Abstract
We have developed an integrative transformation system for metabolic engineering of the tetraacetyl phytosphingosine (TAPS)-secreting yeast Pichia ciferrii. The system uses (i) a mutagenized ribosomal protein L41 gene of P. ciferrii as a dominant selection marker that confer resistance to the antibiotic cycloheximide and (ii) a ribosomal DNA (rDNA) fragment of P. ciferrii as a target for multicopy gene integration into the chromosome. A locus within the nontranscribed region located between 5S and 26S rDNAs was selected as the integration site. A maximum frequency of integrative transformation of approximately 1,350 transformants/μg of DNA was observed. To improve the de novo synthesis of sphingolipid, the LCB2 gene, encoding a subunit of serine palmitoyltransferase, which catalyzes the first committed step of sphingolipid synthesis, was cloned from P. ciferrii and overexpressed under the control of the P. ciferrii glyceraldehyde-3-phosphate dehydrogenase promoter. After transformation of an LCB2 gene expression cassette, several transformants that contained approximately five to seven copies of transforming DNA in the chromosome and exhibited about 50-fold increase in LCB2 mRNA relative to the wild type were identified. These transformants were observed to produce approximately two times more TAPS than the wild type.
Sphingolipids are integral membrane components found in mammalian cells. They are also distributed widely in plants and microbes (14). Sphingolipids constitute a diverse group of complex membrane lipids, all containing a long-chain amino acid alcohol, usually of 18 carbons, connected by an amide linkage to a fatty acid to form ceramide. Sphingolipids have structural functions in maintaining cell membrane integrity, and they act as anchors for proteins. Metabolites of sphingolipids, such as ceramide, sphingosine, and sphingosine 1-phosphate, are involved as bioeffector molecules and second messengers in cell growth, differentiation, cell senescence, apoptosis, and stress responses (for reviews, see references 9, 29, and 20). Ceramide is a major end product of differentiation through the keratinization process in the mammalian epidermis and is secreted into the extracellular space to form a mantle surrounding individual keratinized cells (19). Extracellular ceramide is a major part of the permeability barrier and the water reservoir of the skin (11). Therefore, ceramide has attracted great attention as a specialty ingredient for moisture retention and protection of the skin in the cosmetics industry. Extraction of ceramides from natural sources, however, is difficult, and in some cases the resulting product is not acceptable for cosmetic use. Chemically synthesized ceramides are also available as a mixture of stereoisomers that must be separated prior to use. Biological production of sphingoid base, such as sphinganine and phytosphingosine, can provide the base materials for conversion to various ceramides. This sphingoid base can be obtained from several yeasts and easily converted to ceramides by N acylation with fatty acids.
The yeast Pichia ciferrii produces large quantities of sphingoid base that are secreted into the extracellular medium as acetylated bases. These secreted bases consist mostly of tetraacetyl phytosphingosine (TAPS), which is a precursor of sphingolipid (1). Phytosphingosine that is obtained by deacetylation of TAPS produced from P. ciferrii is better than chemically synthesized phytosphingosine because the phytosphingosine obtained by yeast fermentation is a d-ribo isomer that also occurs in human skin. Biochemical synthesis of phytosphingosine in the yeast Saccharomyces cerevisiae begins with the condensation of l-serine and palmitoyl coenzyme A to yield a C18 carbon unit, 3-ketosphinganine (3-ketodihydrosphingosine). This essentially irreversible reaction is catalyzed by the serine palmitoyltransferase (SPT) (22) encoded by the LCB1 (2, 7) and LCB2 (24, 25) genes. 3-Ketosphinganine is converted to sphinganine (dihydrosphingosine), which is further converted to phytosphingosine by sphinganine hydroxylase (encoded by SUR2/SYR2). For improvement of the yield of TAPS in P. ciferrii, metabolic engineering of these synthetic steps in P. ciferrii may prove to be useful.
Currently, genetic manipulation and transformation methods do not exist for P. ciferrii. Thus, we first needed to develop a transformation system for the yeast. Due to the polyploidic nature of P. ciferrii, auxotrophic mutants are not easily obtainable. Thus, a dominant selection marker system is necessary. As a dominant selection marker, a component of the large ribosomal subunit, RPL41 (encoded by L41), that confers resistance against cycloheximide after changing amino acid residue 56 from Pro to Gln has been used in several yeasts (3, 16, 18). Using the site-directed mutagenized RPL41 gene of P. ciferrii (PcL41) as a selection marker and the ribosomal DNA (rDNA) of P. ciferrii as a locus for multiple gene integration, we have succeeded in establishing a useful transformation system for this yeast. We have also cloned two genes, P. ciferrii LCB2 (PcLCB2) and PcGAPDH, which encode a subunit of SPT and glyceraldehyde-3-phosphate dehydrogenase (GAPDH), respectively, of P. ciferrii. TAPS production in recombinant P. ciferrii is improved, with an increase in the gene copy number and expression level of PcLCB2 under the control of the GAPDH promoter.
MATERIALS AND METHODS
Strains and media.
P. ciferrii (ATCC 14091 and ATCC 32292) and phytosphingosine-overproducing P. ciferrii KFCC-10937 derived from P. ciferrii (ATCC 14091) were provided by C. S. Park (Doosan Corporation Biotech BU, Yong im, Korea). S. cerevisiae L3262 (MATα ura3-52 leu2-3,112 his4-34) was used as a template for PCR cloning. P. ciferrii and S. cerevisiae were grown on YEPD medium (1% yeast extract, 2% Bacto Peptone, and 2% glucose). P. ciferrii transformants were selected in YEPD medium containing 20 μg of cycloheximide per ml. Escherichia coli DH5α [endA1 recA1 hsdR17 supE44 thi-1 gyrA96 re1A1 lacU169 (φ80dlacΔ(lacZ)M15)] was used for general recombinant techniques.
