Abstract
Oncofetal RNA-binding IMPs have been implicated in mRNA localization, nuclear export, turnover and translational control. To depict the cellular actions of IMPs, we performed a loss-of-function analysis, which showed that IMPs are necessary for proper cell adhesion, cytoplasmic spreading and invadopodia formation. Loss of IMPs was associated with a coordinate downregulation of mRNAs encoding extracellular matrix and adhesion proteins. The transcripts were present in IMP RNP granules, implying that IMPs were directly involved in the post-transcriptional control of the transcripts. In particular, we show that a 5.0 kb CD44 mRNA contained multiple IMP-binding sites in its 3′UTR, and following IMP depletion this species became unstable. Direct knockdown of the CD44 transcript mimicked the effect of IMPs on invadopodia, and we infer that CD44 mRNA stabilization may be involved in IMP-mediated invadopodia formation. Taken together, our results indicate that RNA-binding proteins exert profound effects on cellular adhesion and invasion during development and cancer formation.
Keywords: CD44, IMP, invadopodia, RNA-binding proteins, RNP granules
Introduction
The zipcode-binding proteins IMP1, IMP2 and IMP3 (human), ZBP1 (chicken), Vg1-RBP/Vera (Xenopus) and CRD-BP (mouse) are orthologous and paralogous members of the same vertebrate RNA-binding protein family, consisting of two RNA recognition motifs (RRM) and four K-homology (KH) domains (Nielsen et al, 2001; Yaniv and Yisraeli, 2002). IMP1, IMP2 and IMP3 are capable of strong and specific RNA binding, but so far only a handful of RNA targets, including β-actin, Vg1, c-myc, IGF2, H19 and tau, have been reported (Ross et al, 1997; Deshler et al, 1998; Doyle et al, 1998; Havin et al, 1998; Nielsen et al, 1999; Runge et al, 2000; Atlas et al, 2004).
IMPs mainly reside in the cytoplasm, where they are present in large motile RNP granules. IMPs and their orthologues have been implicated in various post-transcriptional processes, such as mRNA localization (Ross et al, 1997; Deshler et al, 1998; Havin et al, 1998; Runge et al, 2000), turnover (Doyle et al, 1998) and translational control (Nielsen et al, 1999; Liao et al, 2005). Vg1-RBP/Vera mediates localization of Vg1 mRNA to the vegetal pole of the Xenopus oocyte and CRD-BP stabilizes the c-myc transcript. ZBP1 has been implicated in localization of β-actin mRNA to the leading edge of fibroblasts (Farina et al, 2003) and in neuronal outgrowth (Tiruchinapalli et al, 2003), and finally IMPs play a role in H19 and tau mRNA transport (Runge et al, 2000; Atlas et al, 2004) and IGF2 mRNA translation (Nielsen et al, 1999; Liao et al, 2005). Moreover, IMPs and their orthologues contain two nuclear export signals and exhibit nuclear transit (Nielsen et al, 2003; Oleynikov and Singer, 2003), so it is conceivable that IMPs are loaded onto their mRNA targets in the nucleus.
IMPs are primarily expressed during early embryogenesis and at mid-gestation in the mouse (Hansen et al, 2004). Disruption of IMP1 expression in mice leads to dwarfism, impaired development of the gut and increased perinatal lethality (Hansen et al, 2004). Also, the IMP3/KOC orthologue Vg1-RBP/Vera may be required for normal development, since it has been demonstrated to promote migration of neural crest cells (Yaniv et al, 2003). Moreover, upregulation of IMP3/KOC is followed by increased proliferation and metaplasia of pancreatic acinar cells (Wagner et al, 2003). Finally, IMPs are frequently overexpressed in various cancers and the proteins are considered to be oncofoetal factors (Doyle et al, 2000; Ioannidis et al, 2001; Gu et al, 2004).
To characterize the cellular functions of the IMPs, we performed a loss-of-function study of IMP1 and IMP3 using RNA interference.
