Abstract
The endochitinase gene chiA74 from Bacillus thuringiensis serovar kenyae strain LBIT-82 was cloned in Escherichia coli DH5αF′. A sequence of 676 amino acids was deduced when the gene was completely sequenced. A molecular mass of 74 kDa was estimated for the preprotein, which includes a putative 4-kDa signal sequence located at the N terminus. The deduced amino acid sequence showed high degree of identity with other chitinases such as ChiB from Bacillus cereus (98%) and ChiA71 from Bacillus thuringiensis serovar pakistani (70%). Additionally, ChiA74 showed a modular structure comprised of three domains: a catalytic domain, a fibronectin-like domain, and a chitin-binding domain. All three domains showed conserved sequences when compared to other bacterial chitinase sequences. A ca. 70-kDa mature protein expressed by the cloned gene was detected in zymograms, comigrating with a chitinase produced by the LBIT-82 wild-type strain. ChiA74 is active within a wide pH range (4 to 9), although a bimodal activity was shown at pH 4.79 and 6.34. The optimal temperature was estimated at 57.2°C when tested at pH 6. The potential use of ChiA74 as a synergistic agent, along with the B. thuringiensis insecticidal Cry proteins, is discussed.
Bacillus thuringiensis is an insecticidal bacterium whose activity is based on the effect of single or mixed Cry or Cyt proteins, acting additively or synergistically, although antagonism has also been reported (9). Despite reports of more than 3,000 insect species, within 16 orders and susceptible to different B. thuringiensis toxins, important pests are highly susceptible to only a few toxins (14). Additionally, development of resistance to Cry proteins, their slow mode of action, and the requirement of ingestion, along with some other limitations, justify the search for new approaches to improve the conventional use of B. thuringiensis.
The insect midgut is internally coated by the peritrophic membrane, whose structure is basically composed of proteins and reinforced with chitin fibers. This is a physical barrier to bacterial or viral infections but allows the flow of digested nutrients, minerals, and water towards the midgut epithelium. It is known that the approach of δ-endotoxins of B. thuringiensis to the microvillus receptor is highly facilitated when the peritrophic membrane is damaged or degraded, causing an increase in its insecticidal activity (25). Factors that damage the peritrophic membrane, such as the enhancin of granuloviruses, promote the insecticidal activity of commercial formulations of B. thuringiensis, especially on those pests with relatively low natural susceptibility, such as Helicoverpa zea (Boddie) and Spodoptera exigua (Hübner) (16). Likewise, the same effect is observed when Cry proteins are tested along with wild-type (31) or recombinant (11) chitinolytic bacteria, as well as with unpurified chitinase (Chi) (25, 40).
On the other hand, chitinases from B. thuringiensis are poorly studied (2, 7, 15), although interest has slowly increased due to their potential role as control agents for insects and plant-pathogenic fungi, as well as a means to exploit shrimp wastes (2, 26). The role of endogenous chitinases in B. thuringiensis pathogenicity has recently been demonstrated, tested both indirectly (29) and with Chi− mutants (32). Furthermore, chitinases from B. thuringiensis serovar kurstaki HD-1(G) culture supernatants increase the insecticidal activity against Plutella xylostella when mixed with the spore-crystal complex (40). Until now, the synergistic effect of purified B. thuringiensis chitinases and the Cry proteins has not been quantitatively demonstrated, mostly due to the low levels of expression of these enzymes. This is an important reason to clone and overexpress the endogenous genes so that their possible synergistic effect can be tested. So far, several bacterial chi genes have been cloned and sequenced (6, 12, 21, 33, 36); however, only one chitinase gene from B. thuringiensis has been cloned and this was from B. thuringiensis serovar pakistani (32).
As part of a long-term study aimed at improving the insecticidal activity of B. thuringiensis through its interaction with chitinases, this report deals with the cloning, sequencing, and partial characterization of a chitinase gene from a Mexican B. thuringiensis serovar kenyae strain.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
B. thuringiensis serovar kenyae strain LBIT-82 is part of a native strain collection held at CINVESTAV, Irapuato, México. This strain was cultured at 28°C in Castañeda medium supplemented with 1% (wt/vol) colloidal chitin (2). Strain DH5αF′ of Escherichia coli was transformed with recombinant plasmids, and was cultured in Luria-Bertani (LB) broth (Difco) to select transformants. Plasmid pBluescript II KS(+) (Stratagene) was used as a cloning vector.
Cloning of the chiA74 gene from B. thuringiensis LBIT-82.
Total genomic DNA from strain LBIT-82 was extracted and purified by a technique described elsewhere (18) in order to prepare a genomic library. DNA was partially digested with HindIII. Fragments were ligated in pBluescript II KS(+) that was previously dephosphorylated, and the ligation product was electroporated in E. coli DH5αF′. Transformants were selected in LB agar supplemented with ampicillin (100 μg/ml), 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) (64 μg/ml) and isopropyl-β-d-thiogalactopyranoside (IPTG) (0.2 mM). White colonies were individually cultured in LB broth, and their chitinase activity was resolved with chitin fluorogenic derivatives as described below, except that the reaction volumes were 480 μl of buffer, 100 μl of supernatant, and 20 μl of substrate (0.1 μg/μl). Transformants showing chitinase activity were selected, and one particular strain (E. coli DH5αF′/pCHIBT) was subjected to further analysis. A restriction map was obtained from plasmid pCHIBT, which was used to develop a series of deletions. Each subclone was subjected to chitinase activity analysis with a tetrameric fluorogenic chitin derivative (2).
