Abstract
The use of 1 N HCl for extraction of small, acid-soluble proteins (SASP) from different Bacillus spore species was examined. The extracts were analyzed by high-performance liquid chromatography and matrix-assisted laser desorption mass spectrometry and were found to be both qualitatively and quantitatively superior to extraction by acetonitrile-5% trifluoroacetic acid (70:30, vol/vol). Both major and minor α/β- and γ-type SASP were characterized by their molecular masses or tryptic peptide maps and by searches of both protein and unannotated genome databases. For all but 1 pair (B. cereus T and B. thuringiensis subsp. Kurstaki) among the 11 variants studied the suites of SASP masses are distinctive, consistent with the use of these proteins as potential biomarkers for spore identification by mass spectrometry.
Significant effort has been expended in recent years to develop rapid techniques for identification of spores of Bacillus species, in particular those of B. anthracis. Some of these techniques have utilized mass spectrometry to identify biomarkers characteristic of spores of these organisms. In the latter analyses the biomarkers have most often been released in situ for laser desorption by treating spores with acetonitrile:water:trifluoroacetic acid (TFA), and species of 1.5 to 10 kDa in molecular size have been characterized by using matrix-assisted laser desorption-time of flight mass spectrometry (MALDI-TOF [MS]) (4, 5, 8, 13, 20). Most of these desorbed biomarkers were found to be secondary metabolites (9, 13) synthesized by the microorganism during growth and could be used to differentiate between different Bacillus spore species. Unfortunately, it has been noticed that these secondary metabolites can quantitatively (8) and qualitatively (13) change with culture conditions, which makes them not very reliable biomarkers for spore identification. However, much higher levels of biomarkers were detected when spores, suspended in acetonitrile:water:TFA, were also subjected to corona plasma discharge or sonication (8, 20). Some of these biomarkers were identified as members of the small, acid-soluble spore protein (SASP) family (2, 20).
The SASP (6 to 10 kDa) are of two types, α/β-type and γ-type, named after the predominant protein(s) of these types in B. subtilis spores (3, 23). Both types of SASP are synthesized only in the developing spore late in sporulation, and they comprise 8 to 15% of total spore protein and even more of the spore's soluble protein. The α/β-type SASP are products of a multigene family with 4 to 7 genes in both Bacillus and Clostridium species. While all of these genes (termed ssp) are expressed in parallel, in spores of Bacillus species two proteins make up ≥80% of the α/β-type SASP pool, which is ∼50% of the total SASP. Although the α/β-type SASP exhibit no sequence similarity to other proteins or protein motifs in available databases, these proteins are extremely similar in amino acid sequence both within and across species. However, even among closely related species there are significant differences among residues in both the C-terminal and N-terminal regions of α/β-type SASP. The likely reason for the high amino acid sequence conservation in α/β-type SASP is that these proteins are nonspecific DNA binding proteins that saturate the spore DNA. This α/β-type SASP binding protects the spore DNA against many types of damage, and consequently α/β-type SASP are a major cause of the extreme resistance of spores of Bacillus and Clostridium species to many different treatments (17, 18, 24). In contrast to the α/β-type SASP, spores of Clostridium species lack γ-type SASP and spores of Bacillus species have only a single γ-type SASP. However, in Bacillus species the gene encoding the γ-type SASP is expressed in parallel with those encoding α/β-type SASP. The γ-type SASP are generally larger than α/β-type SASP, and the only significant sequence homology between the two groups of SASP is in a short pentapeptide region that is the recognition site for the cleavage of these proteins by a specific protease early in spore germination. The γ-type SASP do exhibit sequence homology across species, although this is much less than that for the α/β-type SASP. The reason for the more rapid divergence of γ-type than α/β-type SASP sequences in evolution may be because the only role for the γ-type SASP appears to be to provide a large pool of amino acids early in spore germination when all SASP are degraded. The facts that SASP are such abundant proteins in spores and exhibit sequence differences between closely related species make them good candidates as biomarkers for spore identification. However, effective extraction of these SASP is required.
Previous studies (12, 22) have demonstrated that treatment of spores with 2 N HCl resulted in the release of relatively large amounts of SASP as revealed by gel electrophoretic analysis. Consequently, we have tried this approach for efficient SASP extraction from spores with subsequent SASP identification by MALDI (MS). We have also used a number of B. subtilis strains engineered to have different SASP profiles to demonstrate the unambiguous identification of the spores of these strains and the SASP they contain.
MATERIALS AND METHODS
Bacillus strains and spore preparation.
