was expressed in E. coli as described previously (21).
Preparation of cell extracts and enzyme assay.
E. coli JM109 harboring pKKOYE2 or pKKOYE3 cells was grown at 37°C in M9CA medium. When absorbance at 610 nm reached 0.4, isopropyl-β-d-thiogalactopyranoside was added to the culture medium to attain a final concentration of 0.1 mM in order to induce gene expression. After cultivation for 15 h at 37°C, the cells were harvested by centrifugation and suspended in buffer (40 mM Tris-HCl [pH 8.0], 10 mM MgCl2, 10 mM dithiothreitol [DTT], 200 mM KCl, 10% glycerol, 1 mM phenylmethylsulfonyl fluoride). The cell extracts were prepared by sonication under cooling and were then centrifuged. The soluble fractions were used for the enzyme assay and the reduction of ketoisophorone. The enzymatic activity of OYE was determined by spectrophotometrically measuring the 2-cyclohexen-1-one-dependent decrease in the NADPH content as described previously (10, 15). The reaction was done at 37°C for 1 min. The standard 3.0-ml assay mixture contained 1 mM 2-cyclohexen-1-one, 100 mM potassium phosphate buffer (pH 7.0), 120 μM NADPH, and the enzyme. Preparation of the cell extracts and the method of determining the enzymatic activity for LVR were performed as described previously (21). One unit of enzyme activity was defined as the amount of enzyme that catalyzed the oxidation of 1 μmol of NADH or NADPH per min.
Ketoisophorone reduction with cell extract of E. coli expressing OYE genes.
A 10-ml reaction mixture containing 0.66 mmol (1% [wt/vol]) of ketoisophorone, 250 mM Tris-HCl buffer (pH 8.0), 3.1 μmol of NAD+, 125 U of GDH (Amano Enzyme, Nagoya, Japan), 2.2 mmol of glucose, and 26 U of cell extract of E. coli JM109 expressing OYE2 was incubated at 25°C. At 15-min intervals for 2 h, 1 ml of the mixture was extracted and vigorously shaken with the equivalent volume of ethyl acetate. For the comparison of (6R)-levodione productivity in OYE2 and OYE3, 2.5 ml of reaction mixture containing 25 mM Tris-HCl (pH 8.0), 0.1 mM DTT, 1.3 U of cell extract of E. coli JM109 expressing pKKOYE2 or pKKOYE3, 26 mM NADH or 22 mM NADPH, and 16 μmol (0.1%[wt/vol]) of ketoisophorone was used, and the reaction mixture was incubated at 25°C for 30 min. Then, the reaction mixture was extracted using 1 ml of ethyl acetate. The ethyl acetate layer was analyzed in order to determine its levodione and actinol contents using gas chromatography (GC) as described below.
One-step enzymatic conversion from ketoisophorone to actinol.
A 2.5-ml reaction mixture containing 16 μmol (0.1% [wt/vol]) ketoisophorone, 25 mM Tris-HCl (pH 8.0), 0.1 mM DTT, 26 mM NADH, 1.3 U of cell extract of E. coli JM109 expressing OYE2, and 0.1, 0.2, or 0.5 U of LVR was incubated at 25°C for 30 min. After 30 min, the reaction mixture was extracted using 1 ml of ethyl acetate. The ethyl acetate layer was analyzed in order to determine its levodione and actinol contents using GC and GC-mass spectrometry (MS) as described below.
Two-step enzymatic conversion of ketoisophorone to actinol.
A 25-ml reaction mixture containing 1.6 mmol (1% [wt/vol]) of ketoisophorone, 250 mM Tris-HCl buffer (pH 8.0), 8.4 μmol of NAD+, 312 U of GDH, 5.5 mmol of glucose, and 32.9 U of cell extract of E. coli JM109 expressing OYE2 was incubated at 25°C. At 15-min intervals for 45 min, 1 ml of the mixture was extracted and vigorously shaken with the equivalent volume of ethyl acetate. After 45 min, 10 U of LVR was added to the reaction mixture, which was then incubated at 25°C. At 15-min intervals for another 120 min, the mixture was extracted as described above. The pH of the reaction mixture was automatically controlled in the range of 7.0 to 8.0 using 7% (vol/vol) ammonia solution. The ethyl acetate layer was analyzed in order to determine and identify its levodione and actinol contents using GC and GC-MS as described below.
Determination of the product.