Location of chiA74 on the genome of B. thuringiensis LBIT-82.
Chromosomal and plasmid DNAs were purified from strain LBIT-82 by procedures described elsewhere (8, 18). Samples were separated in agarose gels and transferred to nylon membranes. An internal BamHI-HindIII fragment of the chiA74 gene was used as a probe to perform Southern analysis. Nonradioactive labeling and detection procedures were as recommended by the manufacturer of the digoxigenin DNA labeling and detection kit (Boehringer Mannheim).
Chitinase activity analysis.
Recombinant strain E. coli DH5αF′/pCHIBT was inoculated in 5 ml of LB broth supplemented with antibiotic and with and without IPTG. This culture was inoculated into 45 ml of fresh medium, which was incubated to an optical density at 600 nm of 0.9. The culture was centrifuged, and the pellet was crushed with glass beads. Intracellular proteins were dissolved in McIlvaine's buffer (0.1 M citric acid, 0.2 M NaH2PO4 [pH 6]). Both intracellular and secreted (supernatant) proteins were concentrated 50 times in (NH4)2SO4 to 80% saturation and dissolved in 0.02 M sodium phosphate buffer (pH 6), followed by dialysis overnight in the same buffer. Phenylmethylsulfonyl fluoride (0.2 mM) was added to samples, and the protein concentration was estimated with a Bio-Rad protein microassay kit.
Chitinase activity was evaluated by using the following fluorogenic chitin derivatives: 4-methylumbelliferyl-β-d-N,N′,N′′-triacetylchitotriose [4-MU-(GlcNAc)3], 4-methylumbelliferyl-β-d-N,N′-diacetylchitobioside [4-MU-(GlcNAc)2], and 4-methylumbelliferyl-N-acetyl-β-d-glucosaminide (4-MU-GlcNAc) (Sigma). Reaction mixtures were prepared in a total volume of 600 μl composed of 580 μl of buffer, 10 μl of sample, and 10 μl of substrate (0.1 μg/μl), which was incubated at 37°C for at least 25 min; reactions were terminated by adding 1 volume of 0.2 M Na2CO3. Chitinase activity was measured by the mean fluorescence estimated in a Turner fluorometer (model 450; 340-nm interference filter and 415-nm cut filter). Chitinase activity under different pH conditions was measured at 37°C in the following buffers: McIlvaine's buffer adjusted with NaOH for pH 3 to 7.5 and 0.1 M glycine-0.2 M NaH2PO4 adjusted with NaOH for pH 7.5 to 10. One unit of chitinase activity was defined as the amount of enzyme required to release 1 μmol of 4-methylumbelliferone in 1 h (2, 22). Additionally, chitinase activity under different temperature conditions was measured at pH 6 in McIlvaine's buffer at temperatures ranging from 10 to 76°C. In both cases (pH and temperature tests), data were adjusted to multiple Gaussian models with Origin 4.10 software (Micocal).
Zymogram analysis of ChiA74.
The previously concentrated proteins were treated with Laemmli's disruption buffer for 5 min under the following conditions: with and without β-mercaptoethanol and at room temperature, at 55°C, and in boiling water. Proteins were separated by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis, and chitinases were in situ reactivated by removing SDS with casein-EDTA wash buffer (1% casein, 2 mM EDTA, 40 mM Tris-HCl [pH 9]). The gel was then covered with 1% low-melting-point agarose supplemented with 0.2 mM 4-MU-(GlcNAc)3 in 100 mM sodium acetate buffer (pH 6). Activity was detected with a UV lamp after a 20-min incubation period at 37°C (2, 5). The molecular weights of the renatured chitinases were estimated by comparison with the Benchmark protein ladder (Gibco BRL).
Nucleotide sequencing.
Plasmid pCHIBT and subclones were prepared as double-stranded DNA by purifying the corresponding plasmid from the transformant strain with a plasmid purification kit (Qiagen). The sequence of the chiA74 gene was resolved by using the standard T3 and T7 primers as well as two internal primers specifically designed to obtain overlapping sequences. Sequencing was performed by the dideoxy chain termination method (30) with Big Dye terminator (Applied Biosystems) in a ABI PRISM 377 DNA sequencer. DNA and protein sequences were compared with those of other chitinase genes obtained from GenBank, using both the Blast and DNAStar packages.
Nucleotide sequence accession number.
The nucleotide sequence of the chiA74 gene, including the structural gene along with its flanking regions, is registered under accession number AF424979 at the GenBank nucleotide sequence database.