One set of samples comprises the wild-type B. subtilis strain PS832, a trp+ derivative of strain 168, and derivatives of this strain, including PS355, lacking the genes sspA and sspB, which encode the major α/β-type SASP-α and -β, respectively (15), and also carry a chloramphenicol resistance marker; PS223, lacking the gene encoding SASP-β (16); PS260, lacking the gene encoding SASP-α (16); PS483, lacking the sspE gene encoding SASP-γ (7); PS1450, lacking the genes encoding SASP-α and -β and also carrying a high-copy plasmid with the gene encoding the normally minor α/β-type SASP, SspC, under control of the strong forespore-specific sspB promoter (26); and PS3019, lacking the genes encoding SASP-α and -β and also carrying a high-copy plasmid carrying a modified sspC gene encoding SspCΔ11D13K under the control of the strong forespore-specific sspB promoter (10). B. cereus strain T, originally obtained from H. O. Halvorson, Bacillus globigii, originally obtained from the U.S. Army Laboratories at Dugway Proving Ground, and B. thuringiensis subsp. Kurstaki HD-1 (ATCC 33679) were also studied. B. anthracis Sterne is a nonpathogenic veterinary vaccine strain. Even though B. anthracis Sterne used in this study is a nonpathogenic veterinary vaccine strain, its culture and handling was carefully carried out by using a class II laminar flow hood.
Spores of B. subtilis strains were prepared by growth at 37°C on 2× Schaeffer’s glucose medium agar plates without antibiotics, and the spores were harvested and purified as described previously (19). B. thuringiensis subsp. Kurstaki HD-1 and B. anthracis Sterne spores were prepared in new sporulation medium (NSM) containing 0.3% tryptone, 0.3% yeast extract, 0.2% Bacto-agar, 2.3% Lab-Lemco agar, and 0.001% MnCl2. B. globigii spores were grown on NSM, chemically defined sporulation medium, and casein acid digest medium (8). B. cereus T spores were prepared by growth at 30°C on supplemented nutrient broth agar plates (6), and the spores were harvested and purified with the same method used for the B. subtilis spores. All spore preparations were free (>98%) of growing cells, germinated spores, and cell debris. Spores were routinely stored in water at 10°C protected from light.
Assay of spore viability and DPA release.
For determination of spore killing, spores (∼0.2 mg [dry weight]) were incubated at room temperature in 1 ml of 70% acetonitrile, with the other 30% being various concentrations of TFA in water. At various times aliquots were diluted 1/10 in 25 mM KPO4 (pH 7.4) and then were serially diluted 10-fold in this same buffer. Aliquots of the dilutions were spotted on 2× yeast extract-tryptone medium agar plates (per liter: NaCl, 5 g; tryptone, 16 g; yeast extract, 10 g), the plates were incubated overnight at 30 to 37°C, and colonies were enumerated. In other experiments, spores (∼0.2 mg [dry weight]) in 1 ml of 70% acetonitrile, with the other 30% being various concentrations of TFA, were centrifuged after incubation at room temperature, and the pellet fraction was washed two times by centrifugation with 1 ml of water. Pyridine-2,6-dicarboxylic acid (dipicolinic acid [DPA]) remaining in the spore pellet after this treatment was extracted and analyzed as described previously (19).
Extraction of SASP from dry spores.
About 4 mg of dry spores was suspended in 400 μl of 1 N HCl, the suspension was left to stand at room temperature for 90 min with occasional vortexing, and the suspension was pelleted by centrifugation at 10,000 × g for 5 min. The supernatant fluid was recovered and stored at −80°C until analysis.
In other experiments, spores (4 mg) were suspended in 400 μl of acetonitrile-5% TFA in water (70:30, vol/vol) for 90 min at room temperature and were centrifuged at 10,000 × g for 5 min. The supernatant fluid was transferred to a clean microcentrifuge tube, and the acetonitrile was evaporated under a stream of argon and was replaced with 5% TFA in water. The sample was then stored as described above.
Chromatographic analysis of spore extracts.
High-performance liquid chromatography (HPLC) analysis of spore extracts was performed by using an Aquapore C-8 column (RP-300; 7 μ; 250 by 4.6 mm; Applied Biosystems, San Jose, Calif.). The column was connected to a Shimadzu liquid chromatography system equipped with LC-600 delivery pumps, an SPD-6A UV detector, and a C-P6A chromatogram recorder. The solvent system consisted of 0.1% TFA in water (solvent A) and 0.08% TFA in acetonitrile (solvent B) at a flow rate of 1 ml/min. Aliquots of 350 μl of the spore extracts were injected into the column and were washed first with solvent A for 6 min followed by linear gradient of solvent B, developed from 10 to 80% in 30 min. The column was then reequilibrated with solvent A for 15 min before another injection. Peaks were detected at 215 nm, collected in microcentrifuge tubes, and then freeze-dried.