Results
IMP1 and IMP3 depletion
During development, IMPs are abundantly expressed in epithelial layers, and carcinomas frequently exhibit high levels of IMPs (Yisraeli, 2005). We therefore examined the function of the proteins in HeLa adenocarcinoma cells. IMPs may associate with RNA in a redundant manner (Nielsen et al, 2004), so we first characterized the expression of the individual IMPs in the cells. Whereas no IMP2 was detected, significant amounts of both IMP1 and IMP3 were present in the cells (Figure 1A). Consequently, cellular IMPs were knocked down by transfection with short interfering RNAs (siRNAs) against both IMP1 and IMP3 mRNA, respectively (Figure 1B and C). To avoid erroneous off-target inactivation, all experiments were performed with two different sets of siRNA (IMP(1,3)A and IMP(1,3)B). Negative controls included a nonsilencing scrambled siRNA (Scr) or mock transfection (Mock). Moreover, a lamin A/C siRNA was used to detect possible effects caused by activation of the RNAi system in selected experiments. The transfection efficiency was determined by addition of FITC-conjugated siRNA, and this showed that more than 90% of the cells were transfected (data not shown). Treatment with siRNA pairs IMP(1,3)A or IMP(1,3)B led to an almost complete depletion of IMP1 and IMP3 mRNA after 24 h (Figure 1B). This was followed by a gradual reduction of IMP1 and IMP3 proteins, and after 72 h the IMP1 and IMP3 protein levels were approximately 15% of that found in the Scr and mock control cells (Figure 1C). Unless otherwise indicated, all comparisons were performed 72 h after transfection. During each of the siRNA experiments, Western blot analysis was performed on recovered lysate or on lysate from a parallel transfection, to verify the knockdown efficiency.
Figure 1.
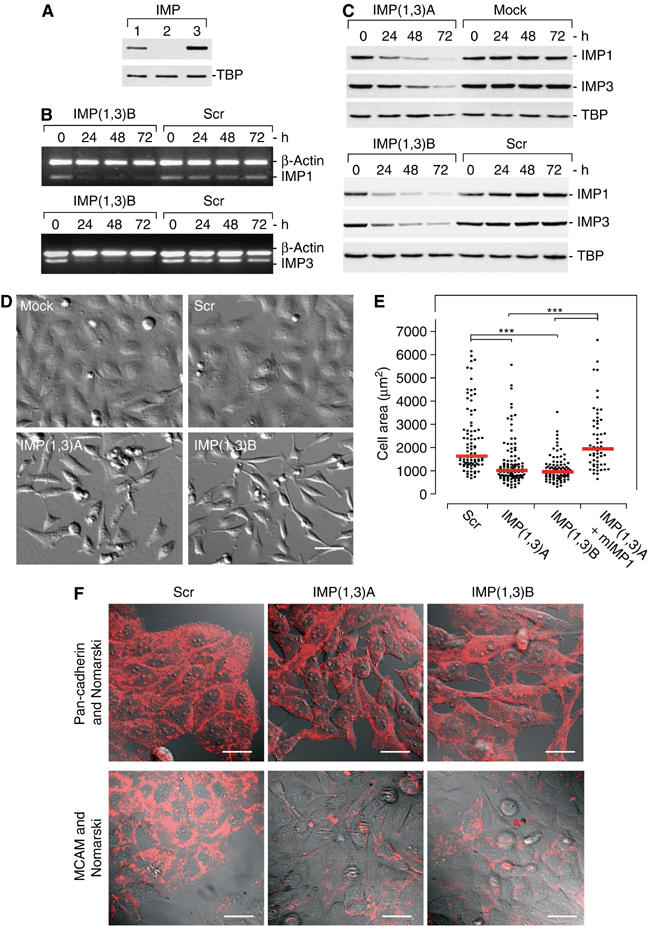
RNAi-mediated knockdown of IMP proteins. (A) IMP expression in HeLa cells was determined by Western blot analysis using rabbit polyclonal anti-IMP1 (lane 1), anti-IMP2 (lane 2) or anti-IMP3 (lane 3) antibodies. TATA-box binding protein (TBP) was used as loading control. (B) siRNA-mediated downregulation of IMP1 and IMP3 mRNA in IMP(1,3)B- or Scr siRNA-treated cells was examined by multiplex RT–PCR. RT–PCR was performed at the indicated times using IMP1 and β-actin-specific primers (upper panel) or IMP3 and β-actin-specific primers (lower panel), respectively. The PCR fragments were resolved on a 1% agarose gel and visualized by ethidium bromide staining. (C) siRNA-mediated downregulation of IMP1 and IMP3 proteins in IMP(1,3)A-, IMP(1,3)B- or Scr siRNA-treated cells was examined by Western blot analysis. The cells were harvested 24, 48 or 72 h after transfection and proteins were detected with anti-IMP1, anti-IMP3 or anti-TBP antibodies. Blots were quantified using a LAS-1000 luminescent imager analyser (Fuji). (D) Morphology of IMP-depleted cells. HeLa cells were mock treated or transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs. At 72 h after transfection, the living cells were photographed (× 400, scale bar, 50 μm). (E) Scatter plot of the 2D area (μm2) of fixed siRNA-treated cells. To verify that the morphological changes were caused by downregulation of IMP, IMP(1,3)A-treated cells were transfected with a plasmid encoding mIMP1. mIMP1-transfected cells were identified by IMP1 immunostaining. Each point represents a cell and the medians are indicated (red line). Asterisks depict statistically significant differences between the indicated groups, by a Mann–Whitney test (***P<0.001). The number of cells measured in the groups were—Scr: n=91, IMP(1,3)A: n=109, IMP(1,3)B: n=88 and IMP(1,3)A+mIMP1: n=57. (F) Expression of the cell–cell adhesion molecule MCAM and pan-cadherin in IMP-depleted cells. Semiconfluent cultures treated with Scr siRNA, IMP(1,3)A or IMP(1,3)B siRNAs were stained with mouse anti-MCAM and anti-pan-cadherin antibody and Texas-Red conjugated anti-mouse IgG. Scale bar, 50 μm.