RESULTS
Cloning of the chiA74 gene from B. thuringiensis LBIT-82.
From ca. 3,000 transformants of the B. thuringiensis gene library that were screened for the production of chitinase, one recombinant strain (E. coli DH5αF′/pCHIBT) was selected. The recombinant plasmid (pCHIBT) contained a HindIII insert of ca. 8 kb in the pBluscript II KS(+) vector. When the recombinant strain was grown in both the presence and absence of IPTG, no significant difference in the chitinase activity was observed, indicating that the transcription of chiA74 was controlled by its own promoter. Once the chiA74 gene was located in the HindIII insert of pCHIBT, a BamHI/HindIII internal fragment was used as a probe in a blot containing separately the plasmid and the chromosomal fragments of strain LBIT-82. Figure 1 shows hybridization to the pCHIBT plasmid (positive control) and the chromosomal fraction, but not the plasmid fraction, in contrast to the usual location of the cry genes of B. thuringiensis.
FIG. 1.
Location of the chiA74 gene in the chromosome of B. thuringiensis LBIT-82. (A) Agarose gel. (B) Southern blot. Lanes: 1, pCHIBT; 2, LBIT-82 plasmids; 3, LBIT-82 chromosome. The arrow indicates hybridization with the chromosomal fraction.
Nucleotide sequence of chiA74.
The ORF of chiA74 consists of 2,031 nucleotides encoding a protein of 676 amino acids with a predicted molecular mass of 74,468 Da and an isoelectric point of 5.886. The putative initiation codon was preceded 5 bp upstream by a potential ribosome-binding (Shine-Dalgarno) sequence (5′-GAAAGG-3′). Putative promoter consensus sequences (−35 TTGAGA and −10 TTAATA) which are identical to those reported for the chiA71 gene of B. thuringiensis serovar pakistani (32) and show high homology with those of E. coli (−35 TTGACA and −10 TATAAT) (27) were found upstream the coding region. These putative promoters also show high homology to those recognized by the primary sigma factor (σA) of vegetative cells of Bacillus subtilis (TTGACA and TATAAT) and the cry3A gene of B. thuringiensis (TTGCAA and TAAGCT) but share no homology with the sporulation-dependent promoters BtI and BtII, which are typical of most of the cry genes of B. thuringiensis (1). Also, a possible transcription terminator was found downstream of the TAG termination codon; this is composed of a 16-bp palindrome corresponding to an mRNA hairpin loop with a ΔG value of −15 kcal/mol (DNAStar, Inc.).
Additionally, the 34 amino acids located at the N terminus of the deduced amino acid sequence show the typical attributes of a signal peptide: a positively charged region, a hydrophobic region, and a signal sequence cleavage site. According to the SignalP site (http://www.cbs.dtu.dk/services/SignalP/), the signal peptide of ChiA74 may be recognized by gram-negative and gram-positive bacteria, as well as eukaryotic organisms. It is presumable that the most likely cleavage sites are between Leu-33 and Ala-34 for gram-negative bacteria, and between Ala-34 and Asp-35 for gram-positive bacteria and eukaryotes, which is compatible with the −3, −1 rule (24).
Comparison of ChiA74 with other chitinases.
Analysis of the deduced amino acid sequence of ChiA74 revealed that the mature protein was a modular enzyme composed of three domains in the following order: a family 18 chitinase domain, two fibronectin-like domains (FLDs), and a chitin-binding domain (CBD). When the deduced amino acid sequence of ChiA74 was compared with entries in the GenBank database, the family 18 catalytic domain (Gly-147 through Ser-222) was found to be identical to that of Bacillus cereus ChiB and highly homologous (98.7%) to that of B. thuringiensis serovar pakistani ChiA71. Further analysis showed some degree of homology with the catalytic domains of other bacterial chitinases, such as Bacillus circulans Chi41 (68.4%), B. circulans ChiA (65.8%), B. subtilis Chi (67.1%), Clostridium paraputrificum ChiA (43.4%), Serratia marcescens ChiA (39.7%), and Enterobacter agglomerans ChiA (39.7%) (Fig. 2). Moreover, amino acid residues Asp-207, Asp-209, and Glu-211 of ChiA74 may correspond to Asp-200, Asp-202, and Glu-204 of B. circulans ChiA, whose role in the catalytic process was previously demonstrated (38, 39).
FIG. 2.
Alignment of the ChiA74 catalytic domain with those of other chitinases. Sequences are from S. marcescens ChiA (Sm), B. circulans Chi41 (Bci), B. circulans ChiA, B. subtilis Chi (Bsub), B. thuringiensis serovar kenyae ChiA74 (Btk) (this report), B. thuringiensis serovar pakistani ChiA71 (Btp), C. paraputrificum ChiA (Cp), E. agglomerans ChiA (Ea), and B. cereus ChiB (Bce). Amino acids conserved in at least three sequences are shaded. Putative proton donor amino acids are indicated by arrows.
