Abstract
A major obstacle in the implementation of the reductive dechlorination process at chloroethene-contaminated sites is the accumulation of the intermediate vinyl chloride (VC), a proven human carcinogen. To shed light on the microbiology involved in the final critical dechlorination step, a sediment-free, nonmethanogenic, VC-dechlorinating enrichment culture was derived from tetrachloroethene (PCE)-to-ethene-dechlorinating microcosms established with material from the chloroethene-contaminated Bachman Road site aquifer in Oscoda, Mich. After 40 consecutive transfers in defined, reduced mineral salts medium amended with VC, the culture lost the ability to use PCE and trichloroethene (TCE) as metabolic electron acceptors. PCE and TCE dechlorination occurred in the presence of VC, presumably in a cometabolic process. Enrichment cultures supplied with lactate or pyruvate as electron donor dechlorinated VC to ethene at rates up to 54 μmol liter−1day−1, and dichloroethenes (DCEs) were dechlorinated at about 50% of this rate. The half-saturation constant (KS) for VC was 5.8 μM, which was about one-third lower than the concentrations determined for cis-DCE and trans-DCE. Similar VC dechlorination rates were observed at temperatures between 22 and 30°C, and negligible dechlorination occurred at 4 and 35°C. Reductive dechlorination in medium amended with ampicillin was strictly dependent on H2 as electron donor. VC-dechlorinating cultures consumed H2 to threshold concentrations of 0.12 ppm by volume. 16S rRNA gene-based tools identified a Dehalococcoides population, and Dehalococcoides-targeted quantitative real-time PCR confirmed VC-dependent growth of this population. These findings demonstrate that Dehalococcoides populations exist that use DCEs and VC but not PCE or TCE as metabolic electron acceptors.
The chlorinated solvents tetrachloroethene (PCE) and trichloroethene (TCE) are among the most abundant groundwater contaminants at numerous industrial, residential, and military sites. Many contaminated subsurface environments are anaerobic, and often stepwise but incomplete reductive dechlorination leads to the accumulation of cis-1,2-dichloroethene (cis-DCE), trans-1,2-dichloroethene (trans-DCE), and vinyl chloride (VC) (1, 5, 8, 26, 32, 33). Due to their toxicity, chlorinated ethenes pose a threat to human health and are regulated by the U.S. Environmental Protection Agency. Of particular concern is the accumulation of VC because this compound is highly toxic and a proven human carcinogen (14), and its maximum concentration level in drinking water was set at 2 ppb. Although the main contributions of VC contamination in subsurface environments is through the incomplete microbial reductive dechlorination of PCE and TCE, other contamination sources exist, and past incidental releases of VC may have led to local groundwater and soil contamination (14). For instance, VC is a precursor in the manufacturing of polyvinyl chloride, and more than 7 million tons of VC was produced in the United States in 1996. In recent years, the estimated annual VC production in the world was 27 million tons (14). Polyvinyl chloride is also a suspected source of VC in landfills, where VC is frequently detected in drainage water (4).
The remediation of groundwater contaminated with chlorinated ethenes is challenging. Traditional “pump-and-treat” systems have proven to be ineffective, time-consuming, and costly, especially at contaminated sites with complex hydrogeology and large plumes. Bioremediation has become an attractive alternative since the discovery of bacterial populations that use chlorinated ethenes as electron acceptors, thus efficiently reducing and detoxifying these compounds. This process, in which bacteria couple the reductive dechlorination process to growth, is known as (de)chlororespiration or chloridogenesis (16, 18, 24). The physiology and phylogeny of several PCE-to-cis-DCE-dechlorinating bacteria are fairly well understood and have received ample review (9, 13, 15, 16, 27, 31). Although the complete microbial reductive dechlorination of chloroethenes to ethene is well documented in microcosms, laboratory cultures, and bioreactors, the nature of the organisms responsible for the final dechlorination step remained elusive. Both Dehalococcoides ethenogenes strain 195 and Dehalococcoides sp. strain FL2 were shown to reduce VC to ethene, however, the reaction was cometabolic, only occurring when the cultures were grown with a higher chlorinated ethene. Neither population grew with VC alone (16, 19, 23, 24). Flynn et al. (7) demonstrated a community shift in response to enrichment with PCE versus cis-DCE or VC, and circumstantial evidence strongly suggested that populations that use VC as a metabolic electron acceptor exist (20, 29). Hendrickson et al. detected Dehalococcoides 16S rRNA gene sequences at 21 chloroethene-contaminated sites that produced ethene (12). Another study demonstrated that five enrichment cultures that were maintained with VC as electron acceptor dechlorinated VC to ethene in the absence of polychlorinated ethenes. All five cultures contained at least one Dehalococcoides population, as demonstrated with 16S rRNA gene-based approaches (K. M. Ritalahti, R. Krajmalnik-Brown, and F. E. Löffler, Abstr. 6th Int. Symp. In Situ On-Site Bioremediation, 2001, session C1). These findings imply that members of the Dehalococcoides cluster with different properties than the PCE/TCE-dechlorinating isolates D. ethenogenes strain 195 and Dehalococcoides sp. strain FL2 are involved in VC reductive dechlorination.
The aim of the present study was to characterize a VC-to-ethene-dechlorinating enrichment culture obtained from the chloroethene-impacted Bachman Road site aquifer, to identify the population(s) catalyzing the critical dechlorination step, and to demonstrate that VC serves as a growth-supporting electron acceptor for the dechlorinating population(s).
MATERIALS AND METHODS
Chemicals.
Chlorinated ethenes and other chemicals were purchased from Aldrich (Milwaukee, Wis.) and Sigma Chemical Co. (St. Louis, Mo.). H2 was obtained from Air Products (Atlanta, Ga.). VC was obtained from Fluka Chemical Corp. (Ronkonkoma, N.Y.), and ethene was purchased from Scott Specialty Gases (Durham, N.C.).
Source of dechlorinating culture.
A sediment-free culture (referred to as the Bachman culture) was derived from PCE-to-ethene-dechlorinating microcosms established with aquifer material from the chloroethene-contaminated Bachman Road site in Oscoda, Mich. (11; J. M. Lendvay, F. E. Löffler, M. Dollhopf, M. R. Aiello, G. Daniels, B. Z. Fathepure, M. Gebhard, R. Heine, J. Shi, R. Krajmalnik-Brown, C. L. Major, Jr., M. J. Barcelona, E. Petrovskis, R. Hickey, J. M. Tiedje, and P. Adriaens, unpublished data).