IMP-depleted cells become spindle-shaped and exhibit reduced adhesion
At 48 h following transfection, a marked change in the cell morphology was apparent in both IMP(1,3)A and IMP(1,3)B siRNA-treated cells. IMP siRNA-treated cells became spindle-shaped, rounded, exhibited fewer cellular extensions and were unable to form proper cell–cell contacts. In contrast, control cells were flat and adherent with convex cellular extensions, lamellipodia and frequent cell–cell adhesion contacts (Figure 1D). To depict cell–cell contacts, we stained nearly confluent cultures with antibodies against MCAM or pan-cadherin, which revealed a general loss of cell–cell adhesions and an overall reduced MCAM staining, in the IMP-depleted cells (Figure 1F). Moreover, a substantial number of cells were floating in the media of IMP siRNA-treated cells, indicating that IMP depletion results in cell detachment. Measurements of the cell area showed that IMP-depleted cells were approximately 40% smaller than Scr siRNA-treated cells (P<0.001) (Figure 1E). To verify that the morphological changes were connected to downregulation of IMP, we transfected IMP(1,3)A-treated cells with a plasmid encoding mouse IMP1 (mIMP1). This rescued the reduction in size, so the median area of the mIMP1-positive cells was 1939 μm2 compared to the 1627 μm2 of Scr and 1009 μm2 of the IMP(1,3)A-treated cells (P<0.001), respectively (Figures 1E and 4C).
Since detachment and size reduction could result from changes in growth rates or apoptosis, we measured the incorporation of bromodeoxyuridine (BrdU) and performed a TUNEL assay. The percentage of BrdU-positive cells were 41.9±3.0% s.d. and 42.0±3.4% s.d. in mock or Scr siRNA transfected cells, respectively, whereas 45.5±4.3% s.d. and 45.4±2.5% s.d. were positive in IMP(1,3)A and IMP(1,3)B siRNA-treated cells, respectively. Moreover, no change in the number of apoptotic cells was detected (data not shown). We therefore infer that the reduced number of cells observed in siRNA-treated cultures results from detachment of cells.
To quantitate the adhesive capacity of the IMP-depleted cells, they were seeded on a laminin-1-coated glass surface. At 1 h after seeding, the dishes were gently washed and fixed, before the number of adherent cells was counted. Compared to the Scr and mock control cells, only ∼50% of the IMP-deficient cells were able to re-attach to the laminin-1-coated surface (Figure 2B). We also noticed that IMP siRNA-treated cells did not spread out their cytoplasm following seeding (Figure 2A). The cytoplasmic spreading was quantified after staining with crystal violet (Figure 2C). After 1 h, ∼70% of the control cells exhibited widened cytoplasm, whereas only 15–25% of the adherent IMP-deficient cells exhibited a spread cytoplasm (P<0.01, in all combinations between control and IMP-depleted cells, calculated by an unpaired, unequal t-test). The cells gradually recovered and after 4 h ∼60% of the siRNA-treated cells had obtained cytoplasmic spreading compared to ∼80% of the control cells. Fewer than 10% of the IMP siRNA-treated cells re-adhered and spread in the same way as control cells, corresponding to the number of untransfected cells in the cultures (see above).
Figure 2.