Mass spectrometry analysis.
The total spore extracts or the HPLC-separated peaks were analyzed by MALDI-TOF (MS) (Kompact MALDI 4; Kratos Analytical, Chestnut Ridge, N.Y.). The instrument, equipped with a nitrogen laser and pulsed extraction, was operated in positive linear mode with an extraction voltage set at −20 kV. Ubiquitin (human) with a molecular mass of 8,565 Da was used as the external calibrant. An aliquot of 0.3 μl of spore extract or of the HPLC-collected peak that was previously redissolved in 10 μl of acetonitrile/0.1% TFA (70:30, vol/vol) was mixed with 0.3 μl of 50 mM sinapinic acid in acetonitrile:0.1% TFA (70:30, vol/vol) and was air dried. Spectra were recorded by accumulating 50 to 100 laser shots across the spot. The mass accuracy obtained in this instrument was typically about ±0.1% of a given measured mass.
Further mass spectrometry analyses of the HPLC-collected fractions were performed on a hybrid quadrupole time of flight (Q-TOF) instrument (QStar/Pulsar; Applied Biosystems, Foster City, Calif.) equipped with a nanospray ion source. Dried samples were typically dissolved in a small volume of methanol/water/acetic acid (50:50:2, vol/vol/vol), and 2 μl of this solution was loaded into a capillary nanospray tip (Protana, Odense, Denmark) and mounted into the source. The spray voltage was typically set at 0.9 kV. The Q-TOF mass spectra were collected between 600 and 2,500 atomic mass units for a period of 1 min, and the mass accuracy was better than 100 ppm.
Genome sequence data.
Preliminary sequence data for B. cereus and B. anthracis were obtained from The Institute for Genome Research website at http://www.tigr.org. The ssp genes were identified by a BLAST search against B. subtilis α/β- and γ-type SASP followed by translation of the located DNA region in DNA Strider to identify complete open reading frames. The initiation codons of these genes were identified in part by homology to the B. subtilis SASP but also, and more importantly, by the presence of a strong potential ribosome binding site at the appropriate spacing just upstream of the initiation codons.
RESULTS AND DISCUSSION
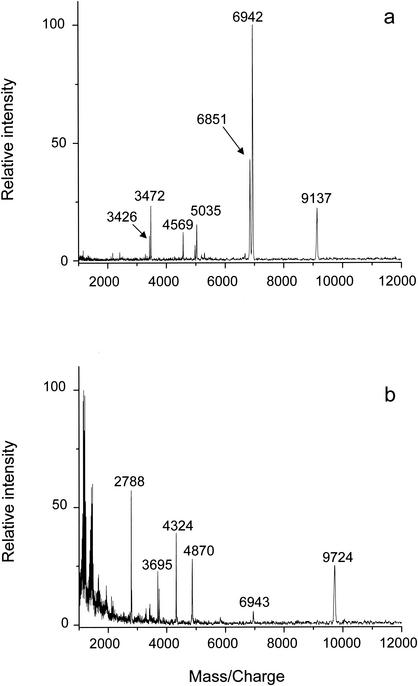
MALDI-TOF mass spectral profiles of extracts of wild-type B. subtilis spores obtained with acetonitrile-5% TFA (70:30, vol/vol) and HCl are shown in Fig. 1. Overall, the HCl extract (Fig. 1a) provided more peaks corresponding to the expected masses of major SASP than the acetonitrile-5% TFA extract (Fig. 1b). The peaks at m/z 6,851, 6,942, and 9,137 detected in the HCl extract of B. subtilis spores are close to the expected masses for the [M + H]+ of the three major SASP of B. subtilis spores, SASP-β, -α, and -γ, respectively (6,849.6, 6,940.6, and 9,137.5 Da without the N-terminal methionine residue that is removed posttranslationally [3]). The masses of these peaks were further confirmed by nanospray Q-TOF analysis and were found to be very close to the theoretical masses of the major SASP in B. subtilis spores (Table 1).
FIG. 1.
MALDI-TOF spectra of extracts of wild-type B. subtilis (strain PS832) spores prepared with 1 N HCl (a) or acetonitrile-5% TFA (70:30, vol/vol) (b). The peaks at m/z 6,851, 6,942, and 9,137 correspond to protonated SASP-β, SASP-α, and SASP-γ, respectively, and the peaks at m/z 3,426, 3,472, and 4,569 are their doubly charged ions, respectively.
TABLE 1.