Following the catalytic domain of ChA74, two FLDs were found: ChiA74-FLD1 (Lys-350 through Tyr-435) and ChiA74-FLD2 (Ile-479 through Thr-574) (Fig. 3A). Regardless of the low homology (ca. 10%) found in ChiA74 FLD1 compared with other FLDs, aromatic residues typical of fibronectin are conserved: Trp-373, Tyr-384, and Tyr-411. ChiA74 FLD2 also showed the conserved aromatic amino acids (Trp-507, Tyr-519, and Tyr-546), but in contrast to the first FLD region, this second region showed higher homology with other FLDs, such as R-1 and R-2 of B. circulans (41 and 40%, respectively) (36) and the ChiA71 FLD of B. thuringiensis serovar pakistani (86%) (32).
FIG. 3.
(A) Alignment of the FLDs. Sequences are from human fibronectin (III-2 and III-3), B. circulans ChiA (R-1 and R-2), B. thuringiensis serovar kenyae ChiA74 (Btk) (this report), B. thuringiensis serovar pakistani ChiA71 (Btp), B. cereus ChiB (Bce), and B. subtilis Chi (Bsub). Aromatic amino acids distinctive for FLD regions are indicated by arrows. (B) Alignment of the CBDs. Sequences are from B. circulans ChiA (Bci), B. circulans ChiD (Bci), Alteromonas sp. strain Chi85 (Al), Bacillus sp. strain CelA (cellulase A) (Bsp), Bacillus sp. strain CelB, Bacillus sp. strain N-186 Cel (Bsp-N186), Bacillus sp. strain N-252 Cel (Bsp-N252), and S. marcescens ChiB (Sm). Diamonds indicate the aromatic amino acids possibly involved in the chitin binding.
Finally, at the C terminus of ChiA74, the CBD (Val-587 through Lys-629) shows homology to several bacterial chitinases (32, 33, 36, 37), as shown in Fig. 3B. Interestingly, the CBD of ChiA74 also shows homology with C termini of bacterial cellulases, such as CelA and CelB of Bacillus sp. strain N-4 (13) and cellulases of Bacillus sp. strain N186-1 and Bacillus sp. strain KSM-N252. Residues Trp-591, Tyr-595, and Trp-626 of ChiA74 are highly conserved.
Chitinase activity of chiA74 expressed in E. coli.
The chitinase secreted by E. coli DH5αF′/pCHIBT showed hydrolytic activity against the chitin tetrameric derivative (4-MU-(GlcNAc)3], just a slight activity against the trimeric derivative [(4-MU-(GlcNAc)2], and practically no activity against the dimeric derivative (4-MU-GlcNAc) (Table 1) when tested at pH 6. These data indicate that ChiA74 is an endochitinase (5, 22). With the purpose of verifying the functionality of the ChiA74 signal peptide in E. coli, chitinase activity in both secreted and intracellular proteins was measured. Although both extracts showed activity, the extracellular proteins were more active (5.98 U/mg) than the intracellular proteins (3.2 U/mg), suggesting the functionality of the signal peptide in E. coli. On the other hand, secreted proteins of B. thuringiensis LBIT-82 showed about twice the chitinase activity of E. coli DH5αF′/pCHIBT secreted proteins when tested on the tetrameric derivative of chitin and about 20 times more activity when tested against the trimeric derivative. No activity was detected with the proteins secreted by the nontransformed strain of E. coli DH5αF′ (Table 1).
TABLE 1.
Analysis of chitinolytic activity
| Strain | Protein | U/mg of protein (mean ± SDa)
|
||
|---|---|---|---|---|
| MU-(GlcNAc)3 | MU-(GlcNAc)2 | MU-GlcNAc | ||
| E. coli DH5αF′ | Secreted | 0.00 ± 0.00d | 0.01 ± 0.01c | 0.01 ± 0.01b |
| E. coli DH5αF′/pCHIBT | Secreted | 5.98 ± 0.16b | 0.29 ± 0.02b | 0.01 ± 0.01b |
| E. coli DH5αF′/pCHIBT | Intracellular | 3.20 ± 0.40c | 0.17 ± 0.01b | 0.02 ± 0.01b |
| B. thuringiensis LBIT-82 | Secreted | 11.02 ± 0.94a | 5.82 ± 0.69a | 0.52 ± 0.03a |
Values with the same letter are not significantly different as determined by Tukey's multiple range test (P < 0.05).
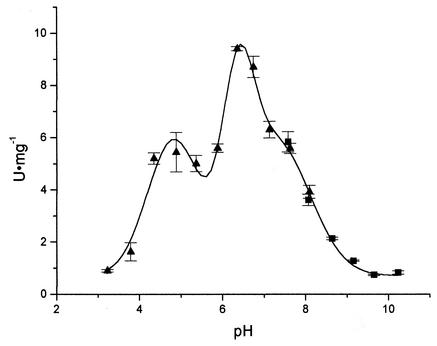
Activity of the recombinant ChiA74 was also measured on the tetrameric derivative at different pH conditions. Activity was observed between pH 4 and 9, but, interestingly, it showed two peaks and one shoulder at estimated pH values of 4.79, 6.34, and 7.45, respectively, after the adjustment of data to a tripartite Gaussian model (Fig. 4). It is important to notice that, from the highest activity detected, more than 40% of the activity is retained at pH 8. Although this pH is still lower than the common pH found in lepidopteran midguts, it is still higher than that for the optimum performance of chitinases previously validated for their synergistic effect with δ-endotoxins (25, 32). Also, it is worth noticing that lepidopteran midguts normally show pH gradients from end to end and from lumen to epithelial microvilli (10, 17), which may enhance the activity of these chitinases at specific midgut areas. This is important if synergism between ChiA74 and Cry proteins is to be measured in future studies.