Plasmids.
The plasmid pBluescript KS(+) (Stratagene) was used for DNA cloning. To construct the PcLCB2 expression vector, prACL2, the ScaI/AflIII 3-kb fragment of PcLCB2 was obtained by digestion of pL2SA with HindIII/Klenow and BamHI. The resulting 3-kb fragment containing the PcLCB2 gene was ligated to Eco47III/BamHI-digested prACH1.9. Plasmid prACL2 contains the P. ciferrii 0.6-kb rDNA fragment as an integration target sequence, the PcL41(P56Q) gene as the dominant selection marker, and the PcLCB2 gene with its own promoter. To replace the original promoter of the PcLCB2 gene with the PcGAPDH promoter, 600 bp of the PcGAPDH promoter was amplified by PCR with the restriction site-containing GHF primer (5′-GAT ATC TAC ATA CA ATT GAC CCA TAG-3′) and the GHR primer (5′-GGA TCC TTA ATT ATT TGT TTG TTT-3′) from pGH1.6, which contains the KpnI/HindIII 1.6-kb fragment with the PcGAPDH open reading frame (ORF) and a promoter. The amplified PcGAPDH promoter was digested with EcoRV/BamHI and then cloned into EcoRV/BamHI-digested pBluescript to construct pGH0.6. Plasmid pL2B2.3, containing the 2.3-kb BamHI fragment of the PcLCB2 gene free of its own promoter, was digested with BamHI. The resulting 2.3-kb fragment was ligated to BamHI-digested pGH0.6 to construct pGAL2, which contains the PcLCB2 gene under the control of the PcGAPDH promoter. The EcoRV/XbaI-digested 2.9-kb fragment of pGAL2 was ligated to Eco47III/XbaI-digested prACH1.9 to construct prACGL2. When the host cell was transformed with either plasmid prACL2 or prACGL2 by single-crossover integration, the resulting transformed cells carried genes from P. ciferrii and other undesirable regions of the bacterial plasmid. To avoid integration of this heterogeneous sequence, plasmids prHECL2 and prHECGL2, which both contain another rDNA fragment (800 bp) downstream of the PcLCB2 gene of prACL2 and prACGL2, were also constructed. Plasmids prACL2 and prACGL2 were linearized with ApaI, and prHECL2 and prHECGL2 were digested with ApaI/SmaI before transformation.
Nucleic acid manipulation and DNA sequencing and analysis.
General DNA manipulations were performed as described by Sambrook et al. (28). The DNA fragments required for subcloning were gel purified with a gel extraction kit (Viogene). Total yeast DNA was isolated according to the method of Holm et al. (10). DNA sequencing was performed with an automatic DNA sequencer (model 373A; Applied Biosystems). Comparison of deduced amino acid sequences was performed by using a BLAST nucleotide sequence similarity search (National Center for Biotechnology Information). General DNA sequence analysis was performed with the DNASTAR program. Oligonucleotides for PCR were obtained from Genotech (Taejon, Korea).
Site-directed mutagenesis.
Site-directed mutagenesis was performed by overlap extension PCR (13) with Pfu DNA polymerase (Stratagene) and the complementary mutagenic primers CHf (5′-GGT CAA ACC AAA CAA GTT TTC-3′), CHr (5′-ATG GAA AAC TTG TTT GGT TTG ACC-3′), T7 (5′-GTA ATA CGA CTC ACT ATA GGG C-3′), and T3 (5′-AAT TAA CCC TCA CTA AAG GG-3′) (mutated codons are underlined.). The 1.9-kb EcoRI fragment containing the PcL41 gene was cloned into the EcoRI site of pBluescript (Stratagene) to construct pCYH1.9. The first PCR was performed with the CHf-T7 and CHr-T3 primers as follows: 94°C for 3 min; 20 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min; and then 72°C for 5 min. The PCR products were purified with a gel extraction kit (Viogene), and an equal amount of the first PCR product was annealed by the second PCR. The second-round PCR was performed with the T7 and T3 primers as follows: 94°C for 3 min; 25 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 2 min; and then 72°C for 5 min.
Transformation of P. ciferrii and E. coli.
The transformation method described by Faber et al. (6) and Kondo et al. (18) was applied to P. ciferrii with a slight modification. Cells of P. ciferrii KFCC-10937 grown in YEPD medium to an optical density at 600 nm value of 1 to 1.5 were collected and resuspended in 0.1 culture volume of 50 mM phosphate buffer (pH 7.5) to which 25 mM dithiothreitol was added. The cells were incubated at 37°C for 15 min, washed twice with a culture volume of ice-cold stabilization solution (270 mM sucrose, 10 mM Tris-HCl [pH 7.5], 1 mM MgCl2), and suspended in 0.01 culture volume of the stabilization solution. Fifty microliters of the resulting cell suspension was mixed with 0.1 μg of plasmid DNA. The mixture was allowed to stand on ice for 10 min, and then the solution was transferred to a 0.2-mm electroporation cuvette (Bio-Rad). Electroporation was performed with a Gene-Pulser II (Bio-Rad) at 500 V, 50 μF, and 700 Ω. After electroporation, the cells were resuspended in 0.5 ml of the stabilization solution and transferred to culture tubes containing 2 ml of YEPD medium. After 6 to 12 h of incubation at 30°C, the culture broth was plated on solid YEPD medium containing 20 μg of cycloheximide per ml. E. coli was transformed by the simple and efficient (SEM) method (12).
Southern blot analysis.
