Abstract
The subseafloor microbial habitat associated with typical unsedimented mid-ocean-ridge hydrothermal vent ecosystems may be limited by the availability of fixed nitrogen, inferred by the low ammonium and nitrate concentrations measured in diffuse hydrothermal fluid. Dissolved N2 gas, the largest reservoir of nitrogen in the ocean, is abundant in deep-sea and hydrothermal vent fluid. In order to test the hypothesis that biological nitrogen fixation plays an important role in nitrogen cycling in the subseafloor associated with unsedimented hydrothermal vents, degenerate PCR primers were designed to amplify the nitrogenase iron protein gene nifH from hydrothermal vent fluid. A total of 120 nifH sequences were obtained from four samples: a nitrogen-poor diffuse vent named marker 33 on Axial Volcano, sampled twice over a period of 1 year as its temperature decreased; a nitrogen-rich diffuse vent near Puffer on Endeavour Segment; and deep seawater with no detectable hydrothermal plume signal. Subseafloor nifH genes from marker 33 and Puffer are related to anaerobic clostridia and sulfate reducers. Other nifH genes unique to the vent samples include proteobacteria and divergent Archaea. All of the nifH genes from the deep-seawater sample are most closely related to the thermophilic, anaerobic archaeon Methanococcus thermolithotrophicus (77 to 83% amino acid similarity). These results provide the first genetic evidence of potential nitrogen fixers in hydrothermal vent environments and indicate that at least two sources contribute to the diverse assemblage of nifH genes detected in hydrothermal vent fluid: nifH genes from an anaerobic, hot subseafloor and nifH genes from cold, oxygenated deep seawater.
Deep-sea hydrothermal vents sustain a variety of microbial habitats, including the warm fluid venting from cracks in the seafloor that contain microorganisms originating from a hotter microbial habitat within the subseafloor. Due to the difficulty of directly sampling the subseafloor associated with unsedimented hydrothermal vent systems, microorganisms found in these diffuse, low-temperature fluids have been used to infer the characteristics of microbial populations living within the subseafloor (28, 31). Many thermophiles and hyperthermophiles have been isolated from hydrothermal vents (6), and these microorganisms can be metabolically versatile or highly specialized and utilize a variety of carbon and energy sources (32, 35, 70). However, the nitrogen sources that support these microbial communities remain unknown (35).
Chemical analyses indicate that nitrate and nitrite are depleted in diffuse hydrothermal vent fluids relative to deep seawater (∼40 μM) and are absent in reduced fluids above 30°C (18, 34, 36, 41). Ammonium concentrations in low-temperature vent fluids are similar to the low (≤1 μM) concentrations in deep seawater (41), with the exception of sedimented hydrothermal vent systems such as Guaymas Basin (37) and the aberrant unsedimented Endeavour Segment on the Juan de Fuca Ridge (42). The largest reservoir of nitrogen in the ocean is dissolved dinitrogen gas (N2), which is abundant in deep seawater (0.59 mM) and slightly elevated in hydrothermal fluids (15). Biological nitrogen fixation was first suggested as a potential source of nitrogen to hydrothermal vent ecosystems based on the 15N/14N ratios of low-trophic-level vent fauna. The nitrogen isotope ratios of vent animals are much lower than the nitrogen isotope ratios of deep-sea organic nitrogen, ammonium, and nitrate but resemble those of deep-oceanic N2 and marine biota associated with nitrogen fixation (57). Heterotrophic and chemolithotrophic microorganisms capable of nitrogen fixation (verified by the acetylene reduction technique) have been cultured from diffuse hydrothermal fluids at the Galapagos Spreading Center (33, 41).
The ability to fix nitrogen is widely distributed among phylogenetically diverse bacteria but appears to be limited to the methanogens within Archaea (43, 60, 63, 77). Methanococcus thermolithotrophicus is currently the most thermophilic microorganism able to fix nitrogen, at 64°C (9), and was isolated from a near-shore submarine hydrothermal vent (29). Nitrogenase, the enzyme complex that catalyzes nitrogen fixation, is composed of two proteins: the iron protein, or dinitrogenase reductase, and the molybdenum iron protein, or dinitrogenase. The nifH gene encodes the iron protein and the nifDK genes encode the molybdenum iron protein. Some microorganisms have alternative nitrogenases in which the dinitrogenase protein contains vanadium instead of molybdenum, encoded by the vnf genes, or only iron, encoded by the anf genes. The iron protein genes (nifH, vnfH, and anfH) are highly conserved among diverse microorganisms, and the NifH phylogenetic tree largely resembles the 16S rRNA phylogenetic tree (26, 51, 77). All nifH genes fall into one of four clusters (16): cluster I includes standard molybdenum nitrogenases from cyanobacteria and proteobacteria (α, β, and γ), as well as γ-proteobacterial vnfH, and cluster II includes methanogen nitrogenases and bacterial anfH. Cluster III includes nitrogenases from diverse anaerobic bacteria such as clostridia (low G+C, gram positive) and sulfate reducers (δ-proteobacteria), which is an example of the NifH phylogeny deviating from the 16S rRNA phylogeny. Cluster IV includes divergent nitrogenases from archaea.
The highly conserved nature of the nifH gene makes it an ideal molecular tool to determine the potential for biological nitrogen fixation in any environment (78). Since the ability to fix nitrogen is distributed widely and inconsistently among prokaryotes (nitrogen fixation is not found in all of the members of any one group), it is difficult to assess the distribution and diversity of nitrogen-fixing microorganisms based solely on 16S rRNA phylogenetic diversity studies. Designing PCR primers that amplify the nifH gene and sequencing the amplified genes can identify the assemblage of microorganisms capable of nitrogen fixation in any environment. This approach has been applied to environments that are nitrogen limited or are known to support nitrogen fixation, such as oligotrophic oceans (81), marine microbial mats (54, 79), modern marine stromatolites (68), tropical seagrass beds (3), rice roots (19, 73), and termite hindguts (53). In order to address the hypothesis that biological nitrogen fixation occurs in nitrogen-poor hydrothermal vent subseafloor habitats, degenerate PCR primers for nifH were designed and used to amplify nifH from diffuse hydrothermal vent fluid. This paper describes the phylogenetic diversity of nifH genes in a nitrogen-poor hydrothermal vent habitat sampled twice over a period of 1 year, during which the fluid temperature decreased; a nitrogen-rich hydrothermal vent habitat; and background deep seawater. These results indicate that the nifH genes present in hydrothermal fluid originate from a diverse nifH assemblage in the subseafloor as well as a phylogenetically distinct nifH assemblage in ambient deep seawater.
MATERIALS AND METHODS
Sampling and DNA extraction.
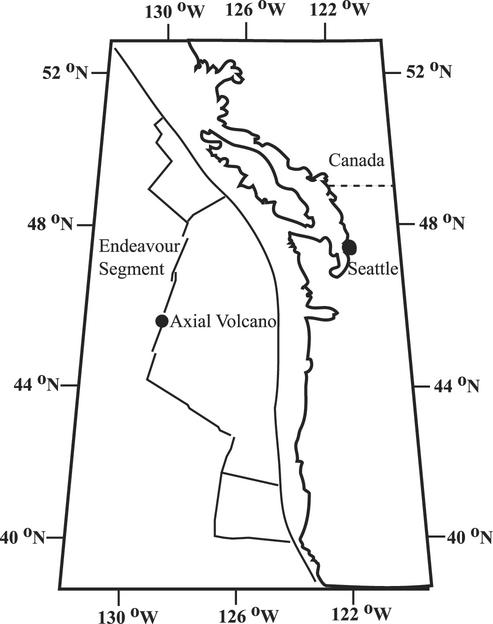
Axial Volcano (46°55′N, 130°00′W; 1,500 m depth) is an active submarine volcano on the Juan de Fuca Ridge (Fig. 1) that most recently erupted in January of 1998 (4). Marker 33, on Axial Volcano, is a site of diffuse venting from a crack in the seafloor that was sampled in July 1999 (dive R485) and in July 2000 (dive R551) with the remotely operated vehicle ROPOS and a hydrothermal fluid and particle sampler (HFPS) (31). Puffer is a high-temperature vent in the main Endeavour hydrothermal field (47°57′N, 129°06′W; 2,200 m depth), which is located on the Endeavour Segment of the Juan de Fuca Ridge (Fig. 1). A diffuse vent on the basalt seafloor near Puffer was sampled in September 2000 (dive ALV3618) with the autonomous underwater vehicle ALVIN, also using the HFPS. During each sampling, the HFPS filtered approximately 1 liter of vent fluid onto a 0.22-μm-pore-size Sterivex-GP filter (Millipore) while recording the temperature and pump rate. On board the research vessel, the filters were placed in sterile 50-ml Falcon tubes (BD Sciences Labware), frozen in liquid nitrogen, and stored at −20°C until DNA was extracted. At each vent, unfiltered vent fluid samples were also obtained by the HFPS and used for chemical analyses, including determination of ammonium and nitrate concentrations, and epifluorescent cell counts. An 18-ml subsample of unfiltered vent fluid was preserved in formaldehyde (3.7% final concentration) in duplicate, stored at 2°C, and counted by epifluorescence microscopy with 4′,6′-diamidino-2-phenylindole (Sigma) (55). A deep-seawater sample with no detectable hydrothermal fluid was collected with a Niskin bottle on a CTD (conductivity, temperature, depth) rosette from 2,200 m depth on Endeavour Segment, and 3 liters was filtered on deck through a 0.22-μm-pore-size Sterivex-GP filter (Millipore). DNA was extracted from 0.22-μm-pore-size Sterivex-GP filters and purified as described previously (31).