SASP extracted from spores of Bacillus species by 1 N HCla
| Name | Sequence | Calculated average mass (Da) | Observed average mass (Da) by:
|
Origin | |
|---|---|---|---|---|---|
| MALDI-TOF | ESI-Q-TOF | ||||
| SASP-α | ANNNSGNSNNLLVPGAAQAIDQMKLEIASEFGVNLGADTTSRANGSVGGEITKRLVSFAQQNMGGGQF | 6,939.6 | 6,941 ± 2 | 6,939 ± 0.3 | B. subtilis |
| SASP-β | ANQNSSNDLLVPGAAQAIDQMKLEIASEFGVNLGADTTSRANGSVGGEITKRLVSFAQQQMGGRVQ | 6,848.6 | 6,850 ± 2 | 6,848 ± 0.1 | B. subtilis |
| SASP-γ | ANSNNFSKTNAQQVRKQNQQSAAGQGQFGTEFASETNAQQVRKQNQQSAGQQGQFGTEFASETDAQQVRQQNQSAE QNKQQNS | 9,136.5 | 9,136 ± 3 | 9,137 ± 0.9 | B. subtilis |
| SASP-2 | SRSTNKLAVPGAESALDQMKYEIAQEFGVQLGADATARANGSVGGEITKRLVSLAEQQLGGYQK | 6,710.5 | 6,709 ± 3 | 6,709 ± 0.5 | B. cereus B. thuringiensis |
| SspD | ASRNKLVVPGVEQALDQFKLEVAQEFGVNLGSDTVARANGSVGGEMTKRLVQQAQSQLNGTTK | 6,572.5 | 6,675 ± 3 | 6,672 ± 0.3 | B. subtilis |
| SspC | AQQSRSRSNNNNDLLIPQAASAIEQMKLEIASEFGVQLGAETTSRANGSVGGEITKRLVRLAQQNMGGQFH | 7,526.5 | 7,626 ± 2 | 7,626 ± 0.7 | B. subtilis |
| SASP-γ | SKKQQGYNKATSGASIQSTNASYGTEFSTETDVQAVKQANAQSEAKKAQASGAQSANASYGTEFATETDVHSVKK QNAKSAAKQSQSSSSNQ | 9,540.1 | 9,540 ± 3 | 9,539 ± 0.8 | B. cereus B. thuringiensis |
| SspCΔ11D13K | AKLLIPQAASAIEQMKLEIASEFGVQLGAETTSRANGSVGGEITKRLVRLAQQNMGGQFH | 6,353.3 | 6,354 ± 2 | 6,353 ± 0.2 | engineered |
| SASPa | ANQNSSNQLVVPGATAAIDQMKYEIAQEFGVQLGADSTARANGSVGGEITKRLVAMAEQSLGGFHK | 6,834.6 | 6,834 ± 2 | 6,834 ± 0.3 | B. anthracis B. cereus B. thuringiensis |
| SASPb | ARSTNKLAVPGAESALDQMKYEIAQEFGVQLGADATARANGSVGGEITKRLVSLAEQQLGGFQK | 6,678.5 | 6,679 ± 2 | 6,678 ± 0.2 | B. anthracis |
| SASPc | ARNRNSNQLASHGAQAALDQMKYEIAQEFGVQLGADTSSRANGSVGGEITKRLVAMAEQQLGGGYTR | 7,080.8 | 7,080 ± 3 | 7,080 ± 0.2 | B. anthracis B. cereus B. thuringiensis |
| SASP-γ | ANSNNKTNAQQVRKQNQQSASGQGQFGTEFASETNVQQVRKQNQQSAAGQGQFGTEFASETDAQQVRQQNQSA EQNKQQNS | 8,889.3 | 8,882 ± 5 | 8,890 ± 0.9 | B. globigii |
| SASP-1 | GKNNSGSRNEVLVRGAEQALDQMKYEIAQEFGVQLGADTTARSNGSVGGEITKRLVAMAEQQLGGRANR | 7,335.1 | 7,334 ± 3 | 7,335 ± 0.6 | B. globigii |
| SASPuk | Unknown | ND | 7,070 ± 3 | 7,067 ± 0.5 | B. globigii |
SASP were identified in HCl extracts by mass spectrometry as described in Materials and Methods. Sequences are shown without the N-terminal methionine, which is removed posttranslationally. The observed masses are reported as an average ± standard deviation of three measurements. SASPa, SASPb, and SASPc sequences were obtained from the The Institute for Genome Research database. All other sequences were obtained from Swiss-Prot and TrEMBL databases. SASPuk, unknown protein; ND, not determined; ESI, electrospray ionization.