The amounts of ketoisophorone, levodione, and actiol were determined by GC under the following conditions. The analysis was performed using a Shimadzu model GC-14B GC equipped with a flame ionization detector by utilizing a type BGB-176 capillary column (0.25 mm by 30 m; BGB ANALYTIK, Anwil, Switzerland). The column temperature program was 107°C for 30 min, 5°C/min to 129°C, 129°C for 15 min, 5°C/min to 180°C, and 180°C for 5 min. Helium was used as a carrier gas at a flow rate of 1 ml/min. Under these conditions, ketoisophorone; (6R)-levodione; (6S)-levodione; actinol; (4R,6S)-, (4S,6R)-, and (4S,6S)-4-hydroxy-2,2,6-trimethylcyclohexanone; and (4R)- and (4S)-4-hydroxy-2,6,6-trimethyl-2-cyclohexenone (phorenol) were eluted at 24.2, 23.4, 29.1, 37.5, 39.0, 39.4, 40.3, and 42.5 min, respectively. For the identification of the reaction product, GC-MS was performed using the HP 5973MSD system (Hewlett-Packard, Palo Alto, Calif.) equipped with an HR-20 M capillary column (0.25 mm by 30 m; Shinwa Chemical Industries, Kyoto, Japan) at 160°C (isothermal). The carrier gas and flow rate were the same as those for GC. The GC-MS was operated with a mass range from m/z 20 to 200 and with an ionization voltage of 70 eV.
Enzymes and chemicals.
Restriction enzymes were purchased from Takara Shuzo Co. Ltd. (Kyoto, Japan) and Toyobo Biochemicals (Osaka, Japan). Ketoisophorone; (6R)- or (6S)-levodione; actinol; (4S,6R)-, (4R,6S)-, and (4S,6S)-4-hydroxy-2,2,6,-trimethylcyclohexenone; and (4R)- or (4S)-phorenol were kindly donated by Nippon-Roche Co., Kamakura, Japan. All other chemicals used in this study were of analytical grade and commercially available.
Other methods.
The molecular masses of the enzyme and protein concentrations were determined as described previously (17).
RESULTS
Expression of S. cerevisiae OYE in E. coli.
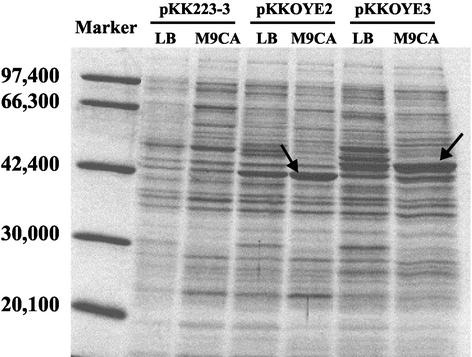
The cell extracts of E. coli JM109-expressed OYE genes were loaded on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2). It has been reported that the molecular masses calculated from amino acid sequences are 45,010 Da (OYE2) (10) and 44,920 Da (OYE3) (15) and that the apparent molecular masses of both OYE proteins on SDS-PAGE are clearly different (15). The apparent molecular masses of these enzymes on SDS-PAGE were consistent with reported values (10, 15). Such anomalies have been reported for a change as small as a single-amino-acid substitution and could be a function of a change in the association of the protein (13). OYE2 seemed to be expressed at almost the same level in the E. coli cell. Several enriched protein bands were observed in an OYE3-overexpressing strain, although the reason for this phenomenon is unknown. As for the medium, better expression was observed in E. coli cultivated in M9CA medium than in E. coli cultivated in LB medium. Thus, M9CA medium was selected for OYE production.
FIG. 2.
SDS-PAGE of OYE2 and OYE3 from soluble fractions of E. coli JM109 harboring pKK223-3 (vector only), pKKOYE2, and pKKOYE3. Two types of cultivation medium, LB and M9CA, were tested. The arrows indicate the positions of the expressed OYE2 (left) and OYE3 (right). The marker standards were phosphorylase b (97,400), bovine serum albumin (66,300), aldolase (42,400), carbonic anhydrase (30,000), and trypsin inhibitor (20,100). The gel was stained for protein using Coomassie brilliant blue R-250 and destained in methanol-acetic acid-water (40:7:53).
Comparison of OYE2 and -3.
In order to evaluate the (6R)-levodione productivities of OYE2 and -3, the enzymes were used for the reduction of ketoisophorone. OYE2 showed higher productivity of (6R)-levodione than did OYE3 (Table 1). In both cases, the optical purity of the (6R)-levodione produced exceeded ∼95.0% enantiomeric excess. This result showed that OYE2 is suitable for the production of (6R)-levodione. The OYE-nonexpressing E. coli strain (pKK223-3) also exhibited a considerable level of ketoisophorone-reducing activity. This activity may have originated from an OYE homologue, the N-ethylmaleimide reductase gene, in the E. coli chromosome (9).
