Abstract
Duplicate anaerobic fermentor systems were used to examine changes in a community of human fecal bacteria supplied with different carbohydrate energy sources. A panel of group-specific fluorescent in situ hybridization probes targeting 16S rRNA sequences revealed that the fermentors supported growth of a greater proportion of Bacteroides and a lower proportion of gram-positive anaerobes related to Faecalibacterium prausnitzii, Ruminococcus flavefaciens-Ruminococcus bromii, Eubacterium rectale-Clostridium coccoides, and Eubacterium cylindroides than the proportions in the starting fecal inoculum. Nevertheless, certain substrates, such as dahlia inulin, caused a pronounced increase in the number of bacteria related to R. flavefaciens-R. bromii and E. cylindroides. The ability of three strictly anaerobic, gram-positive bacteria to compete with the complete human fecal flora was tested in the same experiment by using selective plating to enumerate the introduced strains. The Roseburia-related strain A2-183F was able to grow on all substrates despite the fact that it was unable to utilize complex carbohydrates in pure culture, and it was assumed that this organism survived by cross-feeding. In contrast, Roseburia intestinalis L1-82R and Eubacterium sp. strain A2-194R survived less well despite the fact that they were able to utilize polysaccharides in pure culture, except that A2-194R was stimulated 100-fold by inulin. These results suggest that many low-G+C-content gram-positive obligate anaerobes may be selected against during in vitro incubation, although several groups were stimulated by inulin. Thus, considerable caution is necessary when workers attempt to predict the in vivo effects of probiotics and prebiotics from their effects in vitro.
The human colon is inhabited by a highly complex bacterial community that is dominated by obligately anaerobic bacteria (10, 31). Colonic bacteria obtain energy by fermenting a variety of dietary compounds that are not digested by the host. It is estimated that 15 g or more of nondigestible saccharides per day reaches the colon, and resistant starch, which is not digested by small intestinal amylase, normally provides the largest energy source (20, 34). Elements of the commensal microflora appear to contribute to health through a variety of mechanisms, including the synthesis of fermentation products that provide energy to the colonic epithelium, suppression of pathogenic bacteria, and immune stimulation (5, 7, 41). Manipulation of the human colonic microflora through diet to improve health is therefore a long-term goal. The possible approaches to do this include optimizing the potentially beneficial components of normal diets through the consumption of live beneficial bacteria (probiotics) or of substrates (prebiotics) designed to stimulate desirable groups of bacteria in the intestine. The most widely promoted prebiotics are inulin and fructooligosaccharides (22), neither of which is absorbed in the upper gastrointestinal tract. It has been suggested that these substrates increase the numbers of bifidobacteria detected in feces (14, 19, 21, 24, 34), while the influence of these substrates on other bacterial groups has not been studied.
Recent molecular analyses have emphasized the extreme diversity of the human colonic microflora and have indicated that many major groups are underestimated by cultural approaches. Thus, Suau et al. (39) found that less than 25% of the microbial 16S rRNA sequence diversity present in human feces corresponds to known species. It is important that we consider the heterogeneous microflora both in explaining the effects of diet change on microbial activity and as a source of potential probiotics. The most abundant bacterial groups detected by molecular screening are Bacteroides and the bacteria belonging to the Clostridium coccoides group and the Clostridium leptum group (2, 12, 18, 40, 43). The information available for 16S rRNA sequences has permitted workers to design group-specific oligonucleotide probes that can be used to monitor changes in the gut microbial ecosystem (12, 16).
The first aim of this study was to examine the effects of a range of carbohydrates on the relative levels of different bacterial groups present in the human fecal microflora in an anaerobic continuous fermentor system. To do this, a panel of group-specific oligonucleotides complementary to 16S rRNA sequences (Table 1) was used to enumerate the currently recognized major groups of fecal bacteria (14). The second aim was to examine the use of this system for screening the competitive abilities of potential new probiotic strains. In the same experiment, therefore, we also examined the survival of three marked strains of gram-positive anaerobes following their introduction into the complex fermentor community.
TABLE 1.
Sequences of oligonucleotide probes
| Probe | Sequence (5′-3′) | Targeted bacterial group | Reference |
|---|---|---|---|
| Eub 338 | GCTGCCTCCCGTAGGAGT | Universal eubacterial probe | 2 |
| Bac 303 | CCAATGTGGGGGACCTT | Subgroup of CFB phyluma | 28 |
| Rfla 729 | AAAGCCCAGTAAGCCGCC | R. flavefaciens-like | 16 |
| Rbro 730 | TAAAGCCCAGYAGGCCGC | R. bromii-like | 16 |
| Erec 482 | GCTTCTTAGTCAGGTACCG | Subgroup of cluster XIVa | 12 |
| Fprau 645 | CCTCTGCACTACTCAAGAAAAAC | F. prausnitzii | 39 |
| Bif 164 | CATCCGGCATTACCACCC | Bifidobacterium spp. | 25 |
| Ecyl 387 | CGCGGCATTGCTCGTTCA | E. cylindroides | 16 |
CFB phylum, Cytophaga-Flavobacter-Bacteroides phylum.
MATERIALS AND METHODS
Anaerobic growth of isolated bacteria.