FIG. 1.
Map of the Juan de Fuca Ridge, showing the sampling locations Axial Volcano and Endeavour Segment.
Chemical analysis.
Analytical methods were described previously by Butterfield et al. (14). Fluids collected with HFPS were analyzed shipboard for H2S, pH, ammonia, and dissolved silica. Nitrate was analyzed on filtered, frozen samples by autoanalyzer at the University of Washington. On shore, fluids were also analyzed for major, minor, and trace elements. Precision (±1 standard deviation) of the reported chemical analyses are as follows: pH, 0.05 units; H2S, 4%; Mg, 1%; Cl, 0.5%; silica, 0.5%; nitrate and ammonia, 2%.
Primer design, PCR, and cloning.
The NifH amino acid sequences from divergent nitrogen-fixing microorganisms, including all AnfH, VnfH, and archaeal NifH in the sequence databases, were aligned using ClustalX version 1.81 (72) to find conserved regions in the iron protein sequence. The following degenerate oligonucleotide primers were designed: forward 5′-GGHAARGGHGGHATHGGNAARTC-3′ and reverse 5′-GGCATNGCRAANCCVCCRCANAC-3′, which correspond to the amino acid positions 10 to 17 (GKGGIGKS) and 132 to 139 (VCGGFAMP), respectively (Klebsiella pneumoniae numbering), and amplify approximately 400 bp of the gene. The forward primer corresponds to the ATP-binding domain of the iron protein, and the reverse primer includes one of the cysteine residues that function as a ligand for the 4Fe-4S cluster (11). The nifH genes were amplified from hydrothermal vent and deep-sea DNA extracts in 25 μl of PCR mixtures containing 1× PCR buffer (Promega), 2.5 mM MgCl2, 0.8 mM deoxynucleoside triphosphates, a 0.5 μM concentration of each primer, and 0.5 U of Taq DNA polymerase (Promega). The template DNA was denatured for 2 min at 94°C, followed by 35 cycles of 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. A final extension step was carried out for 10 min at 72°C. The amplified 400-bp bands were excised from 1.5% (wt/vol) agarose gels and gel purified using the QiaQuick gel extraction kit (Qiagen). The purified PCR products were cloned into the pCR4-TOPO Escherichia coli vector by using the TOPO TA cloning kit for sequencing with One Shot TOP10 Competent cells according to the manufacturer's instructions (Invitrogen). Randomly selected colonies were picked, streaked onto agar plates, and grown in 100 μl of Luria-Bertani broth medium in a shaking incubator at 220 rpm for 1 h at 37°C.
Sequencing and phylogenetic analysis.
The nifH inserts were amplified from clones in 50 μl of PCR mixture containing 1 μl of clone culture, 1× PCR buffer (Promega), 3.125 mM MgCl2, 0.4 mM deoxynucleoside triphosphates, 0.4 μM concentrations of M13 reverse and forward primers, and 1.875 U of Taq DNA polymerase (Promega). The cycling conditions were the following: initial denaturation for 2 min at 94°C; 30 cycles of 10 s at 94°C, 30 s at 54°C, and 1 min at 72°C; and a final extension step for 10 min at 72°C. PCR products were cleaned with the QiaQuick PCR purification kit (Qiagen) and sequenced in both directions using the DYEnamic ET dye terminator kit (Amersham Pharmacia Biotech Inc.), T3 forward primer, and T7 reverse primer. Cycle sequencing products were ethanol precipitated according to the manufacturer's instructions (Amersham Pharmacia Biotech Inc.) and analyzed on a MegaBACE 1000 (Molecular Dynamics). Automatic base calls were checked, and forward and reverse nifH sequences were assembled and translated into amino acid sequences using Sequencher (version 4.1.2; Gene Codes Corp.). All diversity and phylogenetic analyses were performed using the deduced amino acid sequences. Sequences closely related to the deep-sea and hydrothermal vent NifH protein sequences were found using the National Center for Biotechnology Information's translating BLAST search tblastn, which compares a protein query sequence against a nucleotide sequence database, dynamically translated in all reading frames (2). NifH protein sequences were aligned using ClustalX version 1.81 (72), and amino acid identity matrices were constructed using BioEdit version 5.0.9 (25). Maximum-likelihood phylogenetic trees were constructed with TREE-PUZZLE version 5.0 (61, 69) using the Whelan and Goldman model of protein evolution (75). All sequences passed the 5% chi-square test, which compares the amino acid composition of each sequence to the frequency distribution assumed in the maximum-likelihood model.
Nucleotide sequence accession numbers.
All 120 nifH sequences obtained in this study have been submitted to the GenBank database at the National Center for Biotechnology Information. The nifH sequences from marker 33, Axial Volcano in 2000 have been assigned the GenBank accession numbers AY120565 to AY120606. The nifH sequences from deep seawater on Endeavour Segment have been assigned the GenBank accession numbers AY120607 to AY120629. The nifH sequences from the diffuse vent near Puffer, on Endeavour Segment, have been assigned the GenBank accession numbers AY120630 to AY120657. The nifH sequences from marker 33, Axial Volcano in 1999 have been assigned the GenBank accession numbers AY120658 to AY120684.
RESULTS
The hydrothermal vent and deep-sea samples that were used to construct nifH clone libraries are described in Table 1. The maximum temperature, pH, maximum H2S concentration, and percent crustal seawater of the samples reflect the amount of mixing between hot source fluid and crustal seawater below the point of venting (31). Marker 33, the diffuse vent sampled on Axial Volcano (Fig. 1), had low ammonium concentrations typical of unsedimented mid-ocean-ridge hydrothermal systems (Table 1). A diffuse vent near Puffer, on Endeavour Segment, had a high ammonium concentration characteristic of Endeavour (42). Nitrate concentrations reach near-zero levels in nearby low-temperature vents at Endeavour at temperatures ranging from 15 to 40°C. Nitrate concentrations measured in marker 33 samples from 1999 and 2000 were depleted relative to deep seawater. The lower nitrate concentration in 2000 indicates increased consumption of nitrate in the shallow low-temperature subseafloor. Cell counts from all three hydrothermal vent samples revealed higher microbial densities than in the background seawater sample.
TABLE 1.
Sampling locations and chemical characteristics of nifH clone libraries
| Sampling location | Yr sampled | Temp (°C) range during sampling | pH | [NH3] (μM) | [NO3−] (μM) | Max. [H2S] (mmol/liter) | % Crustal seawaterb | Cells/ml |
|---|---|---|---|---|---|---|---|---|
| Marker 33, Axial Volcano | 1999 | 45-78a | 4.6a | 2.5 | 28.7 | 2.4a | 76.0a | 1.17 × 105a |
| Marker 33, Axial Volcano | 2000 | 20-32a | 5.8a | 2.2 | 14.5 | 0.2a | 84.7a | 1.15 × 105a |
| Puffer (diffuse) Endeavour | 2000 | 27-39 | 6.1 | 32.5 | No datad | 0.468 | 95.6 | 1.67 × 105 |
| Deep-sea Endeavour | 2000 | 2 | 7.8c | ≤1c | 40c | 0c | 5.0 × 104c |
From Huber et al. (31).