Growth medium and culture conditions.
All experiments were carried out in serum bottles (160-ml nominal volume) containing 100 ml (final volume) of growth medium, which were sealed with black butyl rubber septa (Wheaton, Ochelata, Okla.). Anoxic, bicarbonate-buffered mineral salts medium was prepared as previously described (17, 20) and was amended with pyruvate (10 mM), lactate (5 mM or 10 mM), acetate plus formate (5 mM each), or acetate (5 mM) as substrates. Unless indicated otherwise, H2 was only added to acetate-fed cultures at a partial pressure of 9 kPa. Routinely, l-cysteine and Na2S · 9H2O (0.2 mM each) were used to chemically reduce the medium. dl-Dithiothreitol was tested at a 0.5 mM concentration. Titanium(III) (0.5 mM) was added from a filter-sterilized citrate solution after the medium had been autoclaved (18, 34). Gaseous VC was added as a single dose of 1 to 6 ml per serum bottle, resulting in initial aqueous concentrations ranging from 0.27 to 1.6 mM. Other chloroethenes were added by using Hamilton glass syringes (Hamilton, Reno, Nev.) to initial aqueous concentrations ranging from 0.2 to 0.5 mM. Biogenic cis-DCE was produced from PCE (0.5 mM) by using a culture of “Desulfuromonas michiganensis.” After complete dechlorination to cis-DCE (0.5 mM), the cultures were amended with pyruvate and inoculated with the VC-dechlorinating culture. New cultures were routinely seeded with a 2% (vol/vol) inoculum by using plastic syringes. To minimize the contact of the inoculum with air present in the plastic of the syringes during transfers, the syringes were reduced with a sterile 0.5 mM aqueous sulfide solution for 5 to 10 min prior to use. Gaseous substrates were added with plastic syringes that had been previously flushed with H2- and O2-free N2. All experiments were set up in triplicates, and all results were verified by at least one additional independent experiment. Culture bottles were incubated upside down at room temperature (22 to 25°C) without agitation in the dark, unless indicated otherwise. Duplicate controls (no inoculum or autoclaved inoculum) accompanied each experiment. Ethene formation strictly depended on a viable inoculum, and the loss of VC through the septum was negligible.
Analytical methods and data analysis.
Chloroethenes were measured with a Hewlett-Packard model 6890 gas chromatograph equipped with a HP-624 column (60-m length, 0.32-mm diameter, 1.8-μm film thickness) and a flame ionization detector. Headspace samples of 100 μl were withdrawn with gas-tight 250-μl Hamilton glass syringes with Teflon-lined valves (model 1725) and manually injected into a split injector operated at a split ratio of 2:1. All syringes were flushed with H2- and O2-free N2 gas to prevent contamination of the cultures with these gases. A temperature program that allowed the simultaneous analysis of all chloroethenes and ethene was described previously (11). Standards were prepared as described earlier, and seven-point calibration curves were established for all chloroethenes and ethene at room temperature (10). The instrument detection limits for VC and ethene were 7.3 and 8.6 μM, respectively. For quantification of VC and ethene at different temperatures, concentrations of each compound were determined from three-point calibration curves. Organic acids were monitored by high-pressure liquid chromatography by using a Waters Breeze system (Waters, Milford, Mass.) equipped with a Waters 2487 dual-wavelength absorbance detector (set to 210 nm) and a Waters 717 plus autosampler (50-μl injection volume). The eluent was 5 mM aqueous H2SO4, which was pumped at a flow rate of 0.5 ml min−1 through a heated (60°C) Aminex HPX-87H ion exclusion organic acid analysis column (300 by 7.8 mm; Bio-Rad, Hercules, Calif.). Aqueous samples (1 ml) were periodically withdrawn from the cultures by syringe and frozen immediately at −20°C. Before analysis, solids were removed from the samples by centrifugation in a microcentrifuge (14,000 rpm, 10 min). Samples of the supernatant (475 μl) were transferred to autosampler vials, acidified with 25 μl of 1 M H2SO4, and mixed before analysis. Five-point calibration curves were established for each analyte. H2 was quantified with a RGA3 reduction gas analyzer (Trace Analytical, Menlo Park, Calif.) as described previously (20). The standard errors for all analytical measurements were <15% of the averaged values.
The Monod equation, −(dS/dt) = kXS/(S + KS), was used to describe the substrate reaction kinetics, where S is the chloroethene concentration, KS is the half-saturation coefficient, k is the maximum chloroethene dechlorination rate per unit of biomass, and X is the dechlorinating biomass concentration. It was assumed that Xlag was zero during the lag period due to the small inoculum transferred and that X remained constant during the phase of active dechlorination due to the slow growth of the dechlorinating population(s). Electron donors were added in excess and never became limiting during the period of kinetic data collection. Exploratory experiments demonstrated that chloroethenes were added in concentration ranges not causing inhibitory effects (e.g., prolonged lag time or decreased dechlorination rates). The reduced product ethene had no apparent inhibitory effects on VC dechlorination at the concentrations observed in the cultures. After integration of the Monod equation, nonlinear regression analysis was performed to determine the upper limits for kinetic parameters (i.e., KS and kX).
The distributions of chloroethenes in the gas and liquid phases were calculated according to M = CwVw + CgVg = Cw (Vw + HcVg), where M is the total chloroethene mass (μmol), Cw is the concentration of chloroethene in the liquid phase (in micromoles/liter), Cg is the concentration of chloroethene in the gas phase (in micromoles/liter), Vw is the volume of liquid in the system (in liters), Vg is the headspace volume of the system (in liters), and Hc is the dimensionless Henry's constant (10). The dimensionless Henry coefficients for cis-DCE, trans-DCE, 1,1-DCE, and VC at 25°C are 0.167, 0.384, 1.069, and 1.137, respectively (10).
DNA extraction and PCR.