Integration patterns and the copy number of the transforming DNA were analyzed by Southern hybridization (28). Total chromosomal DNA was isolated and digested with appropriate restriction endonucleases. After size fractionation by gel electrophoresis, DNA was transferred onto a nylon membrane (Schleicher & Schuell, Dassel, Germany) by the capillary transfer method. Nonradioactive labeling of probe DNA was performed with a digoxigenin (DIG) DNA labeling and detection kit (Roche, Mannheim, Germany). Hybridization was performed at 42°C in a hybridization oven (Hybaid) with a hybridization solution (5× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% [wt/vol] N-lauroylsarcosine, 0.02% [wt/vol] sodium dodecyl sulfate, 1% [wt/vol] blocking reagent, and 30 to 50% [vol/vol] formamide) as recommended by the manufacturer. Colorimetric detection of the membrane was performed by reaction of anti-DIG-alkaline phosphatase (Roche).
Northern blot analysis.
The expression patterns of the transformed P. ciferrii LCB2 gene were analyzed by Northern hybridization (28). Total RNA was isolated by the hot-phenol extraction method (5) and fractionated on a 1% formaldehyde-agarose gel, followed by transfer to a nylon membrane (Schleicher & Schuell) by the capillary transfer method. Nonradioactive labeling of probe RNA was performed with a DIG RNA labeling and detection kit (Roche). Hybridization was performed at 68°C in a hybridization oven (Hybaid) with a hybridization solution (5× SSC, 0.1% [wt/vol] N-lauroylsarcosine, 0.02% [wt/vol] SDS, 2% [wt/vol] blocking reagent, and 50% [vol/vol] formamide) as recommended by the manufacturer.
TAPS analysis.
The sphingoid base analysis method described by McNabb et al. (21) was used for analysis of TAPS, with a slightly different mobile-phase system. Cells were grown in YEPD broth medium to the late exponential growth phase at 30°C. Lipids were extracted from a 0.2-ml cell culture with 0.8 ml of chloroform-methanol (2:1, vol/vol) for 15 min at room temperature. Lipid extracts were collected after removal of cell debris by centrifugation. The extracts were dried in vacuo and resuspended in 0.2 ml of high-pressure liquid chromatography (HPLC)-grade chloroform-methanol (9:1, vol/vol). The HPLC system (Agilent 1100 series) consisted of a 3-mm silica column (4.6 by 150 mm) (YMC Silica Pak), a guard column of the same material (4.6 by 25 mm), and a mobile phase of n-chlorohexane-chloroform-methanol (76:20:4, vol/vol). The flow rate was 1 ml/min, and the column was eluted at 25°C. A Eurosep DDL31 evaporative light-scattering detector was used with a nitrogen gas pressure of 1.5 bars, a nebulization temperature of 50°C, an evaporation temperature of 60°C, and a photomultiplier gain setting of 400.
Nucleotide sequence accession numbers.
The DNA sequences of the P. ciferrii ribosomal protein L41, LCB2, and GAPDH genes have been submitted to GenBank under accession numbers AF053457, AF053456, and AF053300, respectively.
RESULTS
Cloning of the ribosomal protein L41 gene (PcL41) and the rRNA gene of P. ciferrii.
Since there is no auxotrophic marker available due to the diploid status of the production strain, P. ciferrii KFCC-10937, we attempted to develop a dominant selection system by using a ribosomal protein L41 (RPL41) gene. P. ciferrii was sensitive to a low concentration of cycloheximide, suggesting that Pro is present as amino acid 56 of RPL41. Two degenerate primers CYH1 (5′-CGC GTA GTT AAY GTN CCN AAR AC-3′ and CYH4 (5-′GCC TGG CCY TTY TGY TTY TTN TC-3′) were used to obtain a genomic DNA fragment coding for a conserved domain within RPL41 of P. ciferrii (PcL41) by using the method of Kondo et al. (18). PCR produced a 300-bp product containing a portion of PcL41. After labeling with DIG, this partial PcL41 was used as a probe to clone a full-length genomic copy containing PcL41.
Southern blots of several restriction enzyme-digested genomic DNAs showed a strong signal from a 1.9-kb EcoRI fragment (data not shown). To clone this genomic fragment, EcoRI-digested P. ciferrii chromosomal DNA fragments of around 1.9 kb were gel eluted and ligated into the EcoRI site of plasmid pBluescript KS(+). From this minilibrary, positive clones hybridizing with the PCR product were isolated and sequenced. Nucleotide sequence analysis revealed an ORF of 737 nucleotides containing a 419-bp intron located just downstream of the initiation codon, which is a common intron pattern reported for other genes encoding the L41 protein (3, 4, 15). The six nucleotides of the 5′ splice site and the three nucleotides of the 3′ splice site of the intron were conserved and were similar to the consensus sequence elements GTPuNGT and PyAG, respectively. The conserved internal sequence element TACTAACA was also found 18 nucleotides upstream of the 3′ end of the intron (33).
The deduced amino acid sequence showed approximately 90% or more sequence similarity with other yeast L41 protein sequences. As expected from the sensitivity of P. ciferrii to cycloheximide, the amino acid 56 of the L41 protein was identified as proline. To change the codon for proline at the position 56 to one for glutamine, the 1.9-kb EcoRI fragment containing the L41-coding region was subjected to site-directed mutagenesis as described in Materials and Methods.
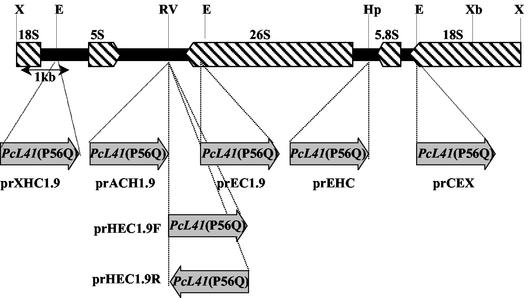
We cloned the P. ciferrii rRNA gene, which is known to be present in up to several hundred copies on the chromosome in yeast, for use as a target for multicopy plasmid integration. A 9-kb XhoI fragment containing an rDNA repeating unit of P. ciferrii was cloned from genomic DNA by using the DIG-labeled PCR product obtained with oligonucleotide primers complementary to the reported partial nucleotide sequence of P. ciferrii rRNA (34). The partial nucleotide sequence of the rRNA gene fragment was determined, and the locations and orientations of the 5S, 26S, 5.8S, and 18S rRNA genes were determined (Fig. 1).