(Lowest Mg content/seawater Mg content) × 100.
Based on standard deep-seawater with no detectable hydrothermal plume signal.
Puffer [NO3−] probably 0 to 5 μM, based on temperature and ammonia concentration.
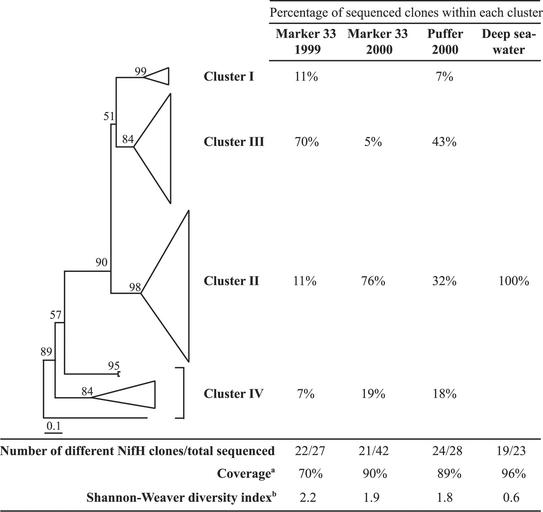
The total number of nifH clones sequenced from each clone library and the number of different NifH amino acid sequences within each clone library are shown in Fig. 2. A total of 120 nifH clones were sequenced from all four libraries combined and, after taking into account identical amino acid sequences, there were 77 different NifH amino acid sequences. Seven out of the 16 groups of identical amino acid sequences contained clones from more than one library, indicating that many NifH sequences were present in more than one of the samples. A maximum-likelihood tree containing all 77 different NifH sequences is shown in Fig. 2, along with the distribution of clones from each library among the four NifH clusters. The 19 phylogenetic groups generated in this study, based on ≥92% amino acid similarity and confirmed by phylogenetic analysis, are listed in Table 2. The coverage and Shannon-Weaver diversity indices shown in Fig. 2 were calculated using these phylogenetic groups. For those phylogenetic groups that are represented by more than one clone (Table 2), each representative clone is 92 to 97% similar to the other representative clones in that group. NifH clones that are ≥97% similar to a representative clone were not included in the corresponding phylogenetic tree, but their quantity is indicated in parentheses following their representative clone.
FIG. 2.
Maximum-likelihood phylogenetic tree of the 77 different deep-sea and hydrothermal vent NifH sequences obtained in this study, showing the phylogenetic relationship between the four NifH clusters. The depth and width of each wedge is proportional to the branch lengths and number of NifH sequences within each cluster, respectively. Cluster I contains α-, β-, and γ-proteobacterial NifH, cluster III contains anaerobic bacterial NifH sequences, cluster II contains methanogen and alternative NifH, and cluster IV contains divergent archaeal NifH. A NifH clone from this study (A37), related to the light-independent protochlorophyllide reductase subunit L gene (bchL) from Chlorobium tepidum, was used as the outgroup. Quartet puzzling support values are shown at each internal branch, and the scale indicates the number of amino acid substitutions per site. Footnotes: a, coverage (C) was calculated according to the equation C = 1 − (n/N), where n is the number of phylogenetic groups that contain only one clone and N is the total number of clones sequenced (23), b, Shannon-Weaver diversity indices (H) were calculated according to the equation H = −Σ(pI) (ln pI), where pI is the number of species in each phylogenetic group divided by the total number of sequences in that library (62).
TABLE 2.
Distribution of hydrothermal vent and deep-sea nifH clones among phylogenetic groupsa
| Phylogenetic group | Phylogenetic affiliation | No. of clones in each phylogenetic group
|
Total no. of clones | |||
|---|---|---|---|---|---|---|
| Marker 33 1999 (E)c | Marker 33 2000 (A) | Puffer 2000 (C) | Deep-seawater (B) | |||
| Cluster I | ||||||
| E27b | α-Proteobacteria | 1 | 1 | |||
| E10 | γ-Proteobacteria | 1 | 1 | |||
| C60, C73, E58 | γ-Proteobacteria | 1 | 2 | 3 | ||
| Total | 3 | 2 | 5 | |||
| Cluster III | Anaerobic bacteria | |||||
| A19, C22, E5, C47, C36 | 7 | 1 | 6 | 14 | ||
| C65, C9, E8 | 5 | 5 | 10 | |||
| E44, E63 | 3 | 3 | ||||
| E12 | 4 | 4 | ||||
| A44 | 1 | 1 | ||||
| C19 | 1 | 1 | ||||
| Total | 19 | 2 | 12 | 33 | ||
| Cluster II | Archaea and anfH | |||||
| B28, C70, A34, C59, B25, B20, B7, B49, B47, B33 | 1 | 16 | 9 | 19 | 45 | |
| A12, E7 | 1 | 8 | 9 | |||
| E59, A16, A11 | 1 | 4 | 3 | 8 | ||
| A2 | 4 | 1 | 5 | |||
| Total | 3 | 32 | 9 | 23 | 67 | |
| Cluster IV | Divergent Archaea | |||||
| A26 | 1 | 2 | 3 | |||
| A8 | 4 | 4 | ||||
| A21 | 1 | 1 | ||||
| C83 | 1 | 1 | ||||
| C61 | 3 | 3 | ||||
| A37 | 1 | 1 | 1 | 3 | ||
| Total | 2 | 8 | 5 | 15 | ||
Phylogenetic groups based on ≥92% amino acid similarity.
Representative clone(s) from each phylogenetic group was used to construct phylogenetic trees.
Letter designations for each clone library are used as prefixes in clone names.
NifH cluster I.
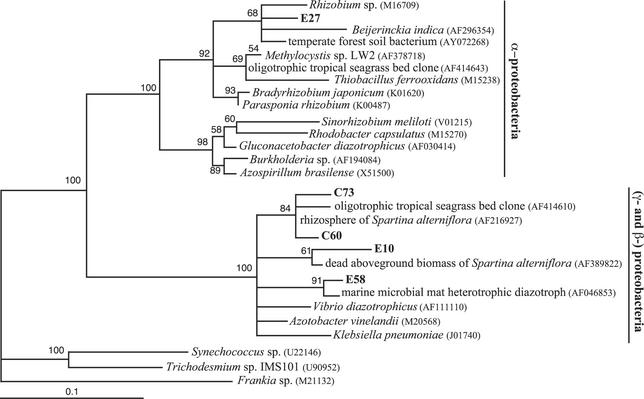
Five hydrothermal vent clones fall within cluster I of the NifH phylogenetic tree, which includes α, β, and γ-proteobacterial and cyanobacterial nitrogenases, and these are shown in Fig. 3. Clone E27, from marker 33 in 1999, is most closely related (95% amino acid similarity) to an isolate from temperate forest soil that was cultured in Azospirilla media (59). Clone E27 is the only hydrothermal vent sequence that belongs to the α-proteobacterial group within cluster I. Clone E10, also from marker 33 in 1999, is most closely related (95% amino acid similarity) to a presumptive γ-proteobacterial clone associated with dead above-ground biomass of the salt marsh smooth cordgrass Spartina alterniflora (45). Clones C60 and C73, from the diffuse vent near Puffer, are 98% similar to a presumptive γ-proteobacterial clone from the rhizosphere of S. alterniflora (46). Clone E58, from marker 33 in 1999, is most closely related (98% amino acid similarity) to a heterotrophic nitrogen-fixing isolate from an intertidal marine microbial mat, presumed to belong to the γ-proteobacteria (54).
FIG. 3.