Culture fluid (20 ml) from cultures that had dechlorinated >90% of the initial dose of VC was filtered through 0.2-μm (pore-size) polycarbonate membranes, and after suspension of the biomass in TE buffer (10 mM Tris, 1 mM EDTA, pH 8.0), genomic DNA was extracted by using the Qiagen Mini Kit (Qiagen, Valencia, Calif.). The purification procedure was performed according to the manufacturer's recommendations, except that 45 μl of proteinase K (25 mg/ml), 20 μl of lysozyme (100 mg/ml), and 10 μl of achromopeptidase (25 mg/ml) were used to improve cell lysis. The quality of the extracted genomic community DNA was verified on 1% agarose gels, and DNA was quantified spectrophotometrically. Dehalococcoides 16S rRNA genes were detected in direct PCR by using the Dehalococcoides-targeted primer pair 5′-GCG GTT TTC TAG GTT GTC-3′ (Dhc 730F) and 5′-CAC CTT GCT GAT ATG CGG-3′ (Dhc 1350R), yielding a 620-bp amplicon as described previously (19; M. Bunge, S. Vogler, H. Al-Fahti, and U. Lechner, Abstr. Annu. Meet. German Soc. Gen. Appl. Microbiol. 2001, abstr. PSA17, p. 64, 2001). Clone libraries of 16S rRNA genes were established by using genomic DNA from VC-dechlorinating cultures and the TOPO TA cloning kit (Invitrogen, Carlsbad, Calif.) as described previously (19). E. coli clones with a 16S rRNA gene insert were screened with the Dehalococcoides-targeted primer pair, and plasmid DNA was extracted from positive E. coli clones by using the QIAprep Spin Miniprep kit (Qiagen) according to the manufacturer's recommendations. Negative controls consisted of PCRs with no added template DNA, genomic DNA, or plasmid DNA with a 16S rRNA gene insert from “Desulfuromonas michiganensis” strain BB1 or an Acetobacterium species. Positive control PCRs had genomic or plasmid DNA containing a 16S rRNA gene insert from Dehalococcoides sp. strain FL2 as a template. Double-stranded sequence analysis of nearly complete 16S rRNA genes was performed by using previously published sequencing primers (19) with an ABI 3100 genetic analyzer (Applied Biosystems, Foster City, Calif.). Sequences were assembled and aligned, and base substitutions and percent similarity values were determined by using the Megalign software of the Lasergene package (DNASTAR, Inc., Madison, Wis.). The presence of the structural gene (tceA) encoding the TCE reductive dehalogenase was tested by using the primer pair 797F and 2490R designed to target the tceA gene of D. ethenogenes (21). Genomic DNA from D. ethenogenes strain 195 (kindly supplied by S. Zinder, Cornell University, via F. von Wintzingerode, Humboldt-Universität zu Berlin), and a Dehalococcoides sp.-containing, TCE-dechlorinating enrichment culture derived from river sediment that produced VC as dechlorination end product (supplied by B. Griffin, Michigan State University) were used as positive controls. For 16S rRNA gene analyses, the 50- to 2,000-bp ladder from Bio-Rad was used for size estimation of amplicons, and the Marker 3 from MBI Fermentas GmbH (St. Leon-Rot, Germany) was used for the analysis of tceA.
Real-time PCR.
Oligonucleotides targeting 16S rRNA gene sequences of D. ethenogenes, Dehalococcoides sp. strain FL2, and the VC-dechlorinating Dehalococcoides population identified in the Bachman culture (GenBank accession no. AF357918, AF004928, and AY165308, respectively) were designed by using Primer Express software (Applied Biosystems). Probe and primer specificities were verified by using the Probe Match program of the RDP-II (Ribosomal Database Project) and BLAST analysis (3, 22). The following oligonucleotides were selected: 5′-CTGGAGCTAATCCCCAAAGCT-3′ (forward primer), 5′-TCCTCAGTTCGGATTGCAGGCTGAA-3′ (probe), and 5′-CAACTTCATGCAGGCGGG-3′ (reverse primer). The probe contained 6-carboxy-fluorescein (FAM) as a reporter fluorochrome on the 5′end, and N,N,N′,N′-tetramethyl-6-carboxy-rhodamine (TAMRA) as quencher on the 3′ end. Each MicroAmp optical tube had a 30-μl reaction volume containing 1× TaqMan Universal PCR Master Mix (including DNA polymerase, deoxynucleoside triphosphates, and MgCl2) (Applied Biosystems); forward primer, reverse primer, and TaqMan probe (300 nM each); and DNA template from each 10-fold-diluted sample. The PCR conditions were as follows: 2 min at 50°C and 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. PCR was carried out in a spectrofluorimetric thermal cycler (ABI Prism 7700 Sequence Detection System; Applied Biosystems).
A calibration curve (log DNA concentration versus an arbitrarily set cycle threshold value [CT]) was obtained by using serial dilutions of DNA of known concentration. The CT values obtained for each sample were compared with the standard curve to determine the initial DNA concentration. Experiments were performed in triplicate along with appropriate controls (e.g., template DNA from “Desulfuromonas michiganensis” and no template DNA). Cell number estimates used the assumptions of an average molecular weight of 660 for a base pair in double-stranded DNA, one 16S rRNA gene operon per Dehalococcoides genome, and a genome size of 1.5 Mbp (www.tigr.org). The following equation was used to calculate the number of Dehalococcoides sp.-derived 16S rRNA gene copies that were present in the DNA obtained from 1 ml of the dechlorinating enrichment culture:
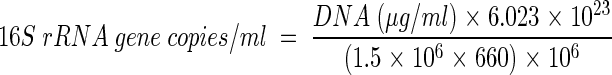 |
(1) |
RESULTS
Sequential transfers from PCE-to-ethene-dechlorinating microcosms to reduced mineral salts medium amended with lactate and VC yielded a sediment-free, ethene-producing enrichment culture. Ethene accumulated as the end product, and no further transformation occurred. Methane formation only occurred at initial VC concentrations of <0.3 mM. After three sequential transfers to medium with aqueous VC concentrations of >0.3 mM, no methane formation from methanogenic substrates occurred, indicating that methanogenic archaea had been diluted out. During the enrichment process, the lag times before dechlorination started became increasingly variable between replicates, ranging from 10 days to several weeks. Transfers were made with plastic syringes, and we occasionally noticed that the inoculum withdrawn from more-enriched cultures turned pink (oxidation of the redox indicator resazurin) inside the syringe during the transfer process. After this observation, all plastic syringes were reduced with a sulfide solution before cell suspensions were transferred. This procedure did not decrease the lag time but significantly reduced the variability between replicate cultures, indicating that the dechlorinating population(s) was very sensitive to O2.