The human fecal butyrate-producing isolates Roseburia sp. strain A2-183, Roseburia intestinalis L1-82, and Roseburia sp. strain A2-194 were isolated from two healthy volunteers (3). R. intestinalis L1-82 was isolated from a fecal sample from an infant (age, 11 months), and this species has also been detected in adult samples (8; unpublished data), while the two Roseburia sp. strains (A2-183 and A2-194) were isolated from a female adult eating a normal diet. Anaerobic rumen fluid-containing medium M2 containing three carbon sources, 0.2% (wt/vol) glucose, 0.2% (wt/vol) soluble starch, and 0.2% (wt/vol) cellobiose (M2GSC medium) (30), was used for routine maintenance of the bacterial isolates. Ten milliliters of YCFA medium containing glucose, soluble starch, and cellulose (8) was used to grow the strains to prepare inocula for the fermentors. YCFA medium contained (per 100 ml) 1 g of Casitone, 0.25 g of yeast extract, 0.4 g of NaHCO3, 0.1 g of cysteine, 0.045 g of K2HPO4, 0.045 g of KH2PO4, 0.09 g of NaCl, 0.009 g of MgSO4 · 7H2O, 0.009 g of CaCl2, 0.1 mg of resazurin, 1 mg of hemin, 1 μg of biotin, 1 μg of cobalamin, 3 μg of p-aminobenzoic acid, 5 μg of folic acid, and 15 μg of pyridoxamine. The final concentrations of the short-chain fatty acids (SCFA) in the medium were 33 mM acetate, 9 mM propionate, 1 mM isobutyrate, 1 mM isovalerate, and 1 mM valerate. Heat-labile vitamins were added after the medium was autoclaved to obtain final concentrations of 0.05 μg of thiamine ml−1 and 0.05 μg of riboflavin ml−1. This medium, which was supplemented with the carbon sources glucose, soluble starch, and cellobiose unless stated otherwise, provided an alternative to rumen fluid medium for cultivation of the strains in this study. The medium was dispensed into Hungate tubes sealed with butyl septum stoppers (Bellco Glass Inc., Vineland, N.J.) by using the anaerobic method, in which the medium is prepared and maintained under O2-free CO2 (4).
Bacterial strains.
Spontaneous antibiotic-resistant mutants of each strain were raised by culturing bacterial suspensions containing approximately 109 CFU in anaerobic M2GSC medium roll tubes (30) containing low concentrations (25 to 50 μg ml−1) of the appropriate antibiotic. Single colonies were isolated and regrown in the presence of antibiotics at final concentrations of 75 to 100 μg ml−1. The specificity of the selection procedure enabled the added strains to be distinguished from each other and from the background microflora and to be enumerated. The mutant Roseburia sp. strain A2-183F was enumerated in M2GSC medium roll tubes containing 75 μg of fusidic acid ml−1; R. intestinalis mutant strain L1-82R was enumerated in M2GSC medium roll tubes containing 100 μg of rifampin ml−1 and 1 μg of tetracycline ml−1; and Roseburia sp. mutant strain A2-194R was enumerated in YCFA medium roll tubes (8) supplemented with 0.5% (wt/vol) inulin and 100 μg of rifampin ml−1.
Collection and preparation of fecal samples.
Fresh fecal samples were provided by a 50-year-old healthy male volunteer. The volunteer did not take any antibiotics or other drugs known to influence the fecal flora for several months before the study commenced. Whole stools were collected, and 5 g was used to inoculate each fermentor. Another portion was used for fluorescent in situ hybridization (FISH), and 0.5 g was suspended in 4.5 ml of filtered (0.2-μm-pore-size membrane) phosphate-buffered saline and vortex mixed for at least 3 min to resuspend the sample. The sample was then centrifuged at 700 × g for 1 min to remove undigested food particles. The supernatant fraction was fixed by mixing it 1:3 with 4% (wt/vol) paraformaldehyde at 4°C for 16 h and was stored at −20°C.
Enumeration of bacteria in the fecal and fermentor samples by FISH analysis.
Single samples from fermentors 1 and 2 were processed individually in the same way as the fecal samples, and FISH analysis was performed as described by Harmsen et al. (16). Depending on the expected number of target cells, samples were diluted 40-to 1,600-fold. The diluted cell suspensions (45-μl aliquots) were added to 5-μl portions of a 50-ng μl−1 solution of the oligonucleotide probes and hybridized overnight at 50°C on glass slides. For total cell counts 4′,6-diamidino-2-phenylindole (DAPI) was added to a final concentration of 100 ng ml−1. To prevent fading of fluorescence 50 μl of Vectashield (Vecto Laboratories, Burlingame, Calif.) was added to each sample. Cells were counted automatically by using image analysis software (21) with a Leica DMRXA epifluorescence microscope, except when the number of cells was less than 10 cells per field of view, in which case the cells were counted visually with an Olympus BH2 epifluorescence microscope. Depending on the number of fluorescent cells, between 10 and 30 microscopic fields were counted. The samples were all assessed with the probes described in Table 1.
Simulated human colonic fermentor studies.