Maximum-likelihood phylogenetic tree of cluster I NifH sequences, which include α-, β-, and γ-proteobacterial and cyanobacterial NifH sequences. GenBank accession numbers in parentheses follow the isolate or clone name. Hydrothermal vent NifH sequences from this study are in bold, followed by the number of additional NifH sequences from this study that are ≥97% similar to it, in parentheses. The letter prefix of each clone name denotes which sample the clone was sequenced from, as follows: A, marker 33, Axial Volcano, 2000; B, deep seawater; C, diffuse vent near Puffer, Endeavour Segment, 2000; E, marker 33, Axial Volcano, 1999. Quartet puzzling support values are shown at each internal branch. The scale indicates the number of amino acid substitutions per site, and the tree is outgroup rooted with Frankia sp.
NifH cluster III.
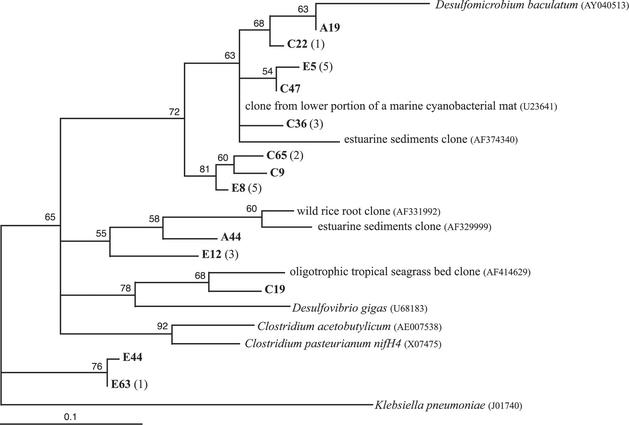
Thirty-three hydrothermal vent NifH sequences fall within cluster III, which includes diverse anaerobes like sulfate reducers (δ-protebacteria), clostridia (gram-positive bacteria), and purple sulfur bacteria (γ-proteobacteria). Representatives from the six cluster III hydrothermal vent phylogenetic groups (Table 2) are shown in Fig. 4. The first group is represented by clones A19, C22, E5, C47, and C36, which are 96 to 99% similar to a clone from the lower, reduced portion of a marine cyanobacterial mat (79) and 90 to 93% similar to Desulfomicrobium baculatum. Clones C65 and C9 are 93 and 94% similar, respectively, to a clone from estuarine sediments (AF374340) (12). Clone E8, despite its location on the cluster III phylogenetic tree (Fig. 4), is most closely related (95%) to the clone from the lower, reduced portion of a marine cyanobacterial mat (79). Clone A44 is 90% similar to a clone from roots of the wild rice Oryza officinalis (19), and clone E12 is only 87% similar to another estuarine sediments clone (AF329999) (12). The phylogenetic group represented by E12 contains NifH sequences from only marker 33 in 1999, when the diffuse vent reached a maximum temperature of 78°C. Clone C19 is 92% similar to an oligotrophic tropical seagrass bed clone (3) and 85% similar to Desulfovibrio gigas. The phylogenetic group represented by clones E44 and E63 does not group with any other sequences, although it lies within cluster III on the phylogenetic tree containing all 77 unique NifH sequences (data not shown). This group is most closely related (85 to 86%) to the clone from the lower, reduced portion of a marine cyanobacterial mat (79) and is 82 to 83% similar to Clostridium pasteurianum nifH4. The deeply branching group represented by clones E44 and E63 is comprised only of NifH sequences from marker 33 in 1999.
FIG. 4.
Maximum-likelihood phylogenetic tree of cluster III NifH sequences, which include NifH sequences from diverse anaerobic bacteria like sulfate reducers and clostridia. GenBank accession numbers in parentheses follow the isolate or clone name. Hydrothermal vent NifH sequences from this study are in bold, followed by the number of additional NifH sequences from this study ≥97% similar to it in parentheses. The letter prefix of each clone name denotes the sample from which the clone was sequenced (A, marker 33, Axial Volcano, 2000; B, deep seawater; C, diffuse vent near Puffer, Endeavour Segment, 2000; and E, marker 33, Axial Volcano, 1999). Quartet puzzling support values are shown at each internal branch. The scale indicates the number of amino acid substitutions per site, and the tree is outgroup rooted with K. pneumoniae.
NifH cluster II.
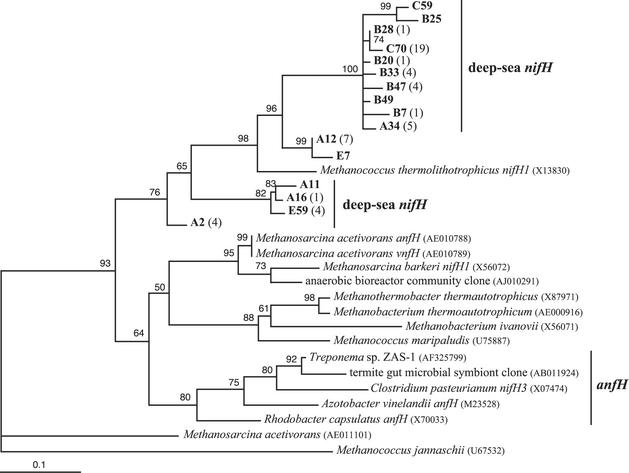
Sixty-seven deep-sea and hydrothermal vent NifH sequences belong to cluster II, which includes methanogens and alternative nitrogenases (anfH, vnfH). Representatives of all four cluster II deep-sea and hydrothermal vent phylogenetic groups (Table 2) are shown in the cluster II phylogenetic tree (Fig. 5). The largest and most diverse phylogenetic group (represented by clones C59, B25, B28, C70, B20, B33, B47, B49, B7, and A34) forms a distinct clade on the cluster II phylogenetic tree. The representative clones from this group are 79 to 83% similar to M. thermolithotrophicus nifH1 (66), which is the copy of nifH that is expressed during nitrogen fixation (67). This phylogenetic group contains 19 out of the 23 deep-seawater NifH clones. The phylogenetic group represented by clones A12 and E7 only contain NifH sequences from marker 33 (Table 2) and are more closely related (87 to 89%) to M. thermolithotrophicus nifH1. Clones A11, A16, and E59 are 77, 78, and 79% similar, respectively, to M. thermolithotrophicus nifH1 and the anaerobic bioreactor community clone shown in Fig. 5 (F. von Wintzingerode, B. Selent, W. Hegemann, and U. B. Goebel, submitted for publication). The deeply branching phylogenetic group represented by clone A2 is 83% similar to M. thermolithotrophicus nifH1.
FIG. 5.
Maximum-likelihood phylogenetic tree of cluster II NifH sequences, which include NifH from methanogens and bacterial AnfH. GenBank accession numbers in parentheses follow the isolate or clone name. Deep-sea and hydrothermal vent NifH sequences from this study are in bold, followed by the number of additional NifH sequences from this study ≥97% similar to it in parentheses. The letter prefix of each clone name denotes the sample from which the clone was sequenced (A, marker 33, Axial Volcano, 2000; B, deep seawater; C, diffuse vent near Puffer, Endeavour segment, 2000; and E, marker 33, Axial Volcano, 1999). Quartet puzzling support values are shown at each internal branch. The scale indicates the number of amino acid substitutions per site, and the tree is outgroup rooted with M. jannaschii.
NifH cluster IV.
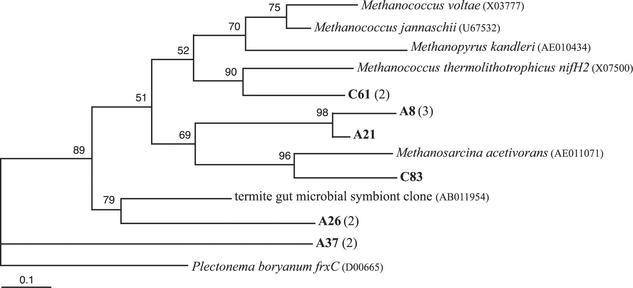
Fifteen hydrothermal vent NifH sequences fall within cluster IV, which includes divergent methanogens, and are shown in Fig. 6. Clones C61, A8, and A21 are 67, 52, and 55% similar, respectively, to M. thermolithotrophicus nifH2. Clone C61 contains an 11-amino-acid insertion at position 78 (K. pneumoniae numbering), as does M. thermolithotrophicus nifH2. Clones A8 and A21 contain a 16-amino-acid insertion at position 78 on NifH (K. pneumoniae numbering). Clone C83 is 69% similar to one of the nifH genes from Methanosarcina acetivorans, an acetate-utilizing methanogen with two nifH genes, an anfH gene, and a vnfH gene (22). Clone A26 is 60% similar to the termite gut symbiont clone shown in Fig. 6, and clone A37 is only 43% similar to frxC of both Methanopyrus kandleri and Plectonema boryanum, a dinitrogenase-like gene involved in the light-independent reduction of protochlorophyllide.