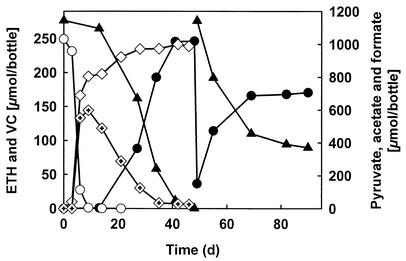
The sediment-free, nonmethanogenic culture was transferred more than 40 times in defined bicarbonate-buffered mineral salts medium with lactate and VC, which was consistently reduced to stoichiometric amounts of ethene within 4 to 8 weeks. Pyruvate or H2 also supported visible growth and dechlorination to ethene. Figure 1 shows the dechlorination of VC to ethene in a culture with pyruvate as the electron donor. Pyruvate was completely consumed before VC reductive dechlorination started, indicating that pyruvate was not the direct electron donor for VC dechlorination. Substantial amounts of H2 (>1 kPa) were formed during the fermentation of pyruvate and the oxidation of formate, which accumulated transiently. After 50 days, the initial amount of 275 μmol of VC was completely reduced to ethene, and the headspaces of triplicate cultures were exchanged with H2-free N2-CO2 (80:20 [vol/vol]). Another 275 μmol of VC, along with 223 μmol of H2, was added, and dechlorination started without an apparent lag time; however, ethene formation slowed down after ca. 20 days. At this time, the H2 concentrations in the cultures had dropped to <0.5 ppm by volume (ppmv), indicating that H2 became limiting. When H2 was added to these cultures, the high rate of dechlorination continued without delay (not shown). Similar observations were made in cultures amended with lactate, which was readily fermented to acetate, propionate, and H2 before VC dechlorination started.
FIG. 1.
Reductive dechlorination of VC (▴) to ethene (•) by the sediment-free, nonmethanogenic enrichment culture in defined mineral salts medium amended with pyruvate. Pyruvate (○) was fermented to acetate (⋄), formate (marked diamond), and H2 before the onset of dechlorination. After the initial dose of VC was consumed, ethene was removed by sparging the headspace with sterile N2-CO2 (80:20 [vol/vol]), and another 275 μmol of VC was added. H2 (223 μmol) was provided as the electron donor. After 65 days, H2 was consumed to 0.5 ppmv, and dechlorination rates decreased. Datum points were averaged from triplicates.
A VC-dechlorinating culture enriched with acetate as the only available electron donor was also derived from the PCE-to-ethene-dechlorinating microcosm. Acetate sustained VC dechlorination for more than 12 consecutive transfers; however, the lag time before dechlorination to ethene occurred was long (3 to 4 weeks), dechlorination to ethene proceeded very slowly, and no visible growth occurred. Both the acetate-enriched and the lactate-enriched cultures were used to explore VC dechlorination in more detail, although the focus was on the high-rate lactate-enriched culture because of its robust VC-dechlorinating activity.
Dechlorination kinetics and lag period.
Table 1 shows the half-velocity coefficients (KS) and maximum aqueous dechlorination rates (kX) for VC and DCEs determined in pyruvate-fed cultures. Similar VC dechlorination rates were measured with lactate as electron donor, and dechlorination at about half these rates were determined with H2 as the electron donor. In acetate-enriched cultures VC dechlorination was sustained at lower rates of 2.4 ± 0.5 μmol liter−1day−1. The experimental data for VC and DCE dechlorination were modeled by using the Monod equation with the parameters KS and kX obtained from nonlinear regression analysis. These models suggested that dechlorination followed zero-order kinetics after the lag period at chloroethene concentrations of >5 μM.
TABLE 1.
Half-velocity coefficients (KS) and maximum dechlorination rates (kX) determined for the VC-enriched culture with pyruvate as the electron donora
| Substrate | Mean ± SD
|
|
|---|---|---|
| KS (μM) | kX (μM day−1) | |
| VC | 5.8 ± 0.4 | 54.4 ± 3.5 |
| cis-DCE | 8.9 ± 0.4 | 23.1 ± 0.8 |
| trans-DCE | 8.5 ± 0.3 | 26.2 ± 0.7 |
Similar values were measured with lactate as a source of reducing equivalents. The data shown represent mean values from three cultures ± one standard deviation.
Similar VC dechlorination rates were observed at temperatures between 22 and 30°C; however, the lag time prior to dechlorination was consistently shorter at 30°C. Dechlorination also occurred at 15°C, but only negligible ethene formation was observed at 4 and 35°C over a 3-month incubation period. The lag time before the onset of VC dechlorination was always at least 10 days, independent of the electron donor added, the initial VC concentration, the reductant used, or the inoculum size (1 to 5% [vol/vol]). No differences in lag periods were observed with commercial cis-DCE or biologically produced cis-DCE. The type and concentrations (0.2 to 0.5 mM) of chemicals used to reduce the medium [i.e., sulfide, DL-dithiothreitol, titanium(III)citrate, l-cysteine] had no effect on the lag periods or the dechlorination rates. The addition of a reductant, however, was essential for dechlorination to occur. To determine whether preconditioned medium would support dechlorination with a shorter lag time, actively dechlorinating cultures amended with pyruvate as the electron donor were autoclaved and then seeded with a second inoculum. Again, a similar lag time of at least 10 days was observed. The addition of filter-sterilized medium from an actively dechlorinating culture had no effect on lag time or culture performance.
Dechlorination of polychlorinated ethenes.
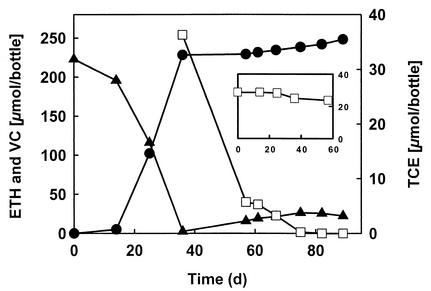
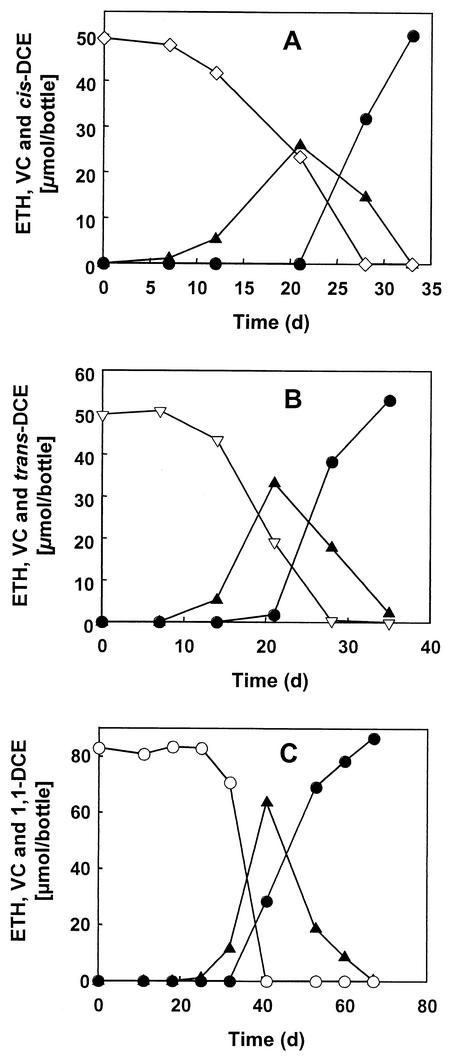
Although the VC-dechlorinating culture was derived from a PCE-to-ethene-dechlorinating microcosm, the culture failed to dechlorinate PCE and TCE after prolonged enrichment with VC. Interestingly, PCE and TCE were dechlorinated to ethene in cultures that simultaneously received VC or when added to VC-grown cultures (Fig. 2). During dechlorination of PCE and TCE, a transient accumulation of VC and small amounts of 1,1-DCE (<9 μmol), cis-DCE (<5 μmol), and trans-DCE (<1 μmol) was observed. A single passage in medium amended with PCE or TCE, but without DCEs or VC, resulted in a complete loss of dechlorinating activity. In contrast, cultures could be repeatedly transferred in medium amended with cis-DCE, trans-DCE, or 1,1-DCE and still maintained the ability to dechlorinate VC with continued production of ethene. As shown in Fig. 3, DCEs were dechlorinated to ethene with the intermediate formation of VC.