FIG. 1.
Effects of the orientation of L41(P56Q) gene transcription and the integration site of rDNA on transformation efficiency. The upper part shows restriction maps of the rDNA repeating unit. The rRNA genes are indicated by hatched arrows. The insertion sites of the PcL41(P56Q) gene are indicated by dotted lines, and the gray arrows indicate the orientation of PcL41(P56Q) gene transcription. Restriction site abbreviations: E, EcoRI; Hp, HpaI; RV, EcoRV; X, XhoI; Xb, XbaI.
Optimization of transformation efficiency.
To test the effect of the integration site and transcriptional orientation of the selection marker gene on the transformation efficiency, the cloned rDNA was fragmented with various enzymes, subcloned into pBluescript, and then ligated to the EcoRI-digested 1.9-kb PcL41(P56Q) gene at the indicated restriction endonuclease sites to make various integration vectors (Fig. 1). All the plasmids were linearized by restriction endonucleases located within the rDNA or the multicloning site of pBluescript before transformation in order to integrate each DNA into the target site by single or double crossover. The results shown in Table 1 reveal that both the integration site and the transcriptional orientation of PcL41(P56Q) were critical for the overall transformation efficiency. The highest transformation efficiency was obtained when the nontranscribed region between the 5S and 26S rRNA structural genes was used as an integration target. The integration vector prHEC1.9F, containing the PcL41(P56Q) gene in the same transcriptional orientation as the 5S rRNA gene, exhibited a significantly higher transformation efficiency than plasmid prHEC1.9R, containing the PcL41(P56Q) gene in the transcriptional orientation opposite to that of the 5S rRNA gene.
TABLE 1.
Transformation efficiency of P. ciferrii
| Plasmid | Crossover | No. of transformants/μg of DNAa |
|---|---|---|
| prEHC | Single | 250 |
| prCEX | Single | 194 |
| prACH1.9 | Single | 1,174 |
| prXHC1.9 | Single | 980 |
| prEC1.9 | Single | 760 |
| prHEC1.9F | Double | 1,260 |
| prHEC1.9R | Double | 54 |
P. ciferrii was transformed with 0.1 μg of linearized plasmid containing different rDNA fragments as indicated in Fig. 1.
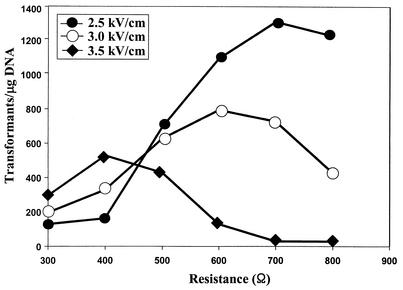
To determine the optimum conditions for electroporation, the effects of field strength and resistance on transformation efficiency were analyzed with linearized prHEC1.9F. The capacitance was set at 50 μF. Transformants were selected after a 4- to 5-day incubation at 30°C. The maximum efficiency was approximately 1,350 transformants per μg of linearized DNA at field strength of 2.5 kV/cm and an internal resistance of 700 Ω (Fig. 2).
FIG. 2.
Effects of field strength on the transformation efficiency of P. ciferrii. The cell suspension was mixed with 0.1 μg of ApaI/SmaI-digested prHEC1.9F, and an electric pulse was applied at a capacitance of 50 μF.
SPT gene of P. ciferrii (PcLCB2).
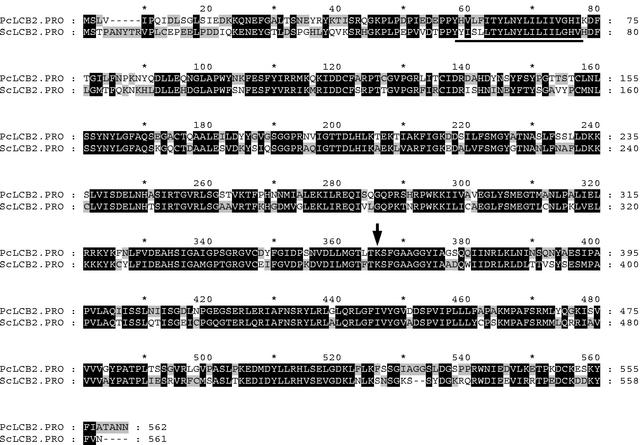
The SPT of S. cerevisiae consists of two subunits, LCB1 (2) and LCB2 (24). In order to clone the gene encoding SPT from P. ciferrii, the PCR-amplified LCB1 and LCB2 genes of S. cerevisiae were used as probes for Southern blotting of a P. ciferrii genomic DNA. Hybridization was performed at various stringencies controlled with different concentrations of formamide ranging from 20 to 50%. A discrete hybridization signal of a HindIII digest at 12 kb was obtained only when the LCB2 gene of S. cerevisiae was used as a probe under hybridization conditions with 30% formamide (data not shown). A minilibrary was constructed by using 12-kb HindIII fragments in pBluescript. After intensive Southern hybridization screening with the labeled LCB2 gene of S. cerevisiae, a positive clone was isolated. The insert fragment containing the PcLCB2 ORF was subcloned to a ScaI/AflIII fragment of 3-kb (pL2SA). The nucleotide sequence of the ScaI/AflIII 3-kb fragment was determined. The PcLCB2 gene comprised 1,688 bp and contained no introns. The putative amino acid sequence showed 62% similarity to S. cerevisiae LCB2. The hydrophobicity pattern (17) suggested a 25-amino-acid transmembrane segment in the N terminus (amino acids 55 to 79), sufficient to span the lipid bilayer of the endoplasmic reticulum membrane (Fig. 3). A putative pyridoxal-5-phosphate binding site near Lys361 contained an amino acid sequence identical to that of S. cerevisiae.