FIG. 6.
Maximum-likelihood phylogenetic tree of cluster IV NifH sequences, which include divergent NifH proteins from methanogens. GenBank accession numbers in parentheses follow the isolate or clone name. Deep-sea and hydrothermal vent NifH sequences from this study are in bold, followed by the number of additional NifH sequences from this study that are ≥97% similar to it in parentheses. The letter prefix of each clone name denotes the sample from which the clone was sequenced (A, marker 33, Axial Volcano, 2000; B, deep seawater; C, diffuse vent near Puffer, Endeavour segment, 2000; and E, marker 33, Axial Volcano, 1999). Quartet puzzling support values are shown at each internal branch. The scale indicates the number of amino acid substitutions per site, and the tree is outgroup rooted with P. boryanum frxC, a dinitrogenase reductase-like protein involved in the light-independent reduction of protochlorophyllide.
DISCUSSION
This is the first description of the diverse community of potential nitrogen fixers that inhabit the subseafloor associated with hydrothermal vents, as well as of the unique nifH genes present in the deep sea. The greatest amount of nifH phylogenetic diversity was found in marker 33 in 1999, whereas the least amount of nifH diversity was found in the deep-seawater sample, according to the Shannon-Weaver diversity indices and coverage estimates (Fig. 2). In general, the distribution of nifH sequences within the clone libraries (Fig. 2) corresponds to the degree of mixing between seawater and hydrothermal fluid in each of the samples. Marker 33 in 2000 and the diffuse vent near Puffer have strong seawater components, based on their temperature and H2S concentration (Table 1), which is consistent with the high proportions of cluster II deep-sea nifH in the venting fluid (Fig. 2). Based on the slightly higher temperature and H2S concentration of the diffuse vent near Puffer compared to marker 33 in 2000, it has a stronger subseafloor component, which is reflected in the higher proportion of cluster III nifH genes in the clone library from the diffuse vent near Puffer (Fig. 2). The cluster III NifH clones from marker 33 in 1999, when the venting fluid reached a maximum temperature of 78°C, best reflect the potential nitrogen-fixing microorganisms in the hot, anaerobic subseafloor.
The presence of nifH genes in the background deep-seawater sample was unanticipated. The nonvent deep sea is not believed to be nitrogen limited, because there is abundant nitrate in cold, oxygenated deep seawater. A study of nifH genetic diversity in picoplankton samples from the euphotic zone of oligotrophic oceans, which is a nitrogen-limited environment, revealed the presence of cyanobacterial, α-, β-, and γ-proteobacterial NifH in cluster I (81). The deep-sea NifH sequences from this study belong to three phylogenetic groups within cluster II (Table 2), but since they are only distantly related (≤83% amino acid similarity) to M. thermolithotrophicus nifH1, it is difficult to assess their phylogenetic affiliation. Many microbial groups inhabit the deep sea, but the most dominant are pelagic crenarchaeota, within the domain Archaea (38). In a study that monitored the archaeal diversity at marker 33 for 3 years, the background deep-seawater sample contained mostly marine group I pelagic crenarchaeota and a few marine group II pelagic euryarchaeota (31). Group II, group III, and the newly described group IV pelagic euryarchaeota form clusters within the methanogen-halophile lineage of the Euryarchaeota and have been found in the deep sea (21, 44, 48, 71). However, these pelagic archaea have resisted cultivation, and it is not known whether they possess nifH.
Diazotrophy has not been reported in obligately halophilic archaea, and the genome sequence of Halobacterium sp. strain NRC-1 does not contain any nif genes, but it is possible that the deep-sea nifH cluster is from halophilic archaea. Methanogens have been detected in seawater particles (“marine snow”) which provide reduced microzones that the strict anaerobes can occupy in oxygenated seawater (47, 64, 74). Despite the divergence of the deep-sea NifH clusters from mesophilic methanogens like Methanosarcina barkeri and Methanococcus maripaludis (Fig. 5), it is possible that the deep-sea NifH sequences belong to methanogens. The previously mentioned study of archaeal diversity at marker 33 revealed a very high abundance of mesophilic methanogens in 1999 (31), whereas only 11% of the nifH clones from marker 33 in 1999 fell within cluster II (Fig. 2), suggesting that the deep-sea clusters are not methanogenic nifH. Archaeal diversity at marker 33 in 2000 showed primarily marine group I crenarchaeota, similar to the background deep-sea sample (31), and 76% of the nifH clones from marker 33 in 2000 fell within cluster II (Fig. 2), suggesting that these cluster II deep-sea NifH belong to uncultivated pelagic archaea. Furthermore, PCR amplification with universal archaeal primers on the same DNA extract from the background deep-seawater sample used in this study has revealed that 30 out of the 36 clones sequenced are marine group I crenarchaeaota and the remainder are marine group II euryarchaeota (M.P. Mehta, J. A. Huber, and J. A. Baross, unpublished data).
All of the cultured organisms in the cluster II phylogenetic tree (Fig. 5) have been shown to fix nitrogen (10, 40, 43), and the cluster II NifH sequences from this study contain no amino acid insertions or deletions. The four cysteine residues that are conserved in all nitrogenase iron proteins (17) occur in all of the deep-sea and hydrothermal vent NifH sequences from this study, with the exception of clone B47 and the four clones ≥97% similar to it, which have arginine substituted for the first cysteine residue. All NifH sequences from this study also have the conserved arginine residue that is the site for the ADP-ribosylation interaction, which provides a reversible enzymatic inactivation of the iron protein in response to environmental factors (51). Given this, it is likely that the deep-sea (and hydrothermal vent) nifH genes encode functional iron proteins, and culturing efforts directed at these pelagic microorganisms will be necessary in order to ascertain the source of the deep-sea nifH genes.
The diffuse fluid sample that showed the least amount of mixing with crustal seawater prior to venting was from marker 33 in 1999, when the fluid reached a maximum temperature of 78°C (Table 1). The majority of clones from that library (70%) fell within cluster III (Fig. 2), indicating that anaerobic bacteria dominated the fluid. Since the hydrothermal fluid from marker 33 in 1999 had a very short residence time in the shallow subseafloor (D.A. Butterfield, M. D. Lilley, J. Huber, K. K. Roe, R. W. Embley, and G. J. Massoth, submitted for publication), these anaerobic bacterial nifH genes probably originated from a hot, reduced, anaerobic subseafloor habitat. The deeply branching phylogenetic groups represented by clones E12, E44, and E63 (Fig. 4) contain only sequences from marker 33 in 1999 and may represent thermophilic or hyperthermophilic anaerobic bacteria. A study that monitored the bacterial diversity at marker 33 for 3 years found clones similar to Thermodesulfobacterium thermophilum, an anaerobic, thermophilic sulfate reducer, and Desulfurobacterium thermolithotrophum, an anaerobic thermophilic, sulfur-reducing bacterium (30). ɛ-Proteobacteria, which are microaerophilic, sulfur-metabolizing bacteria, are also dominant members of the bacterial community at hydrothermal vents (58), and at least two members of the ɛ-proteobacteria are able to fix nitrogen: “Candidatus Arcobacter sulfidicus” (76) and Arcobacter nitrofigilis (49). The observation that unrelated anaerobic bacteria such as sulfate reducers (δ-protebacteria), clostridia (gram-positive bacteria), and purple sulfur bacteria (γ-proteobacteria) have closely related nifH genes points to the possibility of lateral gene transfer having occurred between microorganisms occupying the same anaerobic niche. Cluster III also includes an archaeal nitrogenase gene, M. barkeri nifH2, which is most closely related to the gram-positive bacterium C. pasteurianum nifH1 (16) and may be an example of lateral gene transfer between two organisms that have not shared a common ancestor for over 3 billion years.