Duplicate single-stage fermentor systems based on the model described by Macfarlane et al. (27) were used. The fermentor medium was described previously (27) and was modified and prepared as described by Hillman et al. (17). The carbon sources present in the mixed substrate medium were potato starch (0.5%, wt/vol), xylan (0.06%, wt/vol), pectin (0.06%, wt/vol), amylopectin (0.06%, wt/vol), and arabinogalactan (0.06%, wt/vol). The feed flasks, which initially contained the mixed substrate (see above), were replaced on a 7-day cycle with feed flasks containing (final concentrations, 0.5% [wt/vol]) one of the following specific compounds: amylopectin, pectin, inulin (dahlia), xylan, and inulin (chicory), in that order. The growth medium was maintained under a stream of CO2. The volume of the medium in the fermentor vessel was kept constant at 250 ml, and the flow rate of fresh medium was equivalent to one turnover per day, which gave a dilution rate of 0.042 h−1. At this dilution rate after 7 days a maximum of 0.001% (wt/vol) of the previous substrate remained (32). Significant bacterial population changes normally occur within 5 days under these conditions (1).
Both the sterile-medium feed flask and the fermentor flasks were mixed by using internal stir bars powered by external stirring units. A pH controller delivered sterile solutions of 0.5 M HCl or 0.5 M NaOH to maintain the pH at 6.5 to 6.8, and the temperature was maintained at 37°C by using a thermal jacket. The fermentor vessel was inoculated through a port in the top with a fecal suspension from the same volunteer. Five grams of freshly voided feces was suspended in 20 ml of 50 mM phosphate buffer (pH 6.8) under O2-free CO2 containing 0.05% cysteine, and this was used as the inoculum (which gave a starting concentration of feces in the fermentor of 2% [wt/vol]). Individual bacterial strains were grown overnight in YCFA medium containing no antibiotic. The cells were pelleted and resuspended in 5 ml of YCFA medium prior to inoculation. Samples were obtained from the fermentor regularly by withdrawing 5-ml portions with a sampling device in order to monitor the bacterial counts and SCFA concentrations throughout the investigation.
The substrates used in the fermentor system were obtained from Sigma and were each added at a final concentration of 0.5% (wt/vol) to the fermentor medium. The soluble sugar content of each substrate was analyzed following autoclaving (26). The proportions of soluble sugars (in the order added to the fermentor) were as follows: amylopectin, 1.2%; pectin, 2.3%; inulin (dahlia), 9.3%; xylan (oat), 0.26%; and inulin (chicory), 28.7%.
Fermentation product analysis.
SCFA production was determined in triplicate by using samples obtained from each fermentor at the end of the feeding period (day 7) for each substrate by capillary gas chromatography following conversion to t-butyldimethylsilyl derivatives (33).
RESULTS
Impact of alternative energy sources on human fecal microflora in continuous culture.
Single-stage anaerobic fermentor systems were set up as described previously (27, 37) and were supplied continuously with an anaerobic medium. Duplicate vessels were inoculated with a human fecal microflora suspension. For the first 7 days the medium contained a mixed polysaccharide substrate, and in each successive week thereafter the feed flask was changed to a flask containing a different single polysaccharide substrate (as described in Materials and Methods).
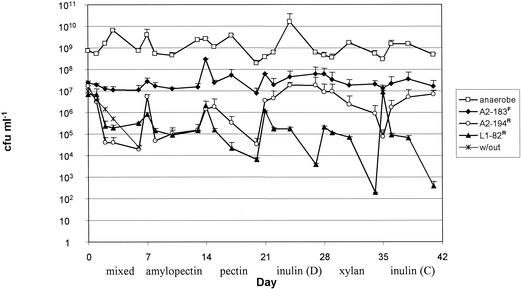
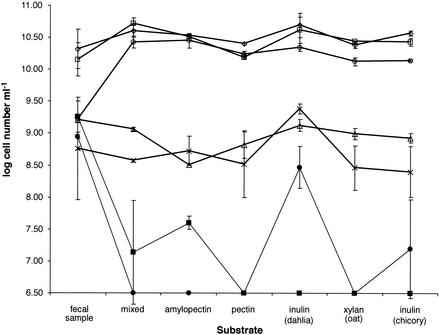
The total numbers of facultative and anaerobic bacteria as determined by cultural methods remained relatively constant during the experiment despite the changes in substrate, ranging from 5 × 107 to 3 × 108 CFU ml−1 for facultative bacteria (data not shown) and from 5 × 108 to 2 × 1010 CFU ml−1 for anaerobes (Fig. 1). The exception was a marked decline in the total anaerobe count to approximately 1 × 108 CFU ml−1 with pectin as the energy source. Changes in the major eubacterial populations were also monitored by FISH for samples taken at the end of each 7-day period by using one universal probe and seven group-specific eubacterial probes (Table 1) that have been developed and extensively validated in previous work (for a summary see reference 14). The counts obtained by DAPI staining and by using the universal eubacterial probe were similar, ranging from 1 × 1010 cells ml−1 with pectin as the substrate to 6 × 1010 cells ml−1 with the initial, mixed growth substrate (Fig. 2). These values are approximately 1 order of magnitude higher than the viable anaerobe counts obtained on M2GSC medium (Fig. 1). This may have been due to bacterial aggregation or to a failure to recover up to 10% of the bacterial diversity by cultivation on a single medium, but it should be noted that both DAPI staining and FISH can detect dead cells prior to lysis.