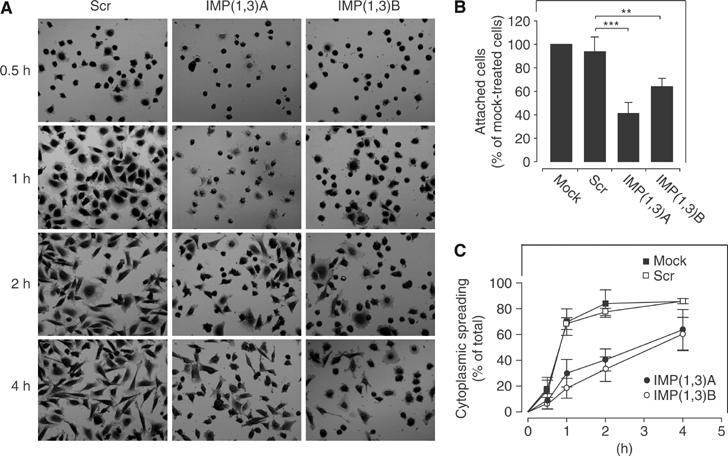
Adhesion and cytoplasmic spreading after IMP depletion. (A) HeLa cells were transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively, before they were trypsinized and allowed to re-seed on a laminin-1-coated surface. The cells were fixed at the indicated times and stained with crystal violet. (B) The number of adhered cells was counted 1 h after seeding. The data are stated as percentage of mock-treated cells (100%) and represent the mean value±s.d. Asterisks depict statistically significant differences between the indicated groups, by an unpaired, unequal t-test (**P<0.01, ***P<0.001) of five independent experiments, where at least 1500 cells were scored in each group, in each experiment. (C) The number of mock-treated (▪), Scr siRNA-treated (□), IMP(1,3)A siRNA-treated (•) and IMP(1,3)B siRNA-treated (○) cells with cytoplasmic spreading were quantitated at the indicated times following seeding. Data are presented as the mean value±s.d. of at least three independent experiments.
Reduced cytoplasmic spreading in IMP-depleted cells
IMP knockdown cells also lacked focal complex (FX)-supported protrusive edges, as depicted by staining with anti-phosphotyrosine antibody and anti-paxillin antibody (Figure 3A and Supplementary Figure S3). Moreover, focal adhesions (FA) were located at the periphery, where they functioned as anchor points for concave-shaped stress fibres outlining the cell. To obtain a dynamic impression of the formation of protrusive edges in IMP-depleted cells, we challenged the cells with 10 μM of the ROCK inhibitor Y-27632, that induces FX-supported membrane ruffles via activation of Rac (Katsumi et al, 2002; Tsuji et al, 2002). Untreated and Scr-transfected HeLa cells responded by disassembly of large stress fibres and FA structures and induction of FX-supported protrusive edges, resulting in round and flat cells with broad protrusive edges supported by FXs (Figure 3B and E). IMP-depleted cells did not attain this morphology. Instead, their fusiform shape with few or no protrusive cell edges became more explicit. The median area of the Scr-treated cells was almost two times larger than the area of IMP-depleted cells (2602 μm2 compared to 1669 and 1245 μm2) after ROCK inhibitor treatment (Figure 3C). Moreover, a scoring of the cells' ‘roundness' (see Figure 3D), where 1 represents a perfect circle, showed that the median score of the Scr-treated cells was 1.4, whereas IMP(1,3)A and IMP(1,3)B siRNA-treated cells had scores of 2.8 and 3.6, reflecting their irregular shape.
Figure 3.