Molecular analyses of the nif genes have not been able to distinguish between the two main theories regarding the evolution of nitrogen fixation (27, 52). The first theory is that nif genes were present in the last common ancestor before the divergence between Archaea and Bacteria and that vertical descent coupled with strong functional constraints, followed by loss of the nif genes in certain lineages, can explain the current distribution of nif genes among the prokaryotes (13, 26, 80). Nitrogenase is widely regarded as an ancient enzyme, but whether or not it was used to fix nitrogen probably depended on the composition of the early earth's atmosphere. In a neutral atmosphere that contained dinitrogen but not ammonia, a primordial nitrogenase would have been necessary, whereas in a reducing atmosphere that contained free ammonia the enzyme might have served to detoxify cyanides or other toxic chemicals present in the primitive atmosphere (65). The second theory invokes relatively recent horizontal gene transfer to explain the sporadic distribution of nitrogen fixation among prokaryotes (60) and is supported by the presence of nif genes on plasmids of marine diazotrophs (8). The most recent molecular analysis of the nif genes concludes that the two theories may not be mutually exclusive but that the most parsimonious hypothesis to explain their results is that the nif genes were present in the last common ancestor (20). Assuming that nitrogenase is an ancient enzyme, its presence and remarkable diversity in hydrothermal vent environments, which have been postulated as sites for the origin of life (7), are especially exciting.
Based on phylogenetic models and geological records, the common ancestor to all extant organisms is envisioned to be hyperthermophilic, and earth's earliest organisms may have been similar to modern thermophiles from hydrothermal environments (5). However, nitrogenase genes are not found in any of the hyperthermophilic and thermophilic bacteria or archaea thus far sequenced, with the exception of methanogens. Euryarchaeotes like Thermoplasma acidophilum and Thermococcus sp. and sulfur-dependent crenarchaeotes like Desulfurococcus mobilis, Sulfolobus solfataricus, and Thermoproteus tenax did not hybridize with nifH DNA (56), and hydrothermal vent Thermococcales isolates did not amplify with the nifH primers used in this study (M. P. Mehta and J. A. Baross, unpublished data). Nitrogen fixation has been reported in members of all three orders of methanogens: the Methanomicrobiales, the Methanococcales, and the Methanobacteriales (43). The phylogenetic group represented by clones A12 and E7 in cluster II (Fig. 5) is comprised only of hydrothermal vent sequences and is more closely related to M. thermolithotrophicus nifH1 than the deep-sea NifH groups. It is possible that this group represents thermophilic methanogens from the subseafloor, which have been documented at marker 33 (31). Divergent nifH genes are also found in thermophilic methanogens that are unable to fix nitrogen, like Methanococcus jannaschii and Methanococcus voltae (see Fig. 6). Their divergent nifH genes could indicate that the methanogens have lost the ability to fix nitrogen due to many years of being cultured in nitrogen-replete media. However, the presence of cluster IV nifH genes in the hydrothermal vent samples (Table 2) as well as other environments, such as termite guts, suggests that methanogens in the environment possess nifH genes that have been subjected to mutation and may no longer encode functional nitrogenases. Given that none of the hydrothermal vent proteobacterial NifH (Fig. 3) were similar to oceanic nifH and that the background deep-seawater sample did not contain any cluster I NifH, it is probable that the proteobacterial NifH from marker 33 in 1999 and the diffuse vent near Puffer in 2000 are from the subseafloor microbial population.
The nitrogen cycle in the subseafloor associated with hydrothermal vents has yet to be described. However, based on the depletion of seawater nitrate at low temperatures in diffuse flow fluids and high microbial cell densities (Table 1; D. A. Butterfield, unpublished data), denitrification is likely to play an important role at vents. Denitrification, as well as autotrophic and heterotrophic nitrification (the aerobic oxidation of either NH4+ or NO2− to nitrate) have been documented at hydrothermal vents, suggesting that all pathways of the nitrogen cycle may be active in different locations within the vent ecosystem (33, 41, 50). The ability of subseafloor microorganisms to assimilate nitrate has not been investigated, but nitrate is the least preferred nitrogen source for marine heterotrophic bacteria (39). Given the low concentrations of nitrate in reduced fluids above 30°C, its assimilation may not be significant in the subseafloor, particularly in the hot, anaerobic zones.
The only differences noted in the nifH populations between the high-ammonia Puffer environment and the low-ammonia marker 33 vent were the presence of the phylogenetic group represented by clones A12 and E7 (Fig. 5), which is comprised of nine sequences from marker 33 and contains no sequences from the diffuse vent near Puffer or the background deep seawater (Table 2). This could be an example of nitrogen availability selecting for different nitrogen-fixing microorganisms in the nitrogen-poor environment. In other nifH genetic studies that reported concentrations of fixed nitrogen, there was no direct correlation between the availability of nitrogen and the population of nifH-possessing microorganisms present (1, 24), indicating that other factors may be responsible for the distribution of nifH genes in the environment.
In conclusion, we have provided the first evidence of nifH genes in diffuse hydrothermal vent fluid and deep seawater, and we have also demonstrated a significant difference between subseafloor and deep-sea nifH populations. In order to determine the importance of these nitrogen-fixing microorganisms in the nitrogen cycle of hydrothermal vent ecosystems, it will be necessary to quantify the rates of nitrogen fixation and study the expression of the nitrogenase genes in situ. Culturing efforts directed at pelagic archaeal diazotrophs and anaerobic subseafloor nitrogen fixers will also be important in more precisely identifying the microorganisms detected in this study.
Acknowledgments
We thank Bob Embley, John Delaney, the crews of the R/V Thompson, R/V Brown, R/V Atlantis, ROV ROPOS, and AUV Alvin for providing the opportunity to collect the samples used in this research, Sheryl Bolton for collecting the samples from Endeavour Segment, and Kevin Roe for performing chemical analyses. We also thank Julie Huber, Matt Schrenk, and Jonathan Kaye for their help as this research evolved.
This research was supported by Washington Sea Grant (NA76RG0119), the National Science Foundation (OCE 9816491), NSF IGERT (DGE-9870713), the NASA Astrobiology Institute through the Carnegie Geophysical Institute, the NOAA/PMEL Vents Program (PMEL contribution no. 2531), and the NOAA West Coast and Polar Undersea Research Center. This publication was also funded by the Joint Institute for the Study of the Atmosphere and Ocean (under NOAA cooperative agreement NA17RJ11232, contribution no. 953).
REFERENCES
- 1.Affourtit, J., J. P. Zehr, and H. W. Paerl. 2001. Distribution of nitrogen-fixing microorganisms along the Neuse River Estuary, North Carolina. Microb. Ecol. 41:114-123. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagwell, C. E., J. R. La Rocque, G. W. Smith, S. W. Polson, M. J. Friez, J. W. Longshore, and C. R. Lovell. 2002. Molecular diversity of diazotrophs in oligotrophic tropical seagrass bed communities. FEMS Microbiol. Ecol. 39:113-119. [DOI] [PubMed] [Google Scholar]
- 4.Baker, E. T., C. G. Fox, and J. P. Cowen. 1999. In situ observations of the onset of hydrothermal discharge during the 1998 submarine eruption of Axial Volcano, Juan de Fuca Ridge. Geophys. Res. Lett. 26:3445-3448. [Google Scholar]
- 5.Baross, J. A. 1998. Do the geological and geochemical records of the early earth support the prediction from global phylogenetic models of a thermophilic cenancestor?, p. 3-18. In J. Wiegel and M. W. W. Adams (ed.), Thermophiles: the keys to molecular evolution and the origin of life? Taylor and Francis Inc., Philadelphia, Pa.
- 6.Baross, J. A., and J. W. Deming. 1995. Growth at high temperatures: isolation and taxonomy, physiology, and ecology, p. 169-217. In D. M. Karl (ed.), The microbiology of deep-sea hydrothermal vents. CRC Press Inc., Boca, Raton, Fla.