FIG. 1.
Effects of changes in polysaccharide substrate on culturable anaerobic bacterial populations. Duplicate fermentors were inoculated with a human fecal inoculum and supplied with anaerobic medium. The polysaccharide substrate supplied was changed every 7 days, as indicated at the bottom. Total anaerobes (□) were counted on M2GSC medium. Also shown is the survival of three marked bacterial strains following introduction into the fermentor. The three strains were coinoculated into the fermentor at the time that the substrate was changed on days 0, 7, 14, 21, 28, and 35. Counts for introduced strains A2-183F (♦), A2-194R (○), and L1-82R (▴) were obtained by using selective media (see text). The counts on days 0, 7, 14, 21, 28, and 35 were obtained immediately after inoculation. Single values are the averages of triplicate counts for duplicate fermentors, and the error bars indicate standard errors. The predicted washout rate (w/out) is also indicated.
FIG. 2.
Changes in a human fecal microbial community monitored by FISH. Fecal samples were compared with samples from the two fermentors shown in Fig. 1. Samples taken at the end of each 7-day incubation period were analyzed by DAPI staining (total count) (⋄) and by FISH by using a panel of seven oligonucleotides. The average counts are shown for total eubacteria (probe Eub 338) (□), the Cytophaga-Flavobacter-Bacteroides group (probe Bac 303) (○), R. bromii-R. flavefaciens (probes Rfla 729 and Rbro 730) (×), the E. cylindroides group (probe Ecyl 387) (•), the E. rectale group (probe Erec 482) (▵), and the F. prausnitzii group (probe Fprau 645) (▪).
The Bacteroides group probe detected 50 to 60% of the eubacterial community in the fermentor samples. The numbers of Bacteroides cells were relatively constant throughout the experiment irrespective of the growth substrate, after a marked approximately 20-fold increase following inoculation (Fig. 2). The numbers of bifidobacteria recognized by the probe Bif 164 were below the levels of detection for FISH analysis (<1 ×107 cells ml−1) for all of the samples (data not shown). Only one of six fecal samples from the donor contained detectable numbers of bifidobacteria when the samples were analyzed by FISH. Significant populations were detected with the Ruminococcus flavefaciens-Ruminococcus bromii probes (Rfla 729 and Rbro 730) and the Clostridium coccoides-Eubacterium rectale group probe (Erec 482) (Table 1) throughout the experiment, although these populations were somewhat smaller than those in the fecal inoculum. There were significant populations of Faecalibacterium prausnitzii-related and Eubacterium cylindroides-related bacteria (detected by Fprau 645 and Ecyl 387, respectively) in the fecal inoculum, but the numbers declined rapidly in the first 2 weeks. The F. prausnitzii population never recovered to detectable levels, but the number of E. cylindroides-related bacteria was increased at least 100-fold by dahlia inulin and was increased to a lesser extent by chicory inulin. Dahlia inulin also increased the sizes of the populations of the R. bromii-R. flavefaciens and C. coccoides-E. rectale groups.
The concentrations of SCFA were monitored throughout the experiment and ranged from 40 to 60 mM, depending on the growth substrate (Table 2). The values showed good reproducibility for the two fermentors. The SCFA proportions were strongly affected by the growth substrate, and the highest proportion of butyrate (13.6%) was found when amylopectin was the substrate. The acetate/propionate/butyrate ratios were approximately 3:3:1 for growth on amylopectin, but at the other extreme the ratios were approximately 10:2:1 for growth on pectin. The high proportion of propionate from growth on the mixed and amylopectin substrates may have been due to the high numbers of Bacteroides spp. present in the fermentors.
TABLE 2.
SCFA concentrations in fermentor samples (at the end of each 7-day feeding period) and molar proportions of the three major SCFA formed
| Substrate | Fermentor | Acetate concn (mM) | Propionate concn (mM) | Butyrate concn (mM) | Total SCFA concn (mM) | Avg acetate mol% | Avg propionate mol% | Avg butyrate mol% |
|---|---|---|---|---|---|---|---|---|
| Mixed | 1 | 24.0 ± 2.3 | 21.0 ± 2.4 | 6.6 ± 0.7 | 54.4 ± 5.4 | 46.2 | 41.2 | 12.6 |
| 2 | 26.0 ± 3.4 | 23.7 ± 1.9 | 6.9 ± 0.1 | 59.7 ± 5.2 | ||||
| Amylopectin | 1 | 22.3 ± 0.8 | 18.5 ± 0.4 | 6.2 ± 0.1 | 50.4 ± 1.3 | 44.2 | 42.2 | 13.6 |
| 2 | 20.7 ± 3.1 | 22.4 ± 1.6 | 7.1 ± 0.1 | 54.8 ± 4.9 | ||||
| Pectin | 1 | 31.8 ± 2.2 | 6.8 ± 0.4 | 3.3 ± 0.1 | 43.8 ± 3.8 | 75.4 | 16.2 | 8.4 |
| 2 | 28.9 ± 4.6 | 6.1 ± 0.5 | 3.6 ± 0.1 | 41.1 ± 5.3 | ||||
| Inulin dahlia | 1 | 22.5 ± 3.8 | 15.9 ± 1.2 | 4.6 ± 0.1 | 47.3 ± 5.1 | 51.1 | 40.0 | 8.9 |
| 2 | 21.3 ± 2.9 | 18.2 ± 0.6 | 3.0 ± 0.1 | 44.9 ± 3.7 | ||||
| Xylan | 1 | 20.7 ± 1.7 | 11.6 ± 0.8 | 4.6 ± 0.1 | 40.9 ± 1.2 | 54.9 | 32.4 | 12.7 |
| 2 | 20.7 ± 0.7 | 12.7 ± 0.2 | 5.0 ± 0.1 | 42.4 ± 0.5 | ||||
| Inulin chicory | 1 | 25.9 ± 3.6 | 17.5 ± 1.5 | 4.1 ± 0.2 | 51.8 ± 5.4 | 53.4 | 37.8 | 8.8 |
| 2 | 27.2 ± 0.3 | 20.1 ± 1.7 | 4.6 ± 0.2 | 54.0 ± 5.6 |
Survival of introduced strains of butyrate-producing bacteria.