FIG. 3.
Alignment of the deduced amino acid sequences of LCB2 proteins. Potential transmembrane segments in the N-terminal regions are underlined. The conserved K residue that forms a Schiff base with the pyridoxal phosphate is indicated by an arrow.
Strong constitutive promoter from the GAPDH gene of P. ciferrii (PcGAPDH).
A strong and constitutive promoter was obtained for overexpression of PcLCB2 in P. ciferrii from the GAPDH gene. GAPDH is a constitutively expressed essential enzyme involved in glycolysis that converts glyceraldehyde-3-phosphate to 1,3-bisphosphoglycerate. Since the GAPDH promoter is little affected by the carbon source and is one of the strongest promoters, it is often used for overexpression of genes in yeasts (27, 31). PcGAPDH was also cloned by homology-based Southern hybridization cloning with the PCR-amplified GAPDH gene of S. cerevisiae as a probe. After screening of a P. ciferrii genomic minilibrary, pGH1.6 containing the PcGAPDH ORF and its promoter was obtained. DNA sequencing of the insert revealed an ORF consisting of 1,004 bp. The nucleotide and putative amino acid sequences showed 69.3% homology and 76.2% similarity to a GAPDH gene from S. cerevisiae, respectively.
Improvement of sphingoid base synthesis in P. ciferrii.
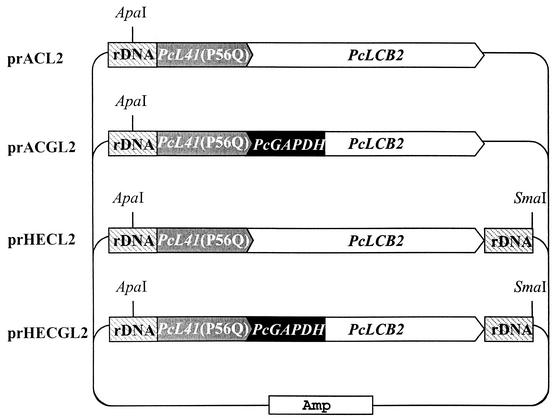
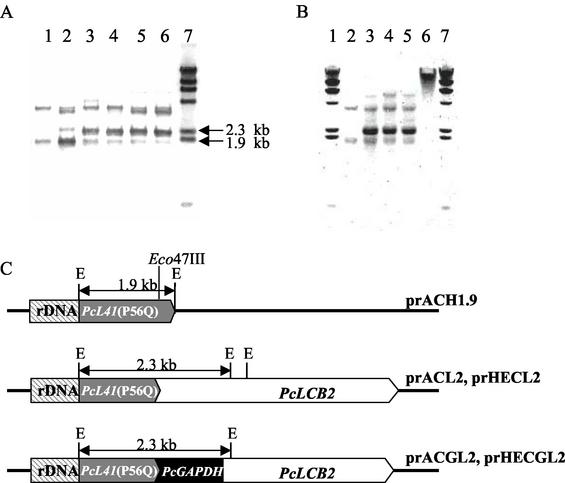
The effect of PcLCB2 overexpression on sphingoid base secretion was tested. PcLCB2 encodes a subunit of SPT that is a possible rate-limiting step for sphingoid base synthesis in P. ciferrii. Four integrative PcLCB2 expression vectors, prACL2, prACGL2, prHECL2, and prHECGL2, were constructed (Fig. 4). PcLCB2 expression was designed to be controlled by the original promoter in prACL2 and prHECL2 and by the PcGAPDH promoter in prACGL2 and prHECGL2. Linearized DNA fragments from each vector (ApaI for prACL2 and prACGL2 and ApaI/SmaI for prHECL2 and prHECGL2) were transformed into P. ciferrii KFCC-10937. Genomic DNAs from single cycloheximide-resistant colonies were prepared and analyzed for a pattern of integration into the chromosome with the PcL41 gene as a probe (Fig. 5A). Southern hybridization of genomic DNA digested with EcoRI resulted in two signals at 3.5 and 1.9 kb in an untransformed control, indicating that P. ciferrii contained two copies of the L41 gene, as these bands originated from the endogenous PcL41 gene (lane 1). An intense 2.3-kb band detected only in transformants indicated the presence of multiple copies of plasmid integrated into the rDNA locus of the P. ciferrii genome. Other bands probably are the result of integration of vectors at other loci (lanes 2 to 6). The copy number of the integrated plasmid was estimated to be five to seven copies per genome by densitometric comparison of the signal intensities of the 2.3-kb band and a 1.9-kb control on the blot in a scanning densitometer. The stability of the integrated vector in the prACGL2 transformant was analyzed after culturing cells in nonselective medium. The copy number did not decrease after prolonged cultivation in YEPD medium, indicating that the integrated plasmid DNA was stably maintained (Fig. 5B). Most plasmid integration appeared to occur by homologous recombination at the rDNA locus where the plasmid was linearized.
FIG. 4.
Physical maps of PcLCB2 gene expression vectors. Only restriction sites used for linearization of the vectors are indicated.
FIG. 5.