- 7.Baross, J. A., and S. E. Hoffman. 1985. Submarine hydrothermal vents and associated gradient environments as sites for the origin and evolution of life. Origins Life 15:327-345. [Google Scholar]
- 8.Beeson, K. E., D. L. Erdner, C. E. Bagwell, C. R. Lovell, and P. A. Sobecky. 2002. Differentiation of plasmids in marine diazotroph assemblages determined by randomly amplified polymorphic DNA analysis. Microbiology 148:179-189. [DOI] [PubMed] [Google Scholar]
- 9.Belay, N., R. Sparling, and L. Daniels. 1984. Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature 312:286-288. [DOI] [PubMed] [Google Scholar]
- 10.Bishop, P. E., and R. Premakumar. 1992. Alternative nitrogen fixation systems, p. 736-762. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
- 11.Burgess, B. K., and D. J. Lowe. 1996. Mechanism of molybdenum nitrogenase. Chem. Rev. 96:2983-3011. [DOI] [PubMed] [Google Scholar]
- 12.Burns, J. A., J. P. Zehr, and D. G. Capone. 2002. Nitrogen-fixing phylotypes of Chesapeake Bay and Neuse River Estuary sediments. Microb. Ecol. 10:1007-1009. [DOI] [PubMed] [Google Scholar]
- 13.Burris, R. H. 1961. Discussion of evolution of biological nitrogen fixation, p. 173-177. In A. I. Oparin (ed.), Proceedings of the 5th International Congress of Biochemistry, vol. 3. Pergamon, Oxford, England.
- 14.Butterfield, D. A., I. R. Jonasson, G. J. Massoth, R. A. Feely, K. K. Roe, R. W. Embley, J. F. Holden, R. E. McDuff, M. D. Lilley, and J. R. Delaney. 1997. Seafloor eruptions and evolution of hydrothermal fluid chemistry. Phil. Trans. R. Soc. London A 355:369-386. [Google Scholar]
- 15.Charlou, J. L., J. P. Donval, E. Douville, P. Jean-Baptiste, J. Radford-Knoery, Y. Fouquet, A. Dapoigny, and M. Stievenard. 2000. Compared geochemical signatures and the evolution of Menez Gwen (37o50′N) and Lucky Strike (37o17′N) hydrothermal fluids, south of the Azores Triple Junction on the Mid-Atlantic Ridge. Chem. Geol. 171:49-75. [Google Scholar]
- 16.Chien, Y.-T., and S. H. Zinder. 1994. Cloning, DNA sequencing, and characterization of a nifD-homologous gene from the archaeon Methanosarcina barkeri 227 which resembles nifD1 from the eubacterium Clostridium pasteurianum. J. Bacteriol. 176:6590-6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dean, D. R., and M. R. Jacobson. 1992. Biochemical genetics of nitrogenase, p. 763-834. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
- 18.Edmond, J. M., C. Measures, B. Mangum, B. Grant, F. R. Sclater, R. Collier, A. Hudson, L. I. Gordon, and J. B. Corliss. 1979. On the formation of metal-rich deposits at ridge crests. Earth Planet. Sci. Lett. 46:19-30. [Google Scholar]
- 19.Engelhard, M., T. Hurek, and B. Reinhold-Hurek. 2000. Preferential occurence of diazotrophic endophytes, Azoarcus spp., in wild rice species and land races of Oryza sativa in comparison with modern races. Environ. Microbiol. 2:131-141. [DOI] [PubMed] [Google Scholar]
- 20.Fani, R., R. Gallo, and P. Lio. 2000. Molecular evolution of nitrogen fixation: the evolutionary history of the nifD, nifK, nifE, and nifN genes. J. Mol. Evol. 51:1-11. [DOI] [PubMed] [Google Scholar]
- 21.Fuhrman, J. A., and A. A. Davis. 1997. Widespread Archaea and novel Bacteria from the deep sea as shown by 16S rRNA gene sequences. Mar. Ecol. Prog. Ser. 150:275-285. [Google Scholar]
- 22.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Good, I. J. 1953. The population frequencies of species and the estimation of population parameters. Biometrika 40:237-262. [Google Scholar]
- 24.Guerinot, M. L., and R. R. Colwell. 1985. Enumeration, isolation, and characterization of N2-fixing bacteria from seawater. Appl. Environ. Microbiol. 50:350-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hall, T. A. 1999. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 26.Hennecke, H., K. Kaluza, B. Thony, M. Fuhrmann, W. Ludwig, and E. Stackebrandt. 1985. Concurrent evolution of nitrogenase genes and 16S rRNA in Rhizobium species and other nitrogen fixing bacteria. Arch. Microbiol. 142:342-348. [Google Scholar]
- 27.Hirsch, A. M., H. I. McKhann, A. Reddy, J. Liao, Y. Fang, and C. R. Marshall. 1995. Assessing horizontal transfer of nifHDK genes in eubacteria: nucleotide sequence of nifK from Frankia strain HFPCcI3. Mol. Biol. Evol. 12:16-27. [DOI] [PubMed] [Google Scholar]
- 28.Holden, J. F., M. Summit, and J. A. Baross. 1998. Thermophilic and hyperthermophilic microorganisms in 3-30oC hydrothermal fluids following a deep-sea volcanic eruption. FEMS Microbiol. Ecol. 25:33-41. [Google Scholar]
- 29.Huber, H., M. Thomm, H. Konig, G. Thies, and K. O. Stetter. 1982. Methanococcus thermolithotophicus, a novel thermophilic lithotrophic methanogen. Arch. Microbiol. 132:47-50. [Google Scholar]
- 30.Huber, J. A., D. A. Butterfield, and J. A. Baross. Bacterial diversity in a subseafloor habitat following a deep-sea volcanic eruption. FEMS Microbiol. Ecol., in press. [DOI] [PubMed]
- 31.Huber, J. A., D. A. Butterfield, and J. A. Baross. 2002. Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68:1585-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jannasch, H. W. 1995. Microbial interactions with hydrothermal fluids, p. 273-296. In S. E. Humphris, R. A. Zierenberg, L. S. Mullineaux, and R. E. Thomson (ed.), Seafloor hydrothermal systems: physical, chemical, biological, and geological interactions. American Geophysical Union, Washington, D.C.
- 33.Jannasch, H. W. 1983. Microbial processes at deep sea hydrothermal vents, p. 677-709. In P. A. Rona, K. Bostrom, L. Laubier, and K. L. Smith (ed.), Hydrothermal processes at seafloor spreading centers. Plenum Publishing Corp., New York, N.Y.
- 34.Johnson, K. S., J. J. Childress, R. R. Hessler, C. M. Sakamoto-Arnold, and C. L. Beehler. 1988. Chemical and biological interactions in the Rose Garden hydrothermal vent field, Galapagos spreading center. Deep-Sea Res. 35:1723-1744. [Google Scholar]
- 35.Karl, D. M. 1995. Ecology of free-living, hydrothermal vent microbial communities, p. 35-124. In D. M. Karl (ed.), The microbiology of deep-sea hydrothermal vents. CRC Press Inc., Boca Raton, Fla.
- 36.Karl, D. M., A. M. Brittain, and B. D. Tilbrook. 1989. Hydrothermal and microbial processes at Loihi Seamount, a mid-plate hot-spot volcano. Deep-Sea Res. 36:1655-1673. [Google Scholar]
- 37.Karl, D. M., G. T. Taylor, J. A. Novitsky, H. W. Jannasch, C. O. Wirsen, N. R. Pace, D. J. Lane, G. J. Olsen, and S. J. Giovannoni. 1988. A microbiological study of Guaymas Basin high temperature hydrothermal vents. Deep-Sea Res. 35:777-791. [Google Scholar]
- 38.Karner, M. B., E. F. DeLong, and D. M. Karl. 2001. Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409:507-510. [DOI] [PubMed] [Google Scholar]
- 39.Kirchman, D. L., and P. A. Wheeler. 1998. Uptake of ammonium and nitrate by heterotrophic bacteria and phytoplankton in the sub-Arctic Pacific. Deep-Sea Res. 45:347-365. [Google Scholar]
- 40.Lilburn, T. G., K. S. Kim, N. E. Ostrom, K. R. Byzek, J. R. Leadbetter, and J. A. Breznak. 2001. Nitrogen fixation by symbiotic and free-living spirochetes. Science 292:2495-2498. [DOI] [PubMed] [Google Scholar]
- 41.Lilley, M. D., J. A. Baross, and L. I. Gordon. 1984. Reduced gases and bacteria in hydrothermal fluids: the Galapagos Spreading Center and 21oN East Pacific Rise, p. 411-449. In P. A. Rona, K. Bostrom, L. Laubier, and K. L. Smith (ed.), Hydrothermal processes at seafloor spreading centers. Plenum Publishing Corp., New York, N.Y.