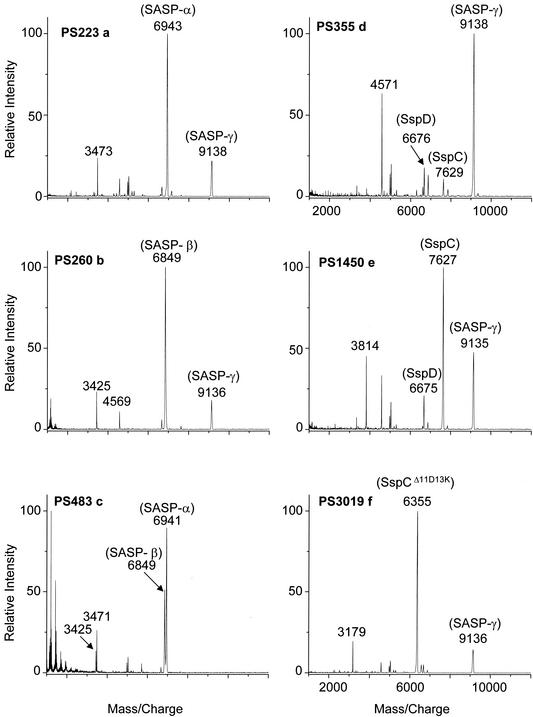
Analysis of HCl extracts of spores engineered to lack SASP-α, -β, or -γ further confirmed the identity of all three peaks, as the appropriate peaks were absent in extracts from spores lacking the genes encoding SASP-α, -β, or -γ (Fig. 2a to c). Analysis of extracts from spores engineered to lack SASP-α and -β was most revealing, since not only did the SASP-α and -β peaks disappear but also the relative intensity of the SASP-γ peak increased, and two other minor peaks that had been dwarfed by those of SASP-α and -β now were easily visible (Fig. 2d). The masses of these two peaks, 6,676 and 7,629 Da, were consistent with these being two minor α/β-type SASP, termed SspD and SspC (6,672.5 Da and 7,626.5 Da, respectively, without the N-terminal methionine that is removed posttranslationally). The assignment of the peak at 7,629 Da as SspC was further confirmed, as spores lacking SASP-α and -β but overexpressing SspC contributed a greatly intensified peak at m/z 7,629, although SspD and SASP-γ were still observed (Fig. 2e). As a final test of the procedure we analyzed an extract from spores lacking SASP-α and -β but overexpressing an engineered version of the α/β-type SASP SspC termed SspCΔ11D13K (10). SspD and SASP-γ were still observed, but the most intense peak (6,354 Da) was close to the mass of SspCΔ11D13K (6,353.3 Da without the N-terminal methionine) (Fig. 2f).
FIG. 2.
MALDI-TOF (MS) analysis of B. subtilis spore extracts. Spores of various B. subtilis strains were extracted with HCl, and aliquots were analyzed by mass spectrometry as described in Materials and Methods. The spores analyzed in the various sections of the figure are PS223, lacking SASP-β (a); PS260, lacking SASP-α (b); PS483, lacking SASP-γ (c); PS355, lacking SASP-α and -β (d); PS1450, lacking SASP-α and -β and overexpressing SspC (e); and PS3019, lacking SASP-α and -β and overexpressing SspCΔ11D13K (f).
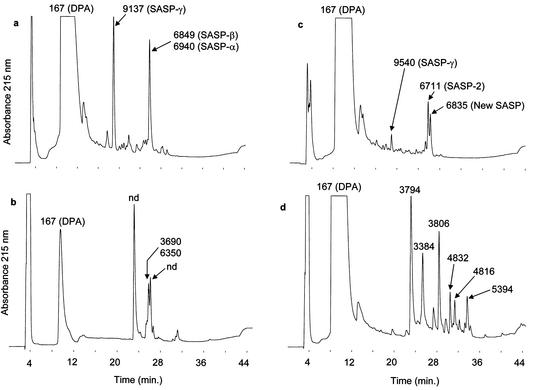
Treatment of B. subtilis spores with 1 N HCl for 1 h resulted in significant killing (>99%) that was accompanied by a large amount (>95%) of DPA release (data not shown), as was expected on the basis of previous results (21). In contrast, a 1-h treatment with acetonitrile-5% TFA (70:30, vol/vol) killed only 5% of these spores and released very little (<15%) DPA (data not shown). Unlike B. subtilis spores, B. cereus spores were killed 99% by a 1-h incubation with acetonitrile-5% TFA (70:30, vol/vol), and this treatment released >98% of the spore's DPA (data not shown). DPA is found in the central region or core, also the site of spore DNA and SASP (3). The fact that there was little DPA extraction and only slow killing upon incubation of B. subtilis spores in acetonitrile-TFA suggested that there would also be little SASP released by this solvent compared to that by 1 N HCl, and HPLC analysis of extracts showed that this was indeed the case (Fig. 3a and b). The HCl extract of B. subtilis spores gave a large peak corresponding to DPA in the chromatogram (Fig. 3a), while this peak was one-tenth as large in the HPLC trace of the acetonitrile-5% TFA extract and SASP were not detected in this extract (Fig. 3b). For B. cereus spores, while the intensity of the peak corresponding to DPA was similar in the two extracts, significantly more SASP was extracted by HCl (Fig. 3c) than by acetonitrile-5% TFA (Fig. 3d), as revealed by mass spectrometry. This latter analysis allowed the identification of two major SASP in spores of B. cereus T as an α/β-type SASP, encoded by a gene originally called SASP-2 (14), as well as SASP-γ (25) (Table 1 and Fig. 4c). The gene for the SASP-2 protein with m/z 6,711 was previously sequenced (14), and the results are in agreement with the sequence reported in Table 1. A third peak with a protonated mass of 6,834 Da was also identified by mass spectrometry in the HCl extract of B. cereus spores (Fig. 3c). This is proposed to be the second major α/β-type SASP in these spores on the basis of preliminary work that used a combination of trypsin digestion, peptide sequencing by mass spectrometry, and a search of the TIGR database (see the sequence in Table 1).