The in vitro fermentor experiment was also used to test the survival of three introduced Roseburia strains, strains A2-194R, A2-183F, and L1-82R, all of which are butyric acid producers (3).
In order to permit enumeration of the organisms, spontaneous antibiotic-resistant mutations were first selected for each of the three strains. In addition, natural resistance to tetracycline in R. intestinalis L1-82 mediated by the tetracycline resistance determinant Tet O (unpublished data) and the unique ability of Roseburia sp. strain A2-194 to use inulin for growth provided additional selection criteria, which enabled each strain to be specifically enumerated in the presence of the other two strains and in the presence of mixed human fecal bacteria (see above). In preliminary experiments in batch cultures, no major selective disadvantage was detected when the mutant strains were subjected to competition with their respective parent strains in antibiotic-free medium following five successive subcultures (data not shown).
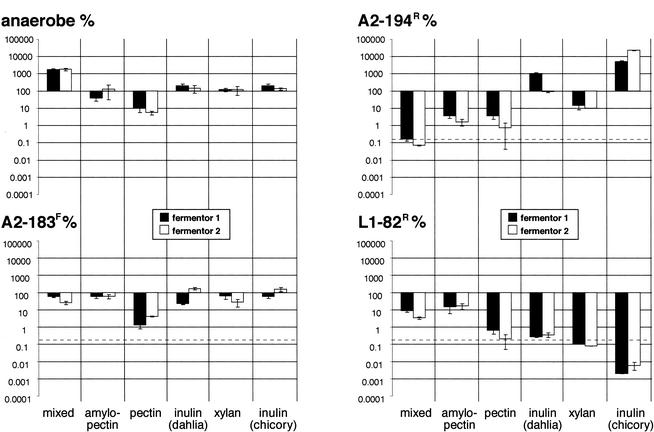
The three bacterial strains were coinoculated into the fermentor at weekly intervals, just before the substrate was changed, as described in Materials and Methods. Counts were obtained for each bacterial species by using selective media at regular intervals (Fig. 1). Figure 3 shows the percentage of the viable count for each strain detectable on day 7 on each substrate relative to the count at the time of inoculation. The results obtained from the duplicate fermentors showed good reproducibility; however, the three introduced strains showed markedly different behaviors. Roseburia sp. strain A2-183F exhibited the best survival, with the number of cells declining to less than 10% of the number in the starting inoculum only on pectin (Fig. 3). As noted above, the total anaerobe and DAPI eubacterial counts also decreased 10-fold after 7 days on pectin (Fig. 1 and 2). Roseburia sp. strain A2-183F was able to thrive on a range of complex substrates in the fermentor but did not grow in pure culture with a single addition of amylopectin, pectin, xylan, or amino acid supplements as the energy source, although slight growth was detected on chicory inulin (Table 3).
FIG. 3.
Data shown in Fig. 1 replotted to show (on the y axis) the percentage of each strain remaining after 7 days of incubation in the fermentor system relative to the starting inoculum with each specific substrate. The results obtained for the two duplicate fermentors are shown separately. The error bars indicate the deviations from the means of triplicate counts in each fermentor system. The proportion of cells projected to remain after washout is indicated by the dotted line.
TABLE 3.
Growth of three Roseburia sp. strains, introduced into the fermentors, on single substrates in batch culture on YCFA medium supplemented with carbon sources and incubated for 24 h at 37°C
| Substratea | Growth ofb:
|
||
|---|---|---|---|
| Roseburia sp. strain A2-194 | Roseburia sp. strain A2-183 | R. intestinalis L1-82 | |
| Amylopectin | +++ | − | +++ |
| Pectin (citrus fruit) | − | − | − |
| Inulin (dahlia) | ++ | − | − |
| Xylan (soluble, oat spelt) | − | − | + |
| Inulin (chicory) | +++ | + | ++ |
Each substrate was added at a concentration of 0.5%.