FIG. 2.
Reductive dechlorination of TCE (□) in VC-grown cultures with pyruvate as the electron donor (VC, ▴; ethene, •). TCE was added on day 36 immediately after the initial amount of VC was dechlorinated. The inset shows that no TCE dechlorination occurred in cultures grown under the same conditions without VC. All datum points were averaged from triplicate cultures.
FIG. 3.
Reductive dechlorination of DCEs by the VC-enriched culture. (A) Dechlorination of cis-DCE (⋄) to VC (▴) and ethene (•). (B) Dechlorination of trans-DCE (▿) to VC and ethene. (C) Dechlorination of 1,1-DCE (○) to VC and ethene. Pyruvate was provided as electron donor to the cis- and trans-DCE-amended cultures, and acetate plus formate was added to the cultures containing 1,1-DCE. Inocula were derived from cultures grown with the same chlorinated electron acceptor, except for the 1,1-DCE experiment, which was initiated with a VC-grown inoculum. All datum points were averaged from triplicate cultures. d, days.
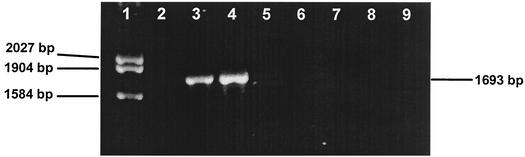
The tceA gene implicated in TCE dechlorination in D. ethenogenes strain 195 was not detected in the VC-dechlorinating Bachman culture, thus supporting the observation that TCE was not used as metabolic electron acceptor (Fig. 4). In contrast, the tceA gene was readily detected in D. ethenogenes and a TCE-to-VC-reducing river sediment enrichment culture containing a Dehalococcoides population.
FIG. 4.
Detection of a structural gene (tceA) implicated in TCE dechlorination in D. ethenogenes. Lane 1, Marker 3; lane 2, no template DNA; lane 3, genomic DNA of D. ethenogenes strain 195; lane 4, genomic DNA of the VC-forming enrichment culture derived from river sediment; lanes 5 to 8, genomic DNA (undiluted and 1:5 diluted) from VC-dechlorinating cultures grown with VC-lactate and VC-H2 (in the presence of ampicillin); lane 9, genomic DNA from “Desulfuromonas michiganensis” strain BB1. The presence of PCR-amplifiable DNA in samples not yielding amplicons (i.e., lanes 5 to 8) with the tceA-targeted primer pair was confirmed in PCR with universal bacterial 16S rRNA gene-targeted primers.
Identification of the dechlorinating population.
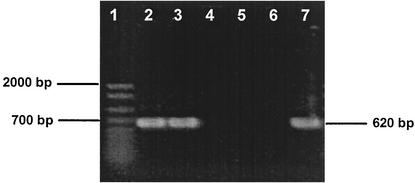
In an effort to identify the VC-dechlorinating population(s), 16S rRNA gene primers targeting known chloroethene dechlorinators were tested on genomic DNA extracted from the enrichment culture. As shown in Fig. 5, amplicons of the expected size (620 bp) were obtained in direct PCR with primers targeting the Dehalococcoides group. The same primer pair identified 5 of 82 clones in 16S rRNA gene clone libraries that also contained 16S rRNA gene inserts most similar to Dehalococcoides 16S rRNA gene sequences. Restriction fragment length polymorphism and partial sequencing (ca. 800 bp analyzed) of clones containing a Dehalococcoides 16S rRNA gene identified two clones with chimeric 16S rRNA gene inserts. The three remaining clones could not be distinguished by restriction fragment length polymorphism and sequencing, suggesting that the 16S rRNA gene inserts were derived from a single Dehalococcoides population. Nearly complete, double-stranded sequence analysis of two cloned 16S rRNA genes yielded identical sequences most similar to Dehalococcoides sequences of the Pinellas subgroup (see Discussion). Table 2 depicts a comparison of the 16S rRNA gene sequence of the VC-dechlorinating Dehalococcoides population present in the Bachman culture with those of known Dehalococcoides isolates and representative environmental clone sequences.
FIG. 5.
Detection of Dehalococcoides populations in the VC-dechlorinating culture with Dehalococcoides 16S rRNA gene-targeted PCR primers. Lane 1, 50- to 2,000-bp ladder (Bio-Rad); lane 2, VC-dechlorinating culture with lactate as electron donor; lane 3, VC-dechlorinating culture with pyruvate as electron donor; lanes 4 to 6, negative controls (Acetobacterium sp., no template DNA, “Desulfuromonas michiganensis” strain BB1); lane 7, Dehalococcoides sp. strain FL2.
TABLE 2.