Southern analysis of transformants probed by the PcL41 gene. (A) Genomic DNA was prepared from wild-type cells (lane 1) and from prACH1.9 (lane 2), prACL2 (lane 3), prACGL2 (lane 4), prHECL2 (lane 5), and prHECGL2 (lane 6) transformants and digested with EcoRI. Lane 7 is DIG-labeled, λ/HindIII-digested DNA. (B). Genomic DNA was isolated from wild-type cells (lane 2) and from the prACGL2 transformant after 10 to 15 generations in YEPD medium (lane 3), 20 to 25 generations in YEPD medium (lane 4), and 40 to 50 generations in YEPD medium (lane 5) and digested with EcoRI. Lane 6 is undigested genomic DNA of the prACGL2 transformant. Lanes 1 and 7 are DIG-labeled λ/HindIII-digested DNA. (C) Schematic representations of the integration of linearized vectors. The 1.9-kb EcoRI fragment of prACH1.9 was used as probe.
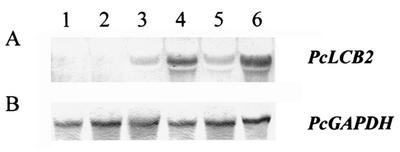
Expression of the PcLCB2 gene in transformants was examined at the transcriptional level by Northern hybridization (Fig. 6). Northern blot analysis of an untransformed control and transformants was performed with DIG-labeled PcLCB2 mRNA as a probe. Endogenous PcLCB2 message levels were low in the wild type and in transformants with prACH1.9, which has no additional PcLCB2 genes (lanes 1 and 2). In contrast, the level of PcLCB2 mRNA increased when multiple copies of PcLCB2 with its own promoter were introduced via prACL2 or prHECL2 (lanes 3 and 5). When the transcription of PcLCB2 was driven with a strong GAPDH promoter by using plasmids prACGL2 and prHECGL2, the level of PcLCB2 mRNA increased approximately 50-fold relative to that in wild-type cells (lanes 4 and 6).
FIG. 6.
Northern analysis of transformants. (A) Total RNAs prepared from wild-type cells (lane 1) and from prACH1.9 (lane 2), prACL2 (lane 3), prACGL2 (lane 4), prHECL2 (lane 5), and prHECGL2 (lane 6) transformants were hybridized with the PcLCB2 mRNA probe. (B) A PcGAPDH probe was used as a control.
Cells with overexpressed PcLCB2 were cultivated in YEPD medium to late exponential phase at 30°C in order to determine the yield of TAPS production. The amount of secreted TAPS was determined by HPLC. All cells transformed with plasmids containing PcLCB2 independent of the promoter produced approximately twofold more TAPS than the parent strain (Table 2).
TABLE 2.
Productivity of TAPS in P. ciferrii transformants
| Plasmid | Total yield of TAPS (mg/liter) |
|---|---|
| Wild type | 2.28 |
| prACH1.9 | 2.45 |
| prACL2 | 4.13 |
| prACGL2 | 4.35 |
| prHECL2 | 4.27 |
| prHECGL2 | 4.04 |
DISCUSSION
We have developed an integrative transformation and metabolic engineering tools for the development of TAPS-overproducing P. ciferrii strains. Genetic transformation systems developed for various yeast species are usually based on the use of auxotrophic mutants and selection marker genes that complement the mutations (30). However, many industrial yeasts, including P. ciferrii, are diploid or polyploid. Therefore, auxotrophic mutants are not easily obtainable. We have cloned the L41 gene, encoding a component of the ribosome large subunit, and used the clone as a dominant selectable marker that confers resistance against cycloheximide upon transformants of P. ciferrii after mutation of the L41 gene such that glutamine is substituted for the proline residue 56. For selection of transformants with multicopy plasmids, the L41 selection marker system has an advantage over an auxotrophic marker system in which a single copy is often enough for full complementation of an auxotrophic mutation. Due to endogenous L41(P56) that is sensitive to cycloheximide, multiple copies of L41(P56Q) that are resistant to cycloheximide are often necessary for development of resistance to cycloheximide (18). Thus, selection of a transformant with the L41(P56Q) marker usually results in selection of a multiple integrant.
We have also cloned an rDNA repeating unit from P. ciferrii for multicopy integration of a plasmid into the host genome by homologous recombination. The rDNA unit exists in multiple copies in eukaryotic chromosomes (26). The use of rDNA as a targeting locus is important for conferring cycloheximide resistance due to the fact that multiple copies of a mutated L41 gene are required for cycloheximide resistance. Southern blot analysis of the transformants showed that multiple integration of plasmid DNA occurred via homologous recombination at the rDNA locus targeted by linearization of a plasmid DNA within the rDNA sequence. No transformants were obtained with an intact plasmid, indicating that no stable autonomously replicating sequence activity existed in the rDNA fragment. The transformation efficiency was significantly correlated with the integration locus and the orientation of PcL41(P56Q) transcription. In general, the nontranscribed sequence between rRNA genes induced higher transformation efficiency than the rRNA coding sequence as an integration locus. The nontranscribed sequence between the 5S and 26S rRNA genes produced the highest transformation when the PcL41(P56Q) gene was integrated with the same orientation as 5S rRNA gene transcription.
SPT is the key enzyme in sphingolipid biosynthesis. It catalyzes the pyridoxal-5′-phosphate-dependent condensation of l-serine and palmitoyl coenzyme A to 3-ketosphinganine. In order to increase SPT activity, we tried to clone the LCB1 and LCB2 genes from P. ciferrii by Southern hybridization with the S. cerevisiae LCB1 and LCB2 genes, respectively, as probes. Despite intensive Southern hybridization under various hybridization conditions, we did not detect any LCB1 signals from the genomic DNA of P. ciferrii. LCB2 showed considerably higher homology and was easily obtained by homology-based Southern hybridization cloning. In the case of S. cerevisiae, overproduction of both the LCB1 and LCB2 genes is essential for an increase in SPT activity (24). However, in the case of human cells (HEK293), SPT activity was increased by overproduction of the mouse LCB2 gene alone (32). It has been reported that overexpression of the LCB2 gene induced rapid transformation of sphingoid base to sphingolipids because a high concentration of sphingoid base affecting many cellular functions is detrimental to cell viability (23). Therefore, an overexpressed PcLCB2 gene probably leads to an increase in the synthesis and secretion of excess TAPS from P. ciferrii. Therefore, we studied the effects of PcLCB2 overexpression on TAPS production.