- 42.Lilley, M. D., D. A. Butterfield, E. J. Olson, J. E. Lupton, S. A. Macko, and R. E. McDuff. 1993. Anomalous CH4 and NH4+ concentrations at an unsedimented mid-ocean-ridge hydrothermal system. Nature 364:45-47. [Google Scholar]
- 43.Lobo, A. L., and S. H. Zinder. 1992. Nitrogen fixation by methanogenic bacteria, p. 191-211. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
- 44.Lopez-Garcia, P., D. Moreira, A. Lopez-Lopez, and F. Rodriguez-Valera. 2001. A novel haloarchaeal-related lineage is widely distributed in deep oceanic regions. Environ. Microbiol. 3:72-78. [DOI] [PubMed] [Google Scholar]
- 45.Lovell, C. R., M. J. Friez, J. W. Longshore, and C. E. Bagwell. 2001. Recovery and phylogenetic analysis of nifH sequences from diazotrophic bacteria associated with dead aboveground biomass of Spartina alterniflora. Appl. Environ. Microbiol. 67:5308-5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lovell, C. R., Y. M. Piceno, J. M. Quattro, and C. E. Bagwell. 2000. Molecular analysis of diazotroph diversity in the rhizosphere of the smooth cordgrass, Spartina alterniflora. Appl. Environ. Microbiol. 66:3814-3822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marty, D. G. 1993. Methanogenic bacteria in seawater. Limnol. Oceanogr. 38:452-456. [Google Scholar]
- 48.Massana, R., E. F. DeLong, and C. Pedros-Alio. 2000. A few cosmopolitan phylotypes dominate planktonic archaeal assemblages in widely different oceanic provinces. Appl. Environ. Microbiol. 66:1777-1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.McClung, C. R., D. G. Patriquin, and R. E. Davis. 1983. Campylobacter nitrofigilis sp. nov., a nitrogen-fixing bacterium associated with roots of Spartina alterniflora. Int. J. Syst. Bacteriol. 33:605-612. [Google Scholar]
- 50.Mevel, G., and D. Prieur. 1998. Thermophilic heterotrophic nitrifiers isolated from Mid-Atlantic Ridge deep-sea hydrothermal vents. Can. J. Microbiol. 44:723-733. [Google Scholar]
- 51.Normand, P., and J. Bousquet. 1989. Phylogeny of nitrogenase sequences in Frankia and other nitrogen-fixing microorganisms. J. Mol. Evol. 29:436-447. [DOI] [PubMed] [Google Scholar]
- 52.Normand, P., M. Gouy, B. Cournoyer, and P. Simonet. 1992. Nucleotide sequence of nifD from Frankia alni strain Arl 3: phylogenetic inferences. Mol. Biol. Evol. 9:495-506. [DOI] [PubMed] [Google Scholar]
- 53.Ohkuma, M., S. Noda, and T. Kudo. 1999. Phylogenetic diversity of nitrogen fixation genes in the symbiotic microbial community in the gut of diverse termites. Appl. Environ. Microbiol. 65:4926-4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Olson, J. B., R. W. Litaker, and H. W. Paerl. 1999. Ubiquity of heterotrophic diazotrophs in marine microbial mats. Aquat. Microb. Ecol. 19:29-36. [Google Scholar]
- 55.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 56.Possot, O., M. Henry, and L. Sibold. 1986. Distribution of DNA sequences homologous to nifH among archaebacteria. FEMS Microbiol. Lett. 34:173-177. [Google Scholar]
- 57.Rau, G. H. 1981. Low 15N/14N in hydrothermal vent animals: ecological implications. Nature 289:484-485. [Google Scholar]
- 58.Reysenbach, A. L., K. Longnecker, and J. Kirshtein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rosch, C., A. Mergel, and H. Bothe. 2002. Biodiversity of denitrifying and dinitrogen-fixing bacteria in an acid forest soil. Appl. Environ. Microbiol. 68:3818-3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruvkun, G. B., and F. M. Ausubel. 1980. Interspecies homology of nitrogenase genes. Proc. Natl. Acad. Sci. USA 77:191-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schmidt, H. A., K. Strimmer, M. Vingron, and A. von Haeseler. 2002. TREE-PUZZLE: maximum likelihood phylogenetic analysis using quartets and parallel computing. Bioinformatics 18:502-504. [DOI] [PubMed] [Google Scholar]
- 62.Shannon, C. E., and W. Weaver. 1949. The mathematical theory of communication. University of Illinois Press, Urbana.
- 63.Sibold, L., D. Pariot, L. Bhatnagar, M. Henriquet, and J.-P. Aubert. 1985. Hybridization of DNA from methanogenic bacteria with nitrogenase structural genes (nifHDK). Mol. Gen. Genet. 200:40-46. [Google Scholar]
- 64.Sieburth, J. M. 1987. Contrary habitats for redox-specific processes: methanogenesis in oxic waters and oxidation in anoxic waters, p. 11-38. In M. A. Sleigh (ed.), Microbes in the sea. Ellis Horwood Limited, Chicester, England.
- 65.Silver, V. S., and J. R. Postgate. 1973. Evolution of asymbiotic nitrogen fixation. J. Theor. Biol. 40:1-10. [DOI] [PubMed] [Google Scholar]
- 66.Souillard, N., M. Magot, O. Possot, and L. Sibold. 1988. Nucleotide sequence of regions homologous to nifH (nitrogen Fe protein) from the nitrogen-fixing archaebacteria Methanococcus thermolithotrophicus and Methanobacterium ivanovii: evolutionary implications. J. Mol. Evol. 27:65-76. [DOI] [PubMed] [Google Scholar]
- 67.Souillard, N., and L. Sibold. 1989. Primary structure, fuctional organization and expression of nitrogenase structural genes of the thermophilic archaebacterium Methanococcus thermolithotrophicus. Mol. Microbiol. 3:541-551. [DOI] [PubMed] [Google Scholar]
- 68.Steppe, T., J. L. Pinckney, J. Dyble, and H. W. Paerl. 2001. Diazotrophy in modern marine Bahamian stromatolites. Microb. Ecol. 41:36-44. [DOI] [PubMed] [Google Scholar]
- 69.Strimmer, K., and A. von Haeseler. 1996. Quartet puzzling: a quartet maximum-likelihood method for reconstructing tree topologies. Mol. Biol. Evol. 13:964-969. [Google Scholar]
- 70.Summit, M. 2000. Ecology, physiology, and phylogeny of subseafloor thermophiles from mid-ocean ridge environments. Ph.D. thesis. University of Washington, Seattle.
- 71.Suzuki, M. T., and E. F. DeLong. 2002. Marine prokaryote diversity, p. 209-234. In J. T. Staley and A.-L. Reysenbach (ed.), Biodiversity of microbial life. Wiley-Liss, Inc., New York, N.Y.
- 72.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ueda, T., Y. Suga, N. Yahiro, and T. Matsuguchi. 1995. Remarkable N2-fixing bacterial diverstiy detected in rice roots by molecular evolutionary analysis of nifH gene sequences. J. Bacteriol. 177:1414-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.van der Maarel, M. J. E. C., W. Sprenger, R. Haanstra, and L. J. Forney. 1999. Detection of methanogenic archaea in seawater particles and the digestive tract of a marine fish species. FEMS Microbiol. Lett. 173:189-194. [DOI] [PubMed] [Google Scholar]
- 75.Whelan, S., and N. Goldman. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18:691-699. [DOI] [PubMed] [Google Scholar]
- 76.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Young, J. P. W. 1992. Phylogenetic classification of nitrogen-fixing organisms, p. 43-86. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, N.Y.
- 78.Zehr, J. P., and D. G. Capone. 1996. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb. Ecol. 32:263-281. [DOI] [PubMed] [Google Scholar]
- 79.Zehr, J. P., M. T. Mellon, S. Braun, W. Litaker, T. Steppe, and H. W. Paerl. 1995. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 61:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zehr, J. P., M. T. Mellon, and W. D. Hiorns. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology 143:1443-1450. [DOI] [PubMed] [Google Scholar]
- 81.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]