TABLE 1.
(6R)-Levodione productivity by the cell extract of E. coli JM109 expressing OYE2 or OYE3 of S. cerevisiae
| Coenzyme plasmid introduced | (6R)-Levodione yield (%)a
|
|
|---|---|---|
| NADH | NADPH | |
| pKK223-3 | 8.4 ± 0.2 | 9.9 ± 0.1 |
| pKKOYE2 | 58.7 ± 3.4 | 34.4 ± 1.9 |
| pKKOYE3 | 12.1 ± 0.9 | 11.5 ± 1.1 |
Added ketoisophorone (0.1% [wt/vol]) was taken as 100%. The values are averages ± standard deviations resulting from three different determinations.
Coenzyme specificity.
In order to evaluate the coenzyme specificities of OYE2 and -3, the reaction was examined with NADH and NADPH. Productivity with NADH is higher than that with NADPH (Table 1.). This result showed that NADH is suitable for the preparative production of (6R)-levodione as a coenzyme. On the other hand, the specificity of the coenzyme in the case of 2-cyclohexenen-1-one reduction has been reported to be the opposite of that in ketoisophorone reduction (10, 15).
Enzymatic conversion from ketoisophorone to levodione.
When the reduction was performed using ketoisophorone as a substrate, OYE2 preferred NADH as a coenzyme. The production of (6R)-levodione with OYE2 and NADH was carried out under optimum conditions. The NADH regeneration system containing NAD+, GDH, and glucose was used for sufficient reduction of ketoisophorone. Using 1% (wt/vol) ketoisophorone as the substrate, 96.4% of the substrate was reduced to (6R)-levodione with a high optical purity of 95.5% enantiomeric excess in 45 min. The reaction product formed after incubation with ketoisophorone, the cell extract, and the NADH regeneration system was analyzed using GC-MS. (6R)-Levodione produced by OYE2 in the presence of NADH had a molecular ion of m/z 154, which corresponds to that of the authentic compound.
One-step enzymatic conversion from ketoisophorone to actinol.
A one-step reaction was carried out with ketoisophorone at a concentration of 0.1% (wt/vol). OYE2 and LVR were added to the reaction mixture at the same time, and the reduction then began. With the increase in LVR concentration, the productivity of actinol increased. However, the content of phorenol (Fig. 1), which contains the hydroxy group at the C-4 position of ketoisophorone, increased too. When 0.5 U of LVR was used, 0.016% (wt/vol) actinol was produced after a 30-min reaction; however, 0.048% (wt/vol) phorenol also accumulated. The accumulated phorenol did not decrease during prolonged incubation, suggesting that OYE2 does not reduce the carbon-carbon double bond of phorenol. This result showed that the one-step reaction was not suitable for actinol production, because coproduced phorenol was not converted to actinol.
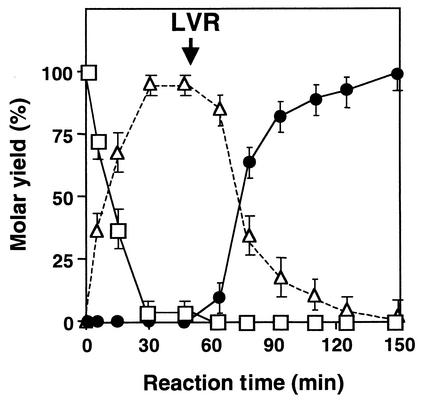
Two-step enzymatic conversion from ketoisophorone to actinol.
A two-step reaction was carried out using an initial ketoisophorone concentration of 1% (wt/vol) (Fig. 3). The NADH regeneration system containing NAD+, GDH, and glucose was used for sufficient reduction of ketoisophorone and levodione. In the first-step reaction from ketoisophorone to levodione, 95.4% of the ketoisophorone added was reduced to (6R)-levodione with a high optical purity of 94.5% enantiomeric excess in 45 min. In the second-step reaction from levodione to actinol, the almost 100% levodione produced by the first-step reaction was reduced to actinol with a high optical purity of 94.2% enantiomeric excess in ∼150 min. The conversion rate of the second step was lower than that of the first step. The reoxidation of actinol may affect the reaction rate because LVR catalyzes reversible oxidoreduction between actinol and levodione (17) or because the substrate concentration is not sufficient for the reduction of levodione.
FIG. 3.