Comparison of the 16S rRNA gene sequences for selected Dehalococcoides isolates and environmental clone sequencesa
| 16S rRNA gene sequence | Groupb | GenBank accession no. | No. | No. of base differences or % similaritya
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | ||||
| D. ethenogenes strain 195 | C | AF004928 | 1 | 99.2 | 98.7 | 98.9 | 98.2 | 98.2 | 98.1 | |
| DCEH2c | C | AJ249262 | 2 | 10 | 98.8 | 99.0 | 98.2 | 98.2 | 98.1 | |
| DHC-vicc | V | AF388550 | 3 | 16 | 15 | 99.8 | 98.8 | 98.8 | 98.7 | |
| DHC-dllc | V | AF388536 | 4 | 14 | 13 | 2 | 98.9 | 98.9 | 98.8 | |
| Dehalococcoides sp. strain CBDB1d | P | AF230641 | 5 | 23 | 23 | 15 | 14 | 100 | 99.9 | |
| Dehalococcoides sp. strain FL2 | P | AF357918 | 6 | 23 | 23 | 15 | 14 | 0 | 99.9 | |
| Dehalococcoides sp. (Bachman culture) | P | AY165308 | 7 | 24 | 24 | 16 | 15 | 1 | 1 | |
The percent similarity values (upper right portion of matrix) were calculated following the manual alignment of 1,296 positions. The numbers of base differences are shown in the lower left portion of the matrix. Numbers 1 through 7 refer to the sequences identified in col. 4.
Group designations are according to Hendrickson et al. (12). C, Cornell; V, Victoria; P, Pinellas (sequence subgroups).
Environmental clone sequence.
Strain CBDB1 utilizes chlorinated benzenes, but not PCE or TCE, as electron acceptors (2).
Evidence for VC-dependent growth.
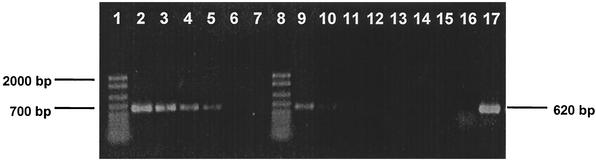
Cultures amended with lactate, pyruvate or H2 showed visible growth before dechlorination started, and no measurable increase in optical density (monitored spectrophotometrically at 600 nm) occurred during the period of VC dechlorination. In order to explore whether the Dehalococcoides population implicated in VC dechlorination grew with VC as a metabolic electron acceptor, 16S rRNA gene-based approaches were used. As shown in Fig. 6, cultures that were fed with pyruvate and VC contained more PCR-amplifiable Dehalococcoides 16S rRNA genes than cultures that were only fed pyruvate. Dehalococcoides 16S rRNA gene targeted real-time PCR confirmed that the increase in Dehalococcoides 16S rRNA gene copies was dependent on the presence of VC. Actively dechlorinating cultures that had dechlorinated 90 μmol of VC contained 51 times ([3.9 ± 0.24] × 105) more Dehalococcoides 16S rRNA gene copies ml−1 than cultures grown under the same conditions without VC ([7.7 ± 0.7] × 103 copies). The cultures received a 2-ml inoculum resulting in 7.8 × 103 Dehalococcoides 16S rRNA gene copies ml of culture fluid−1, thus corroborating that no growth occurred in the absence of VC. Measurements of H2 consumption threshold concentrations provided additional evidence for the metabolic reduction of VC. In the presence of VC, H2 was consumed to concentrations of 0.12 (±0.02) ppmv in lactate- and acetate/H2-fed cultures. In contrast, H2 threshold values in cultures grown under the same conditions but without VC were at least 2 orders of magnitude higher.
FIG. 6.
Increase of Dehalococcoides 16S rRNA genes dependent on the presence of VC as electron acceptor. Lanes 1 and 8, 50- to 2,000-bp ladders (Bio-Rad); lanes 2 to 7, dilution series of template DNA (25 ng μl−1) from pyruvate-VC-grown cultures (1:1, 1:10, 1:50, 1:100, 1:500, and 1:1,000); lanes 9 to 14, dilution series of template DNA (37.5 ng μl−1) from pyruvate-grown cultures; lane 15, H2O; lane 16, “Desulfuromonas michiganensis” strain BB1; lane 17, Dehalococcoides sp. strain FL2.
Dechlorination in the presence of ampicillin.
Reductive dechlorination of DCEs and VC to ethene occurred in the presence of ampicillin, although dechlorination rates were about one-third less than those measured in cultures without the antibiotic. In the presence of ampicillin, reductive dechlorination was strictly dependent on H2 as electron donor, and neither visible growth nor dechlorination occurred with organic electron donors (e.g., lactate, pyruvate, and formate).
DISCUSSION
Sediment-free, nonmethanogenic bacterial enrichment cultures catalyzing the critical final dechlorination step in the reductive dechlorination pathway of chloroethenes leading to complete detoxification (i.e., ethene formation) were obtained from PCE-to-ethene-dechlorinating Bachman microcosms. The 16S rRNA gene-based analysis implicated a Dehalococcoides population in VC reductive dechlorination, a finding that was supported by physiological evidence. First, dechlorination occurred in the presence of ampicillin. Second, dechlorination in ampicillin-amended cultures was strictly dependent on H2, suggesting H2 as the direct electron donor for reductive dechlorination. Both are characteristics of the known Dehalococcoides populations (2, 24). Although H2 was the direct electron donor for reductive dechlorination, higher dechlorination rates were supported in the pyruvate- or lactate-fed enrichment culture, suggesting a supporting role for other populations. This observation may also explain the lower dechlorination rates observed in medium amended with ampicillin, where pyruvate- and lactate-fermenting populations could not grow. The interactions between the dechlorinator and other populations are unclear but might be nutritional in nature. The nutritional requirements of D. ethenogenes are not fully understood, and this organism can only be grown and maintained in medium of undefined composition (24). Hence, further physiological studies leading to an improved understanding of the nutritional requirements of Dehalococcoides species are necessary to facilitate isolation and culturability.
The Dehalococcoides population present in the Bachman culture exhibited relevant physiological differences with regard to electron acceptor utilization patterns to the known chloroethene dechlorinator D. ethenogenes strain 195. D. ethenogenes was reported to utilize PCE, TCE, cis-DCE, and 1,1-DCE as metabolic electron acceptors (zero-order kinetics) but failed to grow with trans-DCE and VC. trans-DCE and VC, however, were cometabolized by strain 195 in the presence of a growth-supporting chloroethene (23, 25). An opposite picture was seen with the Dehalococcoides population present in the Bachman enrichment. All DCEs and VC were readily dechlorinated to ethene but PCE and TCE were only reduced in the presence of lower chlorinated ethenes, presumably in a cometabolic reaction. In support of this observation, tceA, the gene encoding the TCE reductive dechlorinase implicated in dechlorination of TCE and DCEs in D. ethenogenes (21), was not detected in the Bachman culture. These findings demonstrate that Dehalococcoides populations with different substrate specificities toward chlorinated ethenes exist and confirm observations that the complete reductive dechlorination process is most efficiently carried out by more than one population (6, 7, 28).