The number of transcriptional messages was apparently increased, compared to that in untransformed cells, due to introduction of multiple copies of PcLCB2 with both its original promoter (prACL2 and prHECL2) and the PcGAPDH promoter (prACGL2 and prHECGL2). An especially strong constitutive PcGAPDH promoter can significantly increase the number of PcLCB2 messages, by up to 50-fold relative to that for the parental strain. The elevated level of PcLCB2 mRNA, however, did not result in a corresponding increase in TAPS production. In both cases, TAPS production increased approximately twofold compared with that in untransformed cells. Despite the much higher level of PcLCB2 mRNA transcribed under the control of the PcGAPDH promoter compared to the original promoter, there was no significant difference in the TAPS levels. The marked difference between the mRNA and protein levels of the LCB2 gene was also found in human cells transformed with the mouse LCB2 gene. This discrepancy is probably caused by cotranslational down-regulation or posttranslational degradation of the LCB2 protein (32).
For further increase of the TAPS level, it seems necessary to increase the SPT activity through the increase of the LCB2 protein level, which corresponds to its mRNA level. Although it is presently not clear whether the SPT of P. ciferrii is composed of two LCB1 and LCB2 subunits, cooverexpression of LCB1 might be helpful for the improvement of SPT activity in P. ciferrii, as found in S. cerevisiae. Several attempts to find the LCB1 homologue in P. ciferrii were unsuccessful, including complementation of the S. cerevisiae lcb1 mutant with a P. ciferrii cDNA expression library constructed under control of the S. cerevisiae GAPDH promoter in the YEp352 vector and PCR amplification with degenerate primers based on the other yeast and fungal LCB1 genes. Chinese hamster LCB2 protein was purified by affinity peptide chromatography techniques, which use a FLAG- and His6 peptide-tagged version of the hamster LCB1 protein (8). Therefore, it would be possible to purify the P. ciferrii LCB1 protein, if it exists, by using tagged P. ciferrii LCB2 protein. Another approach to improve the SPT activity through the increase of the LCB2 protein level might be by the cytosolic expression of LCB2, which is known as an endoplasmic reticulum membrane protein, after truncation of transmembrane domain. Cloning of the LCB1 gene and cytosolic overexpression of truncated LCB2 to improve TAPS production through the increase of SPT activity are in progress.
In summary, we have developed a transformation system for P. ciferrii that can provide for additional metabolic engineering in the pathway to synthesis of TAPS and for production of useful materials, such as other sphingolipids.
REFERENCES
- 1.Barenholz, Y., N. Gadot, E. Valk, and S. Gatt. 1973. Identification of the enzymatic lesions responsible for the accumulation of acetylated sphingosine bases in the yeast Hansenula ciferri. Biochim. Biophys. Acta 306:341-345. [DOI] [PubMed] [Google Scholar]
- 2.Buede, R., C. Rinker-Schaffer, W. J. Pinto, R. L. Lester, and R. C. Dickson. 1991. Cloning and characterization of LCB1, a Saccharomyces gene required for biosynthesis of the long-chain base component of sphingolipids. J. Bacteriol. 173:4325-4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dehoux, P., J. Davies, and M. Cannon. 1993. Natural cycloheximide resistance in yeast. The role of ribosomal protein L41. Eur. J. Biochem. 213:841-848. [DOI] [PubMed] [Google Scholar]
- 4.Del Pozo, L., D. Abarca, J. Hoenicka, and A. Lmenez. 1993. Two different genes from Schwanniomyces occidentalis determine ribosomal resistance to cycloheximide. Eur. J. Biochem. 213:849-857. [DOI] [PubMed] [Google Scholar]
- 5.Elion, E. A., and J. R. Warner. 1984. The major promoter element of rRNA transcription in yeast lies 2 kb upstream. Cell 39:663-673. [DOI] [PubMed] [Google Scholar]
- 6.Faber, K. N., P. Haima, W. Harder, M. Veenhuis, and G. Ab. 1994. Highly-efficient electrotransformation of the yeast Hansenula polymorpha. Curr. Genet. 25:305-310. [DOI] [PubMed] [Google Scholar]
- 7.Hanada, K., T. Hara, M. Nishijima, O. Kuge, R. C. Dickson, and M. M. Nagiec. 1997. A mammalian homolog of the yeast LCB1 encodes a component of serine palmitoyltransferase, the enzyme catalyzing the first step in sphingolipid synthesis. J. Biol. Chem. 272:32108-32114. [DOI] [PubMed] [Google Scholar]
- 8.Hanada, K., T. Hara, and M. Nishijima. 2000. Purification of the serine palmitoyltransferase complex responsible for sphingoid base synthesis by using affinity peptide chromatography techniques. J. Biol. Chem. 275:8409-8415. [DOI] [PubMed] [Google Scholar]
- 9.Hannun, Y. A. 1996. Functions of ceramide in coordinating cellular responses to stress. Science 274:1855-1859. [DOI] [PubMed] [Google Scholar]
- 10.Holm, C., D. W. Meeks-Wagner, W. L. Fangman, and D. Botstein. 1986. A rapid, efficient method for isolating DNA from yeast. Gene 42:169-173. [DOI] [PubMed] [Google Scholar]