FIG. 3.
HPLC profiles of B. subtilis (strain PS832) and B. cereus T spore extracts. (a) HCl extract of B. subtilis spores; (b) acetonitrile-5% TFA (70:30, vol/vol) extract of B. subtilis spores; (c) HCl extract of B. cereus spores; and (d) acetonitrile-5% TFA (70:30, vol/vol) extract of B. cereus spores. The peaks were collected and their molecular masses were determined by MALDI-TOF (MS) and nanospray-TOF (MS). Average molecular mass values assigned to the SASP proteins are calculated from their primary sequences (see Table 1).
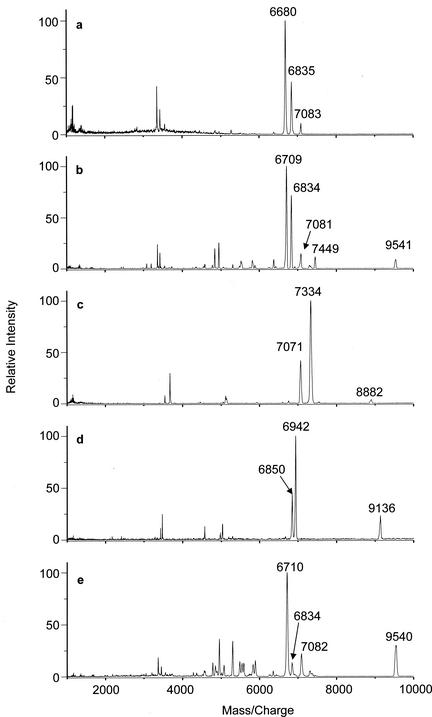
FIG. 4.
MALDI-TOF spectra of HCl extracts of spores of five Bacillus species. (a) B. anthracis Sterne; (b) B. cereus T; (c) B. globigii; (d) B. subtilis (strain PS832); and (e) B. thuringiensis subsp. Kurstaki HD-1.
The fact that DPA is extracted by acetonitrile-5% TFA from B. cereus spores but not from B. subtilis spores is likely due to structural differences between the spores of these two species. The spores of B. cereus and B. subtilis differ significantly in their coat and cortex structures (1, 3), but how these differences contribute to differences in spore killing and extraction by acetonitrile-TFA is not clear. The precise mechanism for extraction of compounds from the spore core by acids is also not clear, although 1 N HCl is known to cause complete spore disruption (21, 27). The specific effects of acetonitrile-TFA on spore structure have not yet been studied, but since this solvent extracted DPA more efficiently than SASP from B. cereus spores, this suggests that acetonitrile:TFA does not disrupt spores in the same way that 1 N HCl does. Previous work has shown that yields of protein biomarkers are low with acetonitrile-TFA extraction of intact spores of several Bacillus species, although yields are increased markedly by treatment of spores in acetonitrile-TFA with corona plasma discharge or sonication (8, 20). Recent studies have also shown that extraction of Bacillus spores with 15% formic acid (C. Afonso and C. Fenselau, Abstr. 50th Am. Soc. Mass Spectrom. Conf., abstr. 313, 2002, and D. N. Dickinson, D. H. Powell, and J. D. Winefordner, Abstr. 50th Am. Soc. Mass Spectrom. Conf., abstr. 314, 2002) or concentrated nitric acid (P. Scholl, personal communication) allows better detection of SASP by MALDI-TOF analysis. However, B. subtilis spores were still resistant to this regimen of 15% formic acid, and only a little SASP was released.