A few studies already provided circumstantial evidence that VC serves as a metabolic electron acceptor and that Dehalococcoides populations are involved in ethene formation (6, 20, 28, 29; Ritalahti et al., Abstr. 6th Int. Symp. In Situ On-Site Bioremediation). The present study provides conclusive evidence that the Dehalococcoides population identified in the Bachman culture grew with VC as a metabolic electron acceptor. Growth of the Dehalococcoides population with VC was confirmed by (i) the VC-dependent increase in the number of Dehalococcoides 16S rRNA genes, (ii) the loss of dechlorinating activity when transferred in the same medium without VC, (iii) the disappearance of VC following zero-order kinetics, and (iv) H2 consumption threshold measurements. After more than 40 transfers with lactate as the source of reducing equivalents, the culture lost the ability to reduce VC with acetate as the only electron donor, presumably because the population(s) implicated in syntrophic acetate oxidation was lost in the enrichment process (11). Hence, no intrinsic H2 formation from acetate occurred, and the measured values represent true H2 consumption threshold concentrations rather than compensation concentrations (20). The H2 consumption threshold concentration determined for the VC-dechlorinating Bachman culture was similar to H2 threshold concentrations determined for other hydrogenotrophic chloridogenic populations (20, 30).
In a recent study, Hendrickson et al. (12) recovered at least one Dehalococcoides 16S rRNA gene sequence from all chloroethene-contaminated sites where complete reductive dechlorination occurred. Based on signature sequences identified in variable regions II and VI of the 16S rRNA gene, these authors distinguished Dehalococcoides spp. into Cornell, Victoria, and Pinellas sequence subgroups. The 16S rRNA gene sequence of the VC-dechlorinating population in the Bachman enrichment is nearly identical with the Pinellas subgroup sequences, except for a transition (G→A) at E. coli position 148 (Table 2). Since Hendrickson et al. (12) performed no physiological characterizations, their study did not distinguish metabolic VC dechlorinators from those Dehalococcoides populations that cannot grow with VC. Additional Dehalococcoides populations that grow with VC as electron acceptor must be identified to determine whether this transition is characteristic for VC-respiring Dehalococcoides strains and is a useful diagnostic tool. Table 2 demonstrates that Dehalococcoides populations share very similar 16S rRNA genes, implying that focusing exclusively on 16S rRNA gene analysis may be insufficient to distinguish Dehalococcoides populations with different dechlorination activities and to predict the potential for complete microbial detoxification of chloroethenes (i.e., ethene formation) at contaminated sites. For instance, Dehalococcoides sp. strain CBDB1 is a member of the Pinellas group and uses chlorobenzenes as electron acceptors but failed to grow with chloroethenes (2).
The physiological characteristics of D. ethenogenes-type populations are reason for concern because VC is not used as metabolic electron acceptor and cometabolic VC reduction requires the presence of higher chlorinated ethenes. Indeed, the reductive dechlorination of polychlorinated ethenes, along with the accumulation of VC, has been observed at numerous sites (12, 33; Lendvay et al., unpublished). Hence, the type of Dehalococcoides population present in the Bachman enrichment culture seems desirable for bioremediation at many chloroethene-contaminated sites because VC serves as a growth-supporting electron acceptor, and dechlorination is sustained in VC plumes not containing PCE, TCE, or DCEs. To overcome the limitations of the 16S rRNA gene approach, future research must (i) focus on the identification of functional genes that are specific for the process of interest and (ii) distinguish Dehalococcoides populations with different dechlorinating activities. These data are critical for providing information on whether biostimulation or bioaugmentation is the most promising approach at any particular chloroethene-contaminated site amenable to bioremediation technologies.
Acknowledgments
This research was supported by the Strategic Environmental Research and Development Program (contract DACA72-00-C-0023) and by a National Science Foundation CAREER award (award 0090496) to F.E.L.
REFERENCES
- 1.Abelson, P. H. 1990. Inefficient remediation of ground-water pollution. Science 250:73. [DOI] [PubMed]
- 2.Adrian, L., U. Szewzyk, J. Wecke, and H. Görisch. 2000. Bacterial dehalorespiration with chlorinated benzenes. Nature 408:580-583. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Coulston, F., and A. C. Kolbye, Jr. 1994. Vinyl chloride and polyvinyl chloride. Regul. Toxicol. Pharmocol. 19:344-346. [Google Scholar]
- 5.DiStefano, T. D., J. M. Gossett, and S. H. Zinder. 1991. Reductive dechlorination of high concentrations of tetrachloroethene to ethene by an anaerobic enrichment culture in the absence of methanogenesis. Appl. Environ. Microbiol. 57:2287-2292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duhamel, M., S. D. Wehr, L. Wu, H. Rizvi, D. Seepersad, S. Dworatzek, E. A. Cox, and E. A. Edwards. 2002. Comparison of anaerobic dechlorinating enrichment cultures maintained on tetrachloroethene, trichloroethene, cis-dichloroethene and vinyl chloride. Water Res. 36:4193-4202. [DOI] [PubMed] [Google Scholar]
- 7.Flynn, S. J., F. E. Löffler, and J. M. Tiedje. 2000. Microbial community changes associated with a shift from reductive dechlorination of PCE to reductive dechlorination of cis-DCE and VC. Environ. Sci. Technol. 34:1056-1061. [Google Scholar]
- 8.Freedman, D. L., and J. M. Gossett. 1989. Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions. Appl. Environ. Microbiol. 55:2144-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gerritse, J., V. Renard, T. M. Pedro Gomes, P. A. Lawson, M. D. Collins, and J. C. Gottschal. 1996. Desulfitobacterium sp. strain PCE1, an anaerobic bacterium that can grow by reductive dechlorination of tetrachloroethene or ortho-chlorinated phenols. Arch. Microbiol. 165:132-140. [DOI] [PubMed] [Google Scholar]
- 10.Gossett, J. M. 1987. Measurement of Henry's law constants for C1 and C2 chlorinated hydrocarbons. Environ. Sci. Technol. 21:202-208. [Google Scholar]