- 11.Imokawa, G., S. Akasaki, M. Hattori, and N. Yoshizuka. 1986. Selective recovery of deranged water-holding properties by stratum corneum lipids. J. Invest. Dermatol. 87:758-761. [DOI] [PubMed] [Google Scholar]
- 12.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 13.Ito, W., H. Ishiguro, and Y. Kurosawa. 1991. A general method for introducing a series of mutations into cloned DNA using the polymerase chain reaction. Gene 102:67-70. [DOI] [PubMed] [Google Scholar]
- 14.Karlsson, K. A. 1970. On the chemistry and occurrence of sphingolipid long-chain bases. Chem. Physiol. Lipids 5:6-43. [DOI] [PubMed] [Google Scholar]
- 15.Kawai, S., S. Murao, M. Mochizuki, I. Shibuya, K. Yano, and M. Takagi. 1992. Drastic alteration of cycloheximide sensitivity by substitution of one amino acid in the L41 ribosomal protein of yeasts. J. Bacteriol. 174:254-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim, I. G., S. K. Nam, J. H. Sohn, S. K. Rhee, G. H. An, S. H. Lee, and E. S. Choi. 1998. Cloning of the ribosomal protein L41 gene of Phaffia rhodozyma and its use a drug resistance marker for transformation. Appl. Environ. Microbiol. 64:1947-1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klein, P., M. Kanehisa, and C. DeLisi. 1985. The detection and classification of membrane-spanning proteins. Biochim. Biophys. Acta 815:468-476. [DOI] [PubMed] [Google Scholar]
- 18.Kondo, K., T. Saito, S. Kajiwara, M. Takagi, and N. Misawa. 1995. A transformation system for the yeast Candida utilis: use of a modified endogenous ribosomal protein gene as a drug-resistant marker and ribosomal DNA as an integration target for vector DNA. J. Bacteriol. 177:7171-7177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lampe, M. A., A. L. Burlingame, J. Whitney, M. L. Williams, B. E. Brown, E. Roitman, and P. M. Elias. 1983. Human stratum corneum lipids: characterization and regional variations. J. Lipid Res. 24:120-130. [PubMed] [Google Scholar]
- 20.Mathias, S., L. A. Pena, and R. N. Kolesnick. 1998. Signal transduction of stress via ceramide. Biochem. J. 335:465-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McNabb, T. J., A. E. Cremesti, P. R. Brown, and A. S. Fischl. 1999. The separation and direct detection of ceramides and sphingoid bases by normal-phase high-performance liquid chromatography and evaporative light-scattering detection. Anal. Biochem. 276:242-250. [DOI] [PubMed] [Google Scholar]
- 22.Merrill, A. H., Jr., and D. D. Jones. 1990. An update of the enzymology and regulation of sphingomyelin metabolism. Biochim. Biophys. Acta 1044:1-12. [DOI] [PubMed] [Google Scholar]
- 23.Merrill, A. H., Jr., A. M. Sereni, V. L. Stevens, Y. A. Hannun, R. M. Bell, and J. M. Kinkade, Jr. 1986. Inhibition of phorbol ester-dependent differentiation of human promyelocytic leukemic (HL-60) cells by sphinganine and other long-chain bases. J. Biol. Chem. 261:12610-12615. [PubMed] [Google Scholar]
- 24.Nagiec, M. M., J. A. Baltisberger, G. B. Wells, R. L. Lester, and R. C. Dickson. 1994. The LCB2 gene of Saccharomyces and the related LCB1 gene encode subunits of serine palmitoyltransferase, the initial enzyme in sphingolipid synthesis. Proc. Natl. Acad. Sci. USA 91:7899-7902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagiec, M. M., R. L. Lester, and R. C. Dickson. 1996. Sphingolipid synthesis: identification and characterization of mammalian cDNAs encoding the Lcb2 subunit of serine palmitoyltransferase. Gene 177:237-241. [DOI] [PubMed] [Google Scholar]
- 26.Petes, T. D. 1979. Yeast ribosomal DNA genes are located on chromosome XII. Proc. Natl. Acad. Sci. USA 76:410-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenberg, S., D. Coit, and P. Tekamp-Olson. 1990. Glyceraldehyde-3-phosphate dehydrogenase-derived expression cassettes for constitutive synthesis of heterologous proteins. Methods Enzymol. 185:341-351. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 29.Spiegel, S., and A. H. Merrill, Jr. 1996. Sphingolipid metabolism and cell growth regulation. FASEB J. 10:1388-1397. [DOI] [PubMed] [Google Scholar]
- 30.Sudbery, P. E. 1994. The non-Saccharomyces yeasts. Yeast 10:1707-1726. [DOI] [PubMed] [Google Scholar]
- 31.Waterham, H. R., M. E. Digan, P. J. Koutz, S. V. Lair, and J. M. Cregg. 1997. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 186:37-44. [DOI] [PubMed] [Google Scholar]
- 32.Weiss, B., and W. Stoffel. 1997. Human and murine serine-palmitoyl-CoA transferase—cloning, expression and characterization of the key enzyme in sphingolipid synthesis. Eur. J. Biochem. 249:239-247. [DOI] [PubMed] [Google Scholar]
- 33.Woolford, J. L., Jr. 1989. Nuclear pre-mRNA splicing in yeast. Yeast 5:439-457. [DOI] [PubMed] [Google Scholar]
- 34.Yamada, Y., K. Maeda, and K. Mikata. 1994. The phylogenetic relationships of the hat-shaped ascospore-forming, nitrate-assimilating Pichia species, formerly classified in the genus Hansenula Sydow et Sydow, based on the partial sequences of 18S and 26S ribosomal RNAs (Saccharomycetaceae): the proposals of three new genera, Ogataea, Kuraishia, and Nakazawaea. Biosci. Biotechnol. Biochem. 58:1245-1257. [DOI] [PubMed] [Google Scholar]