The extraction of SASP from spores by 1 N HCl was tested further in a comparison of spores of five different Bacillus species. MALDI mass spectra (Fig. 4) indicate that distinctive SASP were released from the five different species. For example, B. cereus, B. globigii, and B. subtilis diverge greatly in the molecular masses of their SASP (Fig. 4b, c, and d and Table 1). In particular, the γ-type SASP were found to have large differences in masses between these three species (m/z 9,541, 8,882, and 9,136 for spores of B. cereus, B. globigii, and B. subtilis, respectively). It is worth noting that MALDI spectra of the HCl extracts of spores of B. globigii grown in three different media (NSM, chemically defined sporulation medium, and casein acid digest medium) and of a 34-year-old sample of B. globigii spores contain SASP with the same masses as those shown in Fig. 4c (data not shown).
In contrast to the differences in SASP complement between spores of some species, B. anthracis Sterne, B. cereus T, and B. thuringiensis subsp. Kurstaki, which are closely related genetically (11), gave two SASP with the same masses (6,834 and 7,080 Da) (Fig. 4a, b, and e). These are assigned sequences in Table 1 on the basis of DNA sequences in the TIGR database. In addition, SASP-2 (molecular mass of 6,710 Da) and SASP-γ (9,540 Da) are observed in common in the spectra of B. cereus and B. thuringiensis extracts. Consequently, these two microorganisms have the same SASP profile. However, it has been noticed that the relative abundance of the SASP with a molecular mass of 6,834 was always lower in B. thuringiensis spores than in B. cereus spores; this was true in multiple samples (data not shown). The MALDI spectrum of B. anthracis Sterne contains a peak at m/z 6,680 unique among the set of spores studied here, which is designated SASPb and has a molecular mass of 6,679.5 Da, as deduced from the B. anthracis genome (see Table 1). Surprisingly, the MALDI spectrum of the extract from B. anthracis Sterne spores did not contain an obvious γ-type SASP, although the B. anthracis genome contains a gene encoding a protein that has ∼95% amino acid sequence identity to SASP-γ of B. cereus. The reason for the absence of this protein from the MALDI spectrum of the B. anthracis Sterne spores is unclear.
The results in this communication indicate that extracted SASP can be potential biomarkers for offline identification of spores of Bacillus species by mass spectrometry, with the proviso that spores must be well extracted in order to obtain reliable yields of these proteins. The 1 M HCl extraction procedure used in this study fulfills this criterion. The ensemble of SASP masses revealed in each MALDI spectrum allows genetically distinct species and strains to be differentiated and readily confirms engineering of ssp genes. However, this finding does not rule out the need for systematic identification of Bacillus spores by other means.
Acknowledgments
We are grateful to Federico Tovar for providing the B. cereus spores.
This work was supported by a grant from the NIH (GM19698) to Peter Setlow and by grant GM21248 from the NIH to Catherine Fenselau.
Preliminary sequence data was obtained from The Institute for Genome Research website at http://www.tigr.org.
REFERENCES
- 1.Atrih, A., and S. J. Foster. 2001. Analysis of the role of bacterial endospore cortex structure in resistance properties and demonstration of its conservation amongst species. J. Appl. Microbiol. 91:364-372. [DOI] [PubMed] [Google Scholar]
- 2.Demirev, P. A., J. Ramirez, and C. Fenselau. 2001. Tandem mass spectrometry of intact proteins for characterization of biomarkers from Bacillus cereus T spores. Anal. Chem. 73:5725-5731. [DOI] [PubMed] [Google Scholar]
- 3.Driks, A., and P. Setlow. 1999. Morphogenesis and properties of the bacterial spore, p. 191-218. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, D.C.