Two-step enzymatic conversion from ketoisophorone to actinol. The reaction was carried out as described in Materials and Methods. The symbols indicate the molar residual rate of ketoisophorone (open squares) and the molar yields of (6R)-levodione (open triangles) and actinol (solid circles). The arrow indicates the time of addition of LVR. Each point represents the mean of three replicates and includes standard-error bars.
The reaction product formed after 150 min was analyzed by GC-MS. Actinol, produced by OYE2 and LVR in the presence of NADH, had a molecular ion of m/z 156, which is identical to that of the authentic compound.
DISCUSSION
Microbial production of actinol from ketoisophorone has been reported previously (3, 11, 12, 19); however, in those cases, a racemic mixture of 4-hydroxy-2,2,6-trimethylcyclohexanone was obtained as the reduction product, and the enzymes involved in reduction of ketoisophorone and levodione have not yet been purified. In this study, we have established an overexpression system for S. cerevisiae OYE in E. coli, and the system provides a powerful tool for obtaining optically pure actinol, together with the LVR overexpression system established previously (21).
Many reports have described the asymmetric reduction of ketones to corresponding optically pure secondary alcohol. On the other hand, few reports are available regarding the asymmetric hydrogenation of the carbon-carbon double bond using enzymes. Several enone reductases from S. cerevisiae which can reduce the double bond of ketoisophorone have been reported. These are the enone reductases EI and EII, reported by Wanner and Tressl (18), and OYE, reported by Vaz et al. (16). Kawai et al. (7) also reported a carbon-carbon double-bond reductase from S. cerevisiae; however, its biochemical character remains unknown, and its activity toward ketoisophorone has not been reported. The activity of S. cerevisiae OYE toward ketoisophorone is known (16); however, its application for (6R)-levodione production has not yet been reported. To our knowledge, this is the first report of the application of S. cerevisiae OYE for the practical production of a useful compound. This report also provides the first example of production of the optically pure doubly chiral compound actinol using stepwise enzymatic conversion.
In this system, commercially available GDH should be added to the reaction mixture together with glucose and NAD+ as a cofactor regenerator, because both OYE2 and LVR require NADH for ketoisophorone and levodione reduction. GDH from Bacillus megaterium is stable in various organic solvents (14), and the system with GDH as a cofactor regenerator has been used for many studies of chiral alcohol production (6, 20). It has already been shown that E. coli cells overexpressing the GDH gene from B. megaterium could be used as a cofactor regenerator instead of commercially available GDH in the production system of optically active ethyl 4-chloro-3-hydroxybutanoate (5). Application of this cofactor regeneration system to actinol production is in progress.
In conclusion, using three different enzymes, two-step asymmetric reduction of ketoisophorone to a doubly chiral compound, actinol, was successful for the first time.
Acknowledgments
This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (no. 14760053) to M.W. and by a Sasakawa Scientific Research Grant from the Japan Science Society to A.Y.
REFERENCES
- 1.Britton, G., S. Liaaen-Jensen, and H. Pfander (ed.). 1996. Carotenoids: synthesis. Birkhauser Verlag, Basel, Switzerland.
- 2.Burden, R. S., and H. F. Taylor. 1970. The structure and chemical transformation of xanthoxin. Tetrahedron Lett. 47:4071-4074. [DOI] [PubMed] [Google Scholar]
- 3.Hori, N., T. Hieda, and Y. Mikami. 1984. Microbial conversion of 4-oxoisophorone by the thermophile Thermomonospora curvata. Agric. Biol. Chem. 48:123-129. [Google Scholar]
- 4.Kataoka, M., A. Kotaka, A. Hasegawa, M. Wada, A. Yoshizumi, S. Nakamori, and S. Shimizu. 2002. Old yellow enzyme from Candida macedoniensis catalyzes stereospecific reduction of C=C bond of ketoisophorone. Biosci. Biotechnol. Biochem. 65:2651-2657. [DOI] [PubMed] [Google Scholar]
- 5.Kataoka, M., L. P. S. Rohani, M. Wada, K. Kita, H. Yanase, I. Urabe, and S. Shimizu. 1998. Escherichiacoli transformant expressing the glucose dehydrogenase gene from Bacillus megaterium as a cofactor regenerator in a chiral alcohol production system. Biosci. Biotechnol. Biochem. 62:167-169. [DOI] [PubMed] [Google Scholar]
- 6.Kataoka, M., L. P. S. Rohani, K. Yamamoto, M. Wada, H. Kawabata, K. Kita, H. Yanase, and S. Shimizu. 1997. Enzymatic production of ethyl (R)-4-chloro-3-hydroxybutanoate: asymmetric reduction of ethyl 4-chloro-3-oxobutanoate by an Escherichia coli transformant expressing the aldehyde reductase gene from yeast. Appl. Microbiol. Biotechnol. 48:699-703. [DOI] [PubMed] [Google Scholar]
- 7.Kawai, Y., M. Hayashi, Y. Inaba, K. Saitou, and A. Ohno. 1998. Asymmetric reduction of α,β-unsaturated ketones with a carbon-carbon double bond reductase from baker's yeast. Tetrahedron Lett. 39:5225-5228. [Google Scholar]
- 8.Leuenberger, H. G. W., W. Boguth, E. Widmer, and R. Zell. 1976. Synthesis of optically active natural carotenoids and structurally related compounds. I. Synthesis of chiral key compound (4R,6R)-4-hydroxy-2,2,6-trimethylcyclohexanone. Helv. Chim. Acta 59:1832-1849.