FIG. 4.
Chitinase activity of ChiA74 at different pHs. Error bars indicate standard deviations. Data were adjusted to a tripartite Gaussian model (P < 0.05). ▴, citrate-phosphate buffer; ▪, glycine-phosphate buffer.
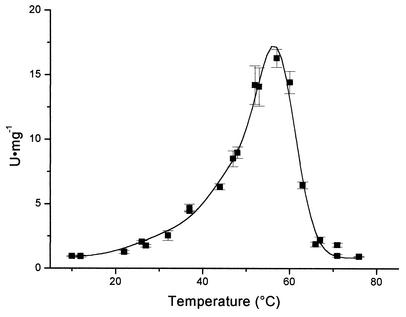
Activity was also measured on the tetrameric derivative at different temperatures. Activity was observed between 21 and 68°C, with an estimated optimum value at 57.2°C, after the adjustment of data to a tripartite Gaussian model (Fig. 5). A slight shoulder was detected at 49.2°C, especially after fitting the secondary Gaussian curve. The third Gaussian curve was used only to obtain the best fit at the left-hand end of the curve.
FIG. 5.
Chitinase activity of ChiA74 at different temperatures. Error bars indicate standard deviations. Data were adjusted to a tripartite Gaussian model (P < 0.05).
Zymogram analysis of ChiA74.
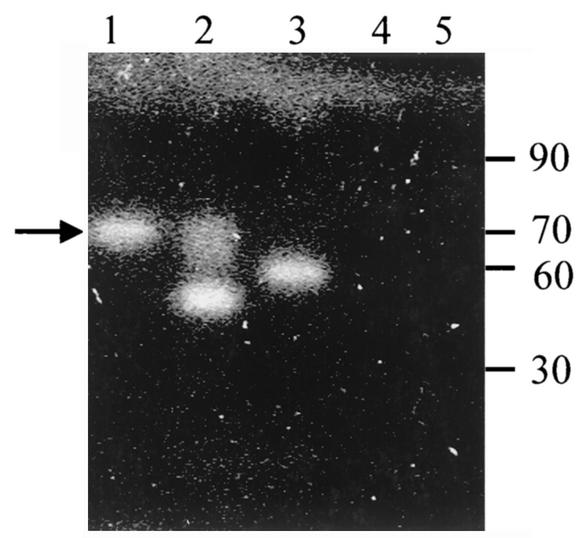
The direct effect of ChiA74, as expressed in E. coli DH5αF′/pCHIBT, was measured in zymograms after extracellular extracts were subjected to heat treatment in β-mercaptoethanol, analyzed by SDS-polyacrylamide gel electrophoresis, renatured, and incubated with [(4-MU-(GlcNAc)3]. Activity was compared with those of protein extracts of B. thuringiensis LBIT-82, E. agglomerans ChiA, and E. coli DH5αF′. Figure 6 shows the activity of a band of ca. 70 kDa from the E. coli DH5αF′/pCHIBT extract, as well as a comigrating band from B. thuringiensis LBIT-82. Another band of ca. 60 kDa from B. thuringiensis LBIT-82 was also detected, which may be a degradation product of ChiA74 or a different endochitinase. Furthermore, similarly to previous reports on the B. thuringiensis LBIT-82 chitinase (2) and on E. agglomerans ChiA (6), recombinant ChiA74 kept its activity even after the heat treatment (55 and 100°C) and the β-mercaptoethanol reducing effect. No activity was detected from E. coli DH5αF′ extracts.
FIG. 6.
Chitinase activity of extracellular proteins after separation in SDS-polyacrylamide gels. 4-MU-(GlcNAc)3 was used as a substrate. Lane 1, E. coli DH5αF′/pCHIBT; lane 2, B. thuringiensis serovar kenyae LBIT-82; lane 3, E. agglomerans ChiA; lane 4, E. coli DH5αF′; lane 5, molecular mass marker (in kilodaltons) (Gibco BRL). The arrow indicates the location of ChiA74.
DISCUSSION
Once the chiA74 gene was cloned and sequenced, several attributes were apparent. For example, its putative promoter showed homology with those active during the vegetative (exponential) growth phase of B. subtilis and with the cry3A gene of B. thuringiensis (1). This observation indicates that expression of chiA74 occurs during the vegetative growth phase of the culture, an observation that was corroborated in our laboratory (data not shown) and by others who have analyzed the expression of chitinases of B. thuringiensis (15).