- 4.Elhanany, E., R. Barak, M. Fisher, D. Kobiler, and Z. Altboum. 2001. Detection of specific Bacillus anthracis spore biomarkers by matrix-assisted laser desorbtion/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15:2110-2116. [DOI] [PubMed] [Google Scholar]
- 5.Fenselau, C., and P. A. Demirev. 2001. Characterization of intact microorganisms by Maldi mass spectrometry. Mass Spectrom. Rev. 20:157-171. [DOI] [PubMed] [Google Scholar]
- 6.Goldrick, S., and P. Setlow. 1983. Expression of a Bacillus megaterium sporulation specific gene in Bacillus subtilis. J. Bacteriol. 155:1459-1462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hackett, R. H., and P. Setlow. 1988. Properties of spores of Bacillus subtilis strains which lack the major small, acid-soluble protein. J. Bacteriol. 170:1403-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hathout, Y., P. A. Demirev, Y.-P. Ho, J. L. Bundy, V. Ryzhov, L. Sapp, J. Stutler, J. Jackman, and C. Fenselau. 1999. Identification of Bacillus spores by matrix-assisted laser desorption ionization-mass spectrometry. Appl. Environ. Microbiol. 65:4313-4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hathout, Y., Y.-P. Ho, V. Ryzhov, P. Demirev, and C. Fenselau. 2000. Kurstakins: a new class of lipopeptides isolated from Bacillus thuringiensis. J. Nat. Prod. 63:1492-1496. [DOI] [PubMed] [Google Scholar]
- 10.Hayes, C. S., and P. Setlow. 2001. An α/β-type small, acid-soluble spore protein which has a very high affinity for DNA prevents outgrowth of Bacillus subtilis spores. J. Bacteriol. 183:2662-2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helgason, E., O. A. Okstad, D. A. Caugant, H. A. Johansen, A. Fouet, M. Mock, I. Hegna, and A. B. Kolsto. 2000. Bacillus anthracis, Bacillus cereus, and Bacillus thuringiensis-one species on the basis of genetic evidence. Appl. Environ. Microbiol. 66:2627-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson, W. C., and D. J. Tipper. 1981. Acid-soluble spore proteins of Bacillus subtilis. J. Bacteriol. 146:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leenders, F., T. H. Stein, B. Kablitz, P. Franke, and J. Vater. 1999. Rapid typing of Bacillus subtilis strains by their secondary metabolites using matrix-assisted laser desorption/ionization mass spectrometry of intact cells. Rapid Commun. Mass Spectrom. 13:943-949. [Google Scholar]
- 14.Loshon, C. A., E. R. Fliss, B. Setlow, H. F. Foerster, and P. Setlow. 1986. Cloning and nucleotide sequence of genes for small, acid-soluble spore proteins of Bacillus cereus, Bacillus stearothermophilus and “Thermoactinomyces thalpophilus.” J. Baceriol. 167:417-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mason, J. M., and P. Setlow. 1986. Evidence for an essential role for small, acid-soluble, spore proteins in the resistance of Bacillus subtilis spores to ultraviolet light. J. Bacteriol. 167:174-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mason, J. M., and P. Setlow. 1987. Different small, acid-soluble proteins of the α/β-type have interchangeable roles in the heat and ultraviolet irradiation resistance of Bacillus subtilis spores. J. Bacteriol. 169:3633-3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McDonnell, G., and A. D. Russell. 1999. Antiseptics and disinfectants: activity, action and resistance. Clin. Microbiol. Rev. 12:147-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson, W. L., N. Munakata, G. Horneck, H. J. Melosh, and P. Setlow. 2000. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol. Mol. Biol. Rev. 64:548-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley and Sons, Chichester, England.
- 20.Ryzhov, V., Y. Hathout, and C. Fenselau. 2000. Rapid characterization of spores of Bacillus cereus group bacteria by matrix-assisted laser desorbtion-ionization time-of-flight mass spectrometry. Appl. Environ. Microbiol. 66:3824-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Setlow, B., C. A. Loshon, P. C. Genest, A. E. Cowan, C. Setlow, and P. Setlow. 2002. Mechanisms of killing of spores of Bacillus subtilis by acid, alkali and ethanol. J. Appl. Microbiol. 92:362-375. [DOI] [PubMed] [Google Scholar]
- 22.Setlow, P. 1978. Purification and characterization of additional low-molecular-weight basic proteins degraded during germination of Bacillus megaterium spores. J. Bacteriol. 136:331-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Setlow, P. 1995. Mechanisms for the prevention of damage to the DNA in spores of Bacillus species. Annu. Rev. Microbiol. 49:29-54. [DOI] [PubMed] [Google Scholar]
- 24.Setlow, P., and E. A. Johnson. 2001. Spores and their significance, p. 33-70. In L. Beuchat, M. Doyle, and T. Montville (ed.), Food microbiology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 25.Sun, D., and P. Setlow. 1988. Cloning and nucleotide sequencing of genes for the second type of small, acid-soluble spore proteins of Bacillus cereus, Bacillus stearothermophilus, and “Thermoactinomyces thalpophilus.” J. Bacteriol. 169:3088-3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tovar-Rojo, F., and P. Setlow. 1991. Analysis of the effects of mutant small, acid-soluble spore proteins from Bacillus subtilis on DNA in vivo and in vitro. J. Bacteriol. 173:4827-4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Warth, A. 1979. Exploding spores. Spore Newslett. 6:4-6. [Google Scholar]