- 9.Miura, K., Y. Tomioka, H. Suzuki, M. Yonezawa, T. Hishinuma, and M. Mizugaki. 1997. Molecular cloning of the nemA gene encoding the N-ethylmaleimide reductase from Escherichia coli. Biol. Pharm. Bull. 20:110-112. [DOI] [PubMed] [Google Scholar]
- 10.Niino, Y. S., S. Chakraborty, B. J. Brown, and V. Massey. 1995. A new old yellow enzyme of Saccharomyces cerevisiae. J. Biol. Chem. 270:1983-1991. [DOI] [PubMed] [Google Scholar]
- 11.Nishii, K., K. Sode, and I. Karube. 1989. Microbial conversion of dihydrooxoisophorone (DOIP) to 4-hydroxy-2,2,6-trimethylcyclohexanone (4-HTMCH) by thermophilic bacteria. J. Biotechnol. 9:117-128. [Google Scholar]
- 12.Nishii, K., K. Sode., and I. Karube. 1990. Sequential two-step conversion of 4-oxoisophorone to 4-hydroxy-2,2,6-trimethylcyclohexanone by thermophilic bacteria. Appl. Microbiol. Biotechnol. 33:245-250. [Google Scholar]
- 13.Noel, D., K. Nikaido., and G. F. Ames. 1979. A single amino acid substitution in a histidine-transport protein drastically alters its mobility in sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry 18:4159-4165. [DOI] [PubMed] [Google Scholar]
- 14.Shimizu, S., M. Kataoka, M. Katoh, T. Morikawa, T. Miyoshi, and H. Yamada. 1990. Stereoselective reduction of ethyl 4-chloro-3-oxobutanoate by a microbial aldehyde reductase in an organic solvent-water diphasic system. Appl. Environ. Microbiol. 56:2374-2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stott, K., K. Saito, D. J. Thiele, and V. Massey. 1993. Old Yellow Enzyme, the discovery of multiple isozymes and a family of related proteins. J. Biol. Chem. 268:6097-6106. [PubMed] [Google Scholar]
- 16.Vaz, A. D. N., C. Sumita, and V. Massey. 1995. Old Yellow Enzyme: aromatization of cyclic enones and the mechanism of a novel dismutation reaction. 34:4246-4256. [DOI] [PubMed]
- 17.Wada, M., A. Yoshizumi, S. Nakamori, and S. Shimizu. 1999. Purification and characterization of monovalent cation-activated levodione reductase from Corynebacterium aquaticum M-13. Appl. Environ. Microbiol. 65:4399-4403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wanner, P., and R. Tressl. 1998. Purification and characterization of two enone reductases from Saccharomyces cerevisiae. Eur. J. Biochem. 255:271-278. [DOI] [PubMed] [Google Scholar]
- 19.Yamazaki, Y., Y. Hayashi, N. Hori, and Y. Mikami. 1988. Microbial conversion of 4-oxoisophorone by Aspergillus niger. Agric. Biol. Chem. 52:2919-2920. [Google Scholar]
- 20.Yasohara, Y., N. Kaizaki, J. Hasegawa, S. Takahashi, M. Wada, M. Kataoka, and S. Shimizu. 1999. Synthesis of optically active ethyl 4-chloro-3-hydroxybutanoate by microbial reduction. Appl. Microbiol. Biotechnol. 51:847-851. [DOI] [PubMed] [Google Scholar]
- 21.Yoshizumi, A., M. Wada, H. Takagi, S. Shimizu, and S. Nakamori. 2001. Cloning, sequence analysis, and expression in Escherichia coli of the gene encoding monovalent cation-activated levodione reductase from Corynebacterium aquaticum M-13. Biosci. Biotechnol. Biochem. 65:830-836. [DOI] [PubMed] [Google Scholar]