Growth was recorded as the change in absorbance over 24 h relative to blanks with no carbohydrates, as follows: −, optical density at 650 nm of ≤0.1; +, optical density at 650 nm of >0.1 and ≤0.5; ++, optical density at 650 nm of >0.5 and ≤1.0; +++, optical density at 650 nm of ≥1.0.
In contrast, the viable counts of Roseburia sp. strain A2-194R actually increased relative to the starting inoculum count on both inulin substrates (Fig. 2 and 3). This might have been predicted from the ability of this strain to utilize inulin in pure culture (Table 3). Although able to use amylopectin in pure culture, Roseburia sp. strain A2-194R competed poorly for this substrate in the mixed community.
The number of R. intestinalis L1-82R declined on most of the substrates used, closely following the washout rate (Fig. 2 and 3) (32). The exceptions were amylopectin and the mixed substrate that contained a high proportion of amylopectin starch, on which the numbers seemed to stabilize and to be maintained after an initial decrease. R. intestinalis L1-82 is able to utilize xylan in pure culture (Table 3) (8) but was apparently unable to compete for this substrate in the fermentor systems.
DISCUSSION
In vitro fermentor systems (13, 27, 29) have obvious attractions for modeling the microbial ecosystem of the intestinal lumen, since they are accessible and since controlled manipulation is possible. The simple single-stage system employed here proved to be remarkably reproducible, as shown by the similarity in the results obtained from duplicate vessels over a 42-day period. It is only recently, however, that molecular tools have been available that allow convenient monitoring of the whole microbial community in such systems. The results of our FISH analyses show that the fermentor conditions favored the growth of Bacteroides strains; the proportion of these organisms increased from 10% in the fecal inoculum to 40 to 60% of the total bacterial count, while the proportion of gram-positive anaerobes fell. The concentrations of bacteria related to F. prausnitzii and E. cylindroides in particular decreased to undetectable levels during the first 2 weeks. In the case of F. prausnitzii this may have been due in part to the absolute requirement for acetate for growth (9). Various factors might explain the dominance of Bacteroides spp., including greater oxygen tolerance and a better ability to compete for substrates and other growth requirements (36), and there is now a good opportunity to use this test system to explore these parameters in future studies. The failure to detect bifidobacteria is not surprising since these organisms were detected in only one of six fecal samples from the volunteer. FISH counts for bifidobacteria were previously found to be low (<3% of the total eubacteria) in adult human fecal samples (16), while in other studies workers have obtained substantial bifidobacterial counts.
Despite the dominance of Bacteroides spp., other bacterial populations were strongly stimulated by certain substrates. In particular, dahlia inulin increased the number of E. cylindroides cells at least 100-fold and the number of ruminococci 10-fold. Inulin and fructooligosaccharides derived from inulin are generally regarded primarily as prebiotics that can be used to stimulate the growth of bifidobacteria (15, 35, 42). Our evidence, along with previous work on rats that have a human-associated flora (23), indicates that inulin is also likely to stimulate many other bacterial groups in vivo. This suggests that bifidobacteria may not always be the most abundant group in the adult flora that responds to inulin.
As observed previously in many batch culture studies (for a review see reference 6), the SCFA proportions were found to change with the growth substrate (Table 2). The highest proportions of butyrate (13.6%) and propionate (42.2%) were found with amylopectin, and the lowest proportions of these SCFA were found with pectin, which appears to be consistent with previous observations made with batch cultures. It seems likely that the bacterial population shifts demonstrated here played some part in determining the shifts in the fermentation pattern.
Another potentially important application of fermentor simulations is in tracking the survival and competitive ability of potential probiotic strains (38). Here we explored the survival of three previously uninvestigated strains of gram-positive strict anaerobes known to be producers of butyric acid (3). FISH probes give precise information on changes in the most abundant bacterial groups, but they provide limited sensitivity and cannot be used to track the survival of individual strains whose population sizes may often be below the detection limit. To do this, we used selective plating, which has been used previously to track strain survival in vivo (11). Although mutations can reduce the fitness of strains in vivo, there was no major reduction in the survival of a mutant when it was coincubated with the parent strain for any of the three strains. The fermentor results demonstrate that there were marked differences in the survival of the three strains, which were not entirely predictable from observations of substrate utilization in pure cultures. R. intestinalis L1-82R did not compete effectively with the other components of the fecal microflora for any of the substrates tested. Roseburia sp. strain A2-183F, on the other hand, was a remarkably successful competitor almost regardless of the energy source, despite the fact that it was unable to degrade any of the complex polysaccharide substrates in pure culture. Presumably, the success of Roseburia sp. strain A2-183F depends on an ability to efficiently utilize degradation products released by other species. Meanwhile, Roseburia sp. strain A2-194R showed a clear dependence on the growth substrate and was stimulated 100-fold by chicory inulin. This finding identifies another group of bacterial strains (not related to ruminococci or E. cylindroides) that may be stimulated by inulin in the human large intestine. It is worth noting that all three strains tested are strict anaerobes that survive for less than 2 min when they are exposed to air (unpublished data), so that their survival in the fermentor on some substrates demonstrates that anaerobic conditions were achieved and maintained.