On the other hand, the difference between the deduced molecular mass of ChiA74 (ca. 74 kDa) and the molecular mass estimated by the zymograms (ca. 70 kDa) may be due to the deletion of the signal peptide (ca. 4 kDa) in the mature enzyme (after processing during its pass through the bacterial membrane). Therefore, if the signal peptide is processed in such a way, the expected N-terminal sequence of the mature ChiA74 is Asp-Ser-Pro-Lys-Gln, which is identical to that of the mature ChiA71 of B. thuringiensis serovar Pakistani (32) and ChiB of B. cereus (19). It is important to notice that a previous report of ChiA74 miscalculated a molecular mass of ca. 68 kDa for the mature protein (2).
According to the sequence analysis, the deduced amino acid sequence of ChiA74 is practically identical (98% identity) to that of the ChiB of B. cereus. This is an interesting observation, as the other known sequence for a chitinase in B. thuringiensis (32) shares only 70% identity with ChiA74. This observation can be explained by the high relationship between these two species, where the only difference is the presence of parasporal bodies in B. thuringiensis, and some authors consider them to be the same species (4). Unfortunately, other than the sequence homology, little can be compared between these two chitinases, as very little information on ChiB is available (20). On the other hand, when sequences of ChiA74 and ChiA71 are compared, their actual difference is the presence of a 118-amino-acid fragment (Val-331 through Glu-448) in ChiA74 which is absent in ChiA71 and the presence of a 93-amino-acid fragment (Tyr-543 through Cys-635) at the C terminus of ChiA71 which is absent in ChiA74. Interestingly, these highly localized differences should explain the radical difference in the chitinase activities of the two enzymes, as ChiA74 was characterized as an endochitinase, while ChiA71 was reported to be an exochitinase (32). Exchange of these regions would be necessary to prove their actual role in each enzyme.
ChiA74 showed a modular structure, which is typical of degrading enzymes of biopolymers such as chitin and cellulose. ChiA74 shows at the catalytic domain an active-site motif between amino acids 203 and 211 (Phe-Asp-Gly-Val-Asp-Leu-Asp-Trp-Glu) which is typical of family 18 of glycosyl hydrolases. It is known that the aromatic amino acids Trp-245 and Phe-232 of ChiA from S. marcescens 2170 are involved in the hydrolysis of β-chitin microfibers (35). Therefore, the highly conserved aromatic amino acids Trp-171, Phe-177, and Trp-210 in the catalytic domain of ChiA74 may have an important role in the hydrolysis of chitin.
In relation to the second domain, this region shows homology to fibronectin type III. This protein was initially found in mammals and forms extracellular, multifunctional matrices which are important in cell bonding (28); however, it is believed that it was horizontally transferred to prokaryotes, and now it is found in bacterial chitinases, cellulases, and amylases (3, 23, 32). Because of the bonding action of fibronectin, its presence in these enzymes may be related to the substrate attachment.
The third module of ChiA74 is constituted by the CBD at the C terminus of the amino acid sequence (residues 587 to 629). This domain is common in many other chitinases, and besides its usual chitin-binding activity, some chitin hydrolysis has been associated with this domain, at least in ChiA and ChiA1 from C. paraputrificum and from B. circulans, respectively (21, 34, 39). Similar to the case for other bacterial chitinases and cellulases, ChiA74 has the conserved aromatic amino acids Trp, Tyr, and Trp in positions 591, 595, and 626, respectively. It is believed that the substrate-binding mechanisms are common among CBDs and cellulose-binding domains, and both Tyr and Trp seem to play an important role during the binding of the enzyme to the pyranosyl rings of N-acetylglucosamine residues in chitin (21).
Finally, the activity analysis of ChiA74 over a range of pH and temperature conditions indicates an interesting enzymatic behavior. First, the bimodal distribution of activity at the pH range tested makes this enzyme very unusual, as normally there is only one optimum pH of activity. This result suggests that there might be two conformational arrangements of the molecule after secretion by the cell, which behave differently at different pHs. On the other hand, the optimal temperature for the enzymatic activity of ChiA74 (57.2°C) is higher than expected, especially if we consider that the most favorable growth temperature for B. thuringiensis is 30°C. These interesting results lead to more exhaustive studies not only on a more detailed characterization of this enzyme but also on its possible use as a synergistic factor acting along with Cry proteins.
Acknowledgments
We thank Guillermo Corona (CINVESTAV-Irapuato) and Gabriel Angeles (Tecnológico de Celaya) for invaluable technical support and B. A. Federici (University of California, Riverside) for his comments.
This research was supported by project J35306-B (CONACYT, Mexico) and project 03/98 (Fundación Guanajuato Produce A.C.).