- 11.He, J., Y. Sung, M. E. Dollhopf, B. Z. Fathepure, J. M. Tiedje, and F. E. Löffler. 2002. Acetate versus hydrogen as direct electron donors to stimulate the microbial reductive dechlorination process at chloroethene-contaminated sites. Environ. Sci. Technol. 36:3945-3952. [DOI] [PubMed] [Google Scholar]
- 12.Hendrickson, E. R., J. A. Payne, R. M. Young, M. G. Starr, M. P. Perry, S. Fahnestock, D. E. Ellis, and R. C. Ebersole. 2002. Molecular analysis of Dehalococcoides16S ribosomal DNA from chloroethene-contaminated sites throughout North America and Europe. Appl. Environ. Microbiol. 68:485-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holliger, C., G. Wohlfarth, and G. Diekert. 1998. Reductive dechlorination in the energy metabolism of anaerobic bacteria. FEMS Microbiol. Rev. 22:383-398. [Google Scholar]
- 14.Kielhorn, J., C. Melber, U. Wahnschaffe, A. Aitio, and I. Mangelsdof. 2000. Vinyl chloride: still a cause for concern. Environ. Health Perspect. 108:579-588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krumholz, L. R. 1997. Desulfuromonas chloroethenica sp. nov. uses tetrachloroethylene and trichloroethylene as electron acceptors. Int. J. Syst. Bacteriol. 47:1262-1263. [Google Scholar]
- 16.Löffler, F. E., J. R. Cole, K. M. Ritalahti, and J. M. Tiedje. Diversity of dechlorinating bacteria. In M. M. Häggblom and I. D. Bossert (ed.), Dehalogenation: microbial processes and environmental applications, in press. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 17.Löffler, F. E., K. M. Ritalahti, and J. M. Tiedje. 1997. Dechlorination of chloroethenes is inhibited by 2-bromoethanesulfonate in the absence of methanogens. Appl. Environ. Microbiol. 63:4982-4985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löffler, F. E., R. A. Sanford, and J. M. Tiedje. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl. Environ. Microbiol. 62:3809-3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Löffler, F. E., Q. Sun, J. Li, and J. M. Tiedje. 2000. 16S rRNA gene-based detection of tetrachloroethene (PCE)-dechlorinating Desulfuromonas and Dehalococcoides species. Appl. Environ. Microbiol. 66:1369-1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Löffler, F. E., J. M. Tiedje, and R. A. Sanford. 1999. The fraction of electrons consumed in electron acceptor reduction (Fe) and hydrogen thresholds as indicators of halorespiratory physiology. Appl. Environ. Microbiol. 65:4049-4056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnuson, J. K., M. F. Romine, D. R. Burris, and M. T. Kingsley. 2000. Trichloroethene reductive dehalogenase from Dehalococcoides ethenogenes: sequence of tceA and substrate range characterization. Appl. Environ. Microbiol. 66:5141-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maidak, B. L., J. R. Cole, T. G. Lilburn, C. T. Parker, Jr., P. R. Saxman, R. J. Farris, G. M. Garrity, G. J. Olsen, T. M. Schmidt, and J. M. Tiedje. 2001. The RDP-II (Ribosomal Database Project). Nucleic Acids Res. 29:173-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maymó-Gatell, X., T. Anguish, and S. H. Zinder. 1999. Reductive dechlorination of chlorinated ethenes and 1,2-dichloroethane by “Dehalococcoides ethenogenes” 195. Appl. Environ. Microbiol. 65:3108-3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maymó-Gatell, X., Y.-T. Chien, J. M. Gossett, and S. H. Zinder. 1997. Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene. Science 276:1568-1571. [DOI] [PubMed] [Google Scholar]
- 25.Maymó-Gatell, X., I. Nijenhuis, and S. H. Zinder. 2001. Reductive dechlorination of cis-dichloroethene and vinyl chloride by “Dehalococcoides ethenogenes.” Environ. Sci. Technol. 35:516-521. [DOI] [PubMed] [Google Scholar]
- 26.Maymó-Gatell, X., V. Tandoi, J. M. Gossett, and S. H. Zinder. 1995. Characterization of an H2-utilizing enrichment culture that reductively dechlorinates tetrachloroethene to vinyl chloride and ethene in the absence of methanogenesis and acetogenesis. Appl. Environ. Microbiol. 61:3928-3933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miller, E., G. Wohlfarth, and G. Diekert. 1997. Comparative studies on tetrachloroethene reductive dechlorination mediated by Desulfitobacterium sp. strain PCE-S. Arch. Microbiol. 168:513-519. [DOI] [PubMed] [Google Scholar]
- 28.Richardson, R. E., V. K. Bhupathiraju, D. L. Song, T. A. Goulet, and L. Alvarez-Cohen. 2002. Phylogenetic characterization of microbial communities that reductively dechlorinate TCE based upon a combination of molecular techniques. Environ. Sci. Technol. 36:2652-2662. [DOI] [PubMed] [Google Scholar]
- 29.Rosner, B. M., P. L. McCarty, and A. M. Spormann. 1997. In vitro studies on reductive vinyl chloride dehalogenation by an anaerobic mixed culture. Appl. Environ. Microbiol. 63:4139-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanford, R. A., J. R. Cole, and J. M. Tiedje. 2002. Characterization and description of Anaeromyxobacter dehalogenans gen. nov., sp. nov., an aryl halorespiring facultative anaerobic Myxobacterium. Appl. Environ. Microbiol. 68:893-900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharma, P. K., and P. L. McCarty. 1996. Isolation and characterization of a facultative bacterium that reductively dehalogenates tetrachloroethene to cis-1,2-dichloroethene. Appl. Environ. Microbiol. 62:761-765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogel, T. M., and P. L. McCarty. 1985. Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions. Appl. Environ. Microbiol. 49:1080-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson, J. T., D. H. Kampbell, J. W. Weaver, B. H. Wilson, T. E. Imbrigiotta, and T. A. Ehlke. 1995. A review of intrinsic bioremediation of trichloroethylene in ground water at Picatinny Arsenal, New Jersey, and St. Joseph, Michigan. U.S. EPA Symposium on Bioremediation of Hazardous Wastes: Research, Development, and Field Evaluations. Document EPA/600/R-95/076. [Online.] http://toxics.usgs.gov/bib/bib-pica-year.html#1995. Office of Research and Development, U.S. Environmental Protection Agency, Washington, D.C.
- 34.Zehnder, A. J. B., and K. Wuhrmann. 1976. Titanium(III) citrate as a nontoxic oxidation-reduction buffering system for the culture of obligate anaerobes. Science 194:1165-1166. [DOI] [PubMed] [Google Scholar]