Studies of the practical possibilities of enriching selected bacterial populations in the human large intestine in order to benefit health have so far concentrated almost exclusively on lactic acid bacteria, specifically bifidobacteria and lactobacilli. Anaerobes related to Eubacterium, Roseburia, or Ruminococcus species or other members of the abundant Clostridium clusters XIVa and IV might, however, also include potentially beneficial strains whose populations could be enhanced through pre- or probiotic strategies. The present work shows that a probiotic strategy might be successful for strains like Roseburia sp. strain A2-183, while stimulation of Eubacterium sp. strain A2-194 by inulin illustrates the potential for prebiosis. Clearly, we need to know considerably more about the ecology of these groups and their interactions with the host before deliberately attempting to boost the populations of such species. However, our work indicates that existing prebiotics and natural dietary components already tend to selectively increase the numbers of certain of these species.
In conclusion, this study revealed that significant population shifts affecting major anaerobic bacterial groups within the colonic microbial community can result from changes in the polysaccharide substrate. The results obtained here are results for a single donor, and we did not investigate the effects of fecal inocula from different donors on strain survival and bacterial community responses. The increasing number of molecular tools that are becoming available to study microbial population dynamics means that future investigations into the effects of pre- and probiotics on gut health can include analysis of the complete microbial ecosystem. This should allow more critical evaluation of likely health benefits.
Acknowledgments
We thank Kenneth Young and Gerwin C. Raangs for technical assistance. This study was funded by the Scottish Executive Environment and Rural Affairs Department (SEERAD). A.G.R. received a BBSRC/CASE Ph.D. studentship supported by Nestlé UK Ltd.
REFERENCES
- 1.Allison, C., C. Mcfarlan, and G. T. Macfarlane. 1989. Studies on mixed populations of human intestinal bacteria grown in single-stage and multistage continuous cultures. Appl. Environ. Microbiol. 55:672-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barcenilla, A., S. E. Pryde, J. C. Martin, S. H. Duncan, C. S. Stewart, C. Henderson, and H. J. Flint. 2000. Phylogenetic relationships of butyrate-producing bacteria from the human gut. Appl. Environ. Microbiol. 66:1654-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant, M. P. 1972. Commentary on the Hungate technique for cultivation of anaerobic bacteria. Am. J. Clin. Nutr. 25:1324-1328. [DOI] [PubMed] [Google Scholar]
- 5.Cummings, J. H., and H. N. Englyst. 1987. Fermentation in the large intestine and the available substrates. Am. J. Clin. Nutr. 45:1243-1255. [DOI] [PubMed] [Google Scholar]
- 6.Cummings, J. H., and G. T. Macfarlane. 1991. A review: the control and consequences of bacterial fermentation in the human colon. J. Appl. Bacteriol. 70:443-459. [DOI] [PubMed] [Google Scholar]
- 7.Cummings, J. H., and G. T. Macfarlane. 1997. Role of intestinal bacteria in nutrient metabolism. Clin. Nutr. 16:3-11. [DOI] [PubMed] [Google Scholar]
- 8.Duncan, S. H., G. L. Hold, A. Barcenilla, C. S. Stewart, and H. J. Flint. 2002. Roseburia intestinalis sp. nov., a novel saccharolytic, butyrate-producing bacterium from human faeces. Int. J. Syst. Evol. Microbiol. 52:1615-1620. [DOI] [PubMed] [Google Scholar]
- 9.Duncan, S. H., G. L. Hold, H. J. M. Harmsen, C. S. Stewart, and H. J. Flint. 2002. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52:2141-2146. [DOI] [PubMed] [Google Scholar]
- 10.Finegold, S. M., V. L. Sutter, and G. E. Mathiesen. 1983. Microflora composition and development, p. 3-119. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 11.Flint, H. J., S. H. Duncan, J. Bisset, and C. S. Stewart. 1988. The isolation of tetracycline-resistant strains of strictly anaerobic bacteria from the rumen. Lett. Appl. Microbiol. 6:113-115. [Google Scholar]
- 12.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces quantified by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freter, R., E. Stauffer, D. Cleven, L. V. Holdeman, and W. E. C. Moore. 1983. Continuous flow cultures as in vitro models of the ecology of large intestinal flora. Infect. Immun. 39:666-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of Bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 15.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 16.Harmsen, H. J. M., G. C., Raangs, T. He, J. E. Degener, and G. W. Welling. 2002. Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl. Environ. Microbiol. 68:2982-2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hillman, K., T. A. Murdoch, R. J. Spencer, and C. S. Stewart. 1994. Inhibition of enterotoxigenic Escherichia coli by the microflora of the porcine ileum, in an in vitro semi-continuous culture system. J. Appl. Bacteriol. 76:294-300. [DOI] [PubMed] [Google Scholar]