REFERENCES
- 1.Agaisse, H., and D. Lereclus. 1995. How does Bacillus thuringiensis produce so much insecticidal crystal protein? J. Bacteriol. 177:6027-6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barboza-Corona, J. E., J. C. Contreras, R. Velásquez-Robledo, M. Bautista-Justo, M. Gómez-Ramírez, R. Cruz-Camarillo, and J. E. Ibarra. 1999. Selection of chitinolytic strain of Bacillus thuringiensis. Biotechnol. Lett. 21:1125-1129. [Google Scholar]
- 3.Candussio, A., G. Schmid, and A. Bock. 1990. Biochemical and genetic analysis of a maltopentose-producing amylase from an alkaliphilic gram-positive bacterium. Eur. J. Biochem. 191:177-185. [DOI] [PubMed] [Google Scholar]
- 4.Carlson, C. R., D. Caugant, and A.-B. Kolstø. 1994. Genotypic diversity among Bacillus cereus and Bacillus thuringiensis strains. Appl. Environ. Microbiol. 60:1719-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chernin, L., Z. Ismailov, S. Haran, and I. Chet. 1995. Chitinolytic Enterobacter agglomerans antagonistic to fungal plant pathogens. Appl. Environ. Microbiol. 61:1720-1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chernin, L., L. de la Fuente, V. Sovoleb, S. Haran, C. E. Vorgias, A. B. Oppenheim, and I. Chet. 1997. Molecular cloning, structural analysis, and expression in Escherichia coli of a chitinase gene from Enterobacter agglomerans. Appl. Environ. Microbiol. 63:834-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cody, M. R. 1989. Distribution of chitinase and chitobiase in Bacillus. Curr. Microbiol. 19:201-205. [Google Scholar]
- 8.Cutting, S. M., and P. B. Vander Horn. 1990. Genetic analysis, p. 65. In S. M. Cutting and P. B. Vander Horn (ed.). Molecular biological methods for Bacillus. John Wiley & Sons, London, United Kingdom.
- 9.Del Rincón-Castro, M. A., J. Barajas-Huerta, and J. E. Ibarra. 1999. Antagonism between Cry1Ac1 and CytA1 toxins of Bacillus thuringiensis. Appl. Environ. Microbiol. 65:2049-2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dow, J. A. 1992. pH gradients in lepidopteran midgut. J. Exp. Biol. 172:355-375. [DOI] [PubMed] [Google Scholar]
- 11.Downing, K., G. Leslie, and J. A. Thomson. 2000. Biocontrol of the sugarcane borer Eldana saccharina by expression of the Bacillus thuringiensis cry1Ac7 and Serratia marcescens chiA genes in sugarcane-associated bacteria. Appl. Environ. Microbiol. 66:2804-2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuchs, R. L., S. A. McPherson, and D. J. Drahos. 1986. Cloning of a Serratia marcescens gene encoding chitinase. Appl. Environ. Microbiol. 51:504-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fukumori, F., N. Sashihara, T. Kudo, and K. Horikoshi. 1986. Nucleotide sequences of two cellulase genes from alkalophilic Bacillus sp. strain N-4 and their strong homology. J. Bacteriol. 168:479-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glare, T. R., and M. O'Callaghan. 2000. Bacillus thuringiensis: biology, ecology and safety, p. 27. John Wiley & Sons, London, United Kingdom
- 15.Gómez-Ramírez, M., L. I. Rojas-Avelizapa, and R. Cruz-Camarillo. 2001. The chitinase of Bacillus thuringiensis, p. 273-282. In R. A. A. Muzzarelli (ed.), Chitin enzymology. Atec Edizioni, Atec, Italy.
- 16.Granados, R. R., Y. Fu, B. Corsaro, and P. R. Hughes. 2001. Enhancement of Bacillus thuringiensis toxicity to lepidopterous species with the enhancin from Trichoplusia ni granulovirus. Biol. Control 20:153-159. [Google Scholar]
- 17.Gringorten, J. L., D. N. Crawford, and W. R. Harvey. 1993. High pH in the ectoperitrophic space of the larval lepidopteran midgut. J. Exp. Biol. 183:353-359. [DOI] [PubMed] [Google Scholar]
- 18.López-Meza, J. E., B. A. Federici, W. J. Poehner, A. M. Martínez-Castillo, and J. E. Ibarra. 1995. Highly mosquitocidal isolates of Bacillus thuringiensis subspecies kenyae and entomocidus from Mexico. Biochem. Syst. Ecol. 23:461-468. [Google Scholar]
- 19.Mabuchi, N., I. Hashizume, and Y. Araki. 2000. Characterization of chitinases excreted by Bacillus cereus CH. Can. J. Microbiol. 46:370-375. [PubMed] [Google Scholar]
- 20.Mabuchi, N., and Y. Araki. 2001. Cloning and sequencing of two genes encoding chitinases A and B from Bacillus cereus CH. Can. J. Microbiol. 47:895-902. [PubMed] [Google Scholar]