- 18.Hold, G. L., S. E. Pryde, V. J. Russell, E. Furrie, and H. J. Flint. 2002. Assessment of microbial diversity in human colonic samples by 16S rDNA sequence analysis. FEMS Microbiol. Ecol. 39:33-39. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins, M. J., J. H. Cummings, and G. T. MacFarlane. 1998. Inter-species differences in maximum specific growth rates and cell yields of bifidobacteria cultured on oligosacccharides and other simple carbohydrate sources. J. Appl. Microbiol. 85:381-386. [Google Scholar]
- 20.Jacobasch, G., D. Schmiedl, M. Kruschewski, and K. Schmehl. 1999. Dietary resistant starch and chronic inflammatory bowel diseases. Int. J. Colorect. Dis. 14:201-211. [DOI] [PubMed] [Google Scholar]
- 21.Jansen, G. J., A. C. Wildeboer-Veloo, R. H. Tonk, A. M. Franks, and G. W. Welling. 1999. Development and validation of an automated, microscopy based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215-221. [DOI] [PubMed] [Google Scholar]
- 22.Jenkins, D. J. A., C. W. C. Kendall, and V. Vuksan. 1999. Inulin, oligofructose, and intestinal function. J. Nutr. 129:1431S-1433S. [DOI] [PubMed] [Google Scholar]
- 23.Kleessen, B., L. Hartmann, and M. Blaut. 2001. Oligofructose and long-chain inulin: influence on the gut microbial ecology of rats associated with a human faecal flora. Br. J. Nutr. 86:291-300. [DOI] [PubMed] [Google Scholar]
- 24.Kleessen, B., B. Sykura, H.-J. Zunft, and M. Blaut. 1997. Effects of inulin and lactose on fecal microflora, microbial activity, and bowel habit in elderly constipated persons. Am. J. Clin. Nutr. 65:1397-1402. [DOI] [PubMed] [Google Scholar]
- 25.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lever, M. 1977. Carbohydrate determination with 3-hydroxybenzoic acid hydrazide (PAHBAH): effect of bismuth on the reaction. Anal. Biochem. 81:21-27. [DOI] [PubMed] [Google Scholar]
- 27.Macfarlane, G. T., S. Hay, and G. R. Gibson. 1989. Influence of mucin on glycosidase, protease and arylamidase activities of human gut bacteria grown in a 3-stage continuous culture system. J. Appl. Bacteriol. 66:407-417. [DOI] [PubMed] [Google Scholar]
- 28.Manz, W., R. Amann, W. Ludwig, M. Vancanneyt, and K. H. Schleifer. 1996. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology 142:1097-1106. [DOI] [PubMed] [Google Scholar]
- 29.Miller, T. L., and M. J. Wolin. 1981. Fermentation by the human large intestine microbial community in an in vitro semi-continuous culture system. Appl. Environ. Microbiol. 42:400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazaki, K., J. C. Martin, R. Marinsek-Logar, and H. J. Flint. 1997. Degradation and utilisation of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe 3:373-381. [DOI] [PubMed] [Google Scholar]
- 31.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal microflora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pirt, S. J. 1975. Chemostat culture, p. 41. In Principles of microbe and cell cultivation. Blackwell Science, Oxford, United Kingdom.
- 33.Richardson, A. J., G. C. Calder, C. S. Stewart, and A. Smith. 1989. Simultaneous determination of volatile and nonvolatile fermentation products of anaerobes by capillary gas chromatography. Lett. Appl. Microbiol. 9:5-8. [Google Scholar]
- 34.Roberfroid, M. B. 1993. Dietary fiber, inulin, and oligofructose: a review comparing their physiological effects. Crit. Rev. Food Sci. Nutr. 33:103-148. [DOI] [PubMed] [Google Scholar]
- 35.Rycroft, C. E., M. R. Jones, G. R. Gibson, and R. A. Rastall. 2001. A comparative evaluation of the fermentation properties of prebiotic oligosaccharides. J. Appl. Microbiol. 91:878-887. [DOI] [PubMed] [Google Scholar]
- 36.Salyers, A. A., J. K. Palmer, and T. D. Wilkins. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott, K. P., D. K. Mercer, L. A. Glover, and H. J. Flint. 1998. The green fluorescent protein as a visible marker for lactic acid bacteria in complex ecosystems. FEMS Microbiol. Ecol. 26:219-230. [Google Scholar]
- 38.Scott, K. P., D. K. Mercer, A. J. Richardson, C. M. Melville, L. A. Glover, and H. J. Flint. 2000. Chromosomal integration of the green fluorescent protein gene in lactic acid bacteria and the survival of marked strains in human gut simulations. FEMS Microbiol. Lett. 182:23-27. [DOI] [PubMed] [Google Scholar]
- 39.Suau, A., R. Bonnet, M. Stutren, G. R. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suau, A., V. Rochet, A. Sghir, G. Gramet, S. Brewaeys, M. Sutren, L. Rigottier-Gois, and J. Doré. 2001. Fusobacterium prausnitzii and related species represent a dominant group within the human fecal flora. Syst. Appl. Microbiol. 24:139-145. [DOI] [PubMed] [Google Scholar]
- 41.Topping, D. L., and P. M. Clifton. 2001. Short-chain fatty acids and human colonic function: roles of resistant starch and nonstarch polysaccharides. Physiol. Rev. 81:1031-1064. [DOI] [PubMed] [Google Scholar]
- 42.Tuohy, K. M., R. K. Finlay, A. G. Wynne, and G. R. Gibson. 2001. A human volunteer study on the probiotic effects of HP-inulin—faecal bacteria enumerated using fluorescent in situ hybridisation (FISH). Anaerobe 7:113-118. [Google Scholar]
- 43.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]