- 21.Marimoto, K., S. Karita, T. Kimura, K. Sakka, and K. Ohmiya. 1997. Cloning, sequence, and expression of the gene encoding Clostridium paraputrificum chitinase ChiB and analysis of the functions of novel cadherin-like domains and a chitin-binding domain. J. Bacteriol. 179:7306-7314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McCreath, K. J., and G. W. Gooday. 1992. A rapid and sensitive microassay for determination of chitinolytic activity. J. Microbiol. Methods 14:229-237. [Google Scholar]
- 23.Meink, A., C. Braum, N. R. Gilkes, D. G. Kiburn, R. C. Miller, and R. A. J. Warren. 1991. Unusual sequence organization in CenB, an inverting endoglucanase from Cellumonas fimi. J. Bacteriol. 173:308-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 25.Regev, A., M. Keller, N. Strizhov, B. Sneh, E. Prudovsky, I. Chet, I. Ginzberg, Z. Koncz-Kalman, Z. Koncz, J. Schell, and A. Zilberstein. 1996. Synergistic activity of a Bacillus thuringiensis δ-endotoxin and a bacterial endochitinase against Spodoptera littoralis. Appl. Environ. Microbiol. 62:3581-3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rojas-Avelizapa, L. I., R. Cruz-Camarillo, M. I. Guerrero, R. Rodríguez-Vázquez, and J. E. Ibarra. 1999. Selection and characterization of a proteo-chitinolytic strain of Bacillus thuringiensis, able to grow in shrimp waste media. World. J. Microbiol. Biotechnol. 15:299-308. [Google Scholar]
- 27.Rosenberg, M., and D. Court. 1979. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu. Rev. Genet. 13:319-353. [DOI] [PubMed] [Google Scholar]
- 28.Ruoslahti, E. 1988. Fibronectin and its receptors. Annu. Rev. Biochem. 57:375-413. [DOI] [PubMed] [Google Scholar]
- 29.Sampson, M. N., and G. W. Gooday. 1998. Involvement of chitinases of Bacillus thuringiensis during pathogenesis in insects. Microbiology 144:2189-2194. [DOI] [PubMed] [Google Scholar]
- 30.Sanger, F., S. Nicklen, and A. R. Coulson. 1977. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74:5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sneh, B., S. Schuster, and S. Gross. 1983. Improvement of the insecticidal activity of Bacillus thuringiensis var. entomocidus on larvae of Spodoptera littoralis (Lepidoptora, Noctuidae) by addition of chitinolytic bacteria, a phagostimulant and a UV protectant. Z. Angew. Entomol. 96:77-83. [Google Scholar]
- 32.Thamthiankul, S., S. Suan-Ngay, S. Tantimavanich, and W. Panbangred. 2001. Chitinase from Bacillus thuringiensis subsp. pakistani. Appl. Microbiol. Biotechnol. 56:395-401. [DOI] [PubMed] [Google Scholar]
- 33.Tsujibo, H., H. Orikoshi, H. Tanno, K. Fujimoto, K. Miyamoto, C. Imada, Y. Okami, and Y. Inamori. 1993. Cloning, sequence, and expression of a chitinase gene from a marine bacterium, Alteromonas sp. strain O-7. J. Bacteriol. 175:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tormo, J., R. Lamed, A. J. Chirino, E. Morag, E. A. Bayer, Y. Shoham, and T. A. Steitz. 1996. Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose. EMBO J. 21:5739-5751. [PMC free article] [PubMed] [Google Scholar]
- 35.Uchiyama, T., F. Katouno, N. Nikaidou, T. Nonaka, J. Sugiyama, and T. Watanabe. 2001. Roles of the exposed aromatic residues in crystalline chitin hydrolysis by chitinase A from Serratia marcescens 2170. J. Biol. Chem. 276:41343-41349. [DOI] [PubMed] [Google Scholar]
- 36.Watanabe, T., K. Susuki, W. Oyanagi, K. Ohnishi, and H. Tanaka. 1990. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J. Biol. Chem. 265:15659-15665. [PubMed] [Google Scholar]
- 37.Watanabe,T., W. Oyanagi, K. Suzuki, K. Ohnishi, and H. Tanaka. 1992. Structure of the gene encoding chitinase D of Bacillus circulans WL-12 and possible homology of the enzyme to other prokaryotic chitinases and class III plant chitinases. J. Bacteriol. 174:408-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Watanabe, T., K. Kobori, K. Miyashita, T. Fujii, M. Sakai, M. Uchida, and H. Tamaka. 1993. Identification of glutamic acid 201 and aspartic 200 in chitinase A1 of Bacillus circulans WL-12 as essential residues for chitinase activity. J. Biol. Chem. 268:18567-18572. [PubMed] [Google Scholar]
- 39.Watanabe, T., M. Uchida, K. Kabori, and H. Tanaka. 1994. Site-directed mutagenesis of the Asp-197 and Asp-202 in the chitinase A1 of Bacillus circulans WL-12. Biosci. Biotechnol. Biochem. 58:2283-2285. [DOI] [PubMed] [Google Scholar]
- 40.Wiwat, C., S. Thaithanum, S. Pantuwatana, and A. Bhumiratana. 2000. Toxicity of chitinase-producing Bacillus thuringiensis ssp. kurstaki HD-1 (G) toward Plutella xylostella. J. Invertebr. Pathol. 76:270-277. [DOI] [PubMed] [Google Scholar]