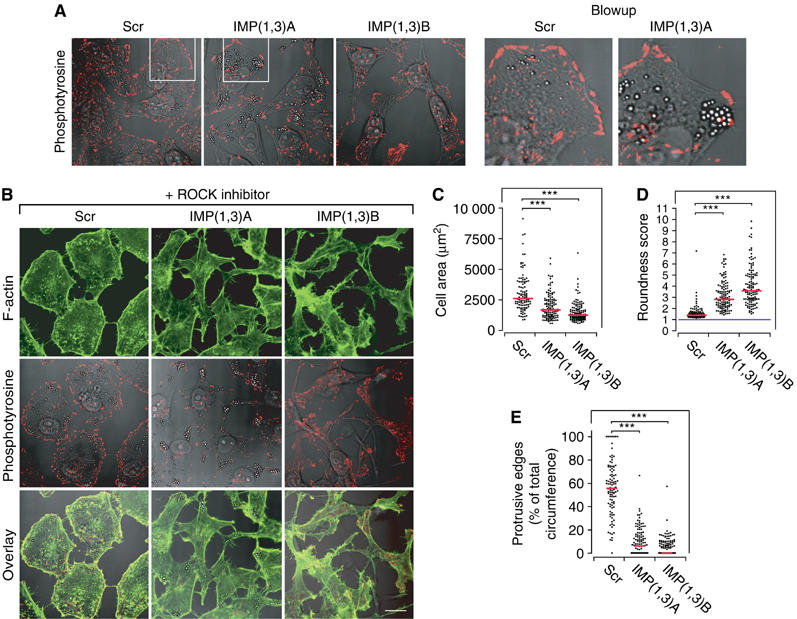
Formation of FX-supported protrusive edges in IMP-depleted cells. (A) HeLa cells were transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively, and stained with anti-phosphotyrosine antibody to reveal FXs and FA (left panel). Blowups (right panel) illustrate the characteristic convex FX-supported edge in an Scr-treated cell and a typical concave FX-depleted edge in an IMP siRNA-treated cell. (B) HeLa cells were transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively, and incubated with 10 μM ROCK inhibitor (Y-27632) for 20 h and stained for F-actin (top row) or for phosphotyrosine (middle row). An overlay of these is shown in the bottom row. Scale bar, 50 μm. (C) Scatter plot of the 2D area of ROCK inhibitor-treated cells following IMP depletion. (D) Scatter plot of the roundness score from ROCK inhibitor-treated cells following IMP depletion. Cells were scored according to the formula (length of circumference)2/(4π*cell area), which is 1 for a circle (blue line). The more a cell differs from a circular shape, the higher the score. (E) Scatter plot showing the percentage of protrusive edge in IMP-depleted cells compared to Scr siRNA-treated cells. Each point in panels (C)–(E) represents a cell and the medians are indicated (red line). Asterisks depict statistically significant differences between the indicated groups, by a Mann–Whitney test (***P<0.001). The number of cells measured or scored in the groups were—Scr: n=107, IMP(1,3)A: n=122 and IMP(1,3)B: n=125.
It is noteworthy that there was no change in the total amount of β-actin and Arp2/3 proteins (see below; Supplementary Figure S1), and by visual inspection it was apparent that IMP-depleted cells contain a significant amount of stress fibres. Neither did we observe any changes in the activation of Rho, Rac and Cdc42, nor altered levels or activity of β1 integrins (see Supplementary Figure S2) or in the levels of α3, α5 and α6 integrin chains.
Loss of IMP1 and IMP3 disrupts invadopodia formation
Staining of the F-actin cytoskeleton revealed that loss of IMP1 and IMP3 was also associated with an almost complete loss of 0.6–1.2 μm actin-rich structures located at the cell–substratum interface, resembling podosomes or invadopodia (Figure 4A and B). By immunohistochemical staining, we found that they were highly enriched in tyrosine-phosphorylated proteins and contained the Arp2/3 complex protein p34-Arc and β1 integrin (Figure 4D and E). A fluorescence recovery after photobleaching (FRAP) experiment on cells transfected with a plasmid encoding EYFP-actin revealed a fast actin turnover, with a 50% recovery 20–25 s after bleaching (Figure 4F). Based on the characteristic spatial distribution, size, protein components and actin dynamics of the structures, we conclude that they meet the criteria for podosomes and/or invadopodia (Buccione et al, 2004). To distinguish between podosomes and invadopodias, we performed a matrix degradation assay. HeLa cells were allowed to attach to a FITC-laminin-1-coated glass surface for 24 h before they were fixed and stained with phalloidin. This demonstrated that the FITC-laminin-1 matrix was degraded directly under the podosome/invadopodia-like structures, and by confocal microscopy it was apparent that they protruded into the FITC-laminin-1 extracellular matrix (ECM) layer (Figure 4G), confirming that they were invadopodia. To verify that the loss of invadopodia was a result of IMP depletion and not an effect of active RNA interference in the cells, we included an additional control siRNA targeting the lamin A/C proteins, but this had no effect on the presence of the invadopodia (Figure 4B). Moreover, it was feasible to partially rescue the invadopodia by cotransfecting with a mIMP1-encoding plasmid (Figure 4C). Taken together, we therefore infer that loss of IMP leads to loss of invadopodia.
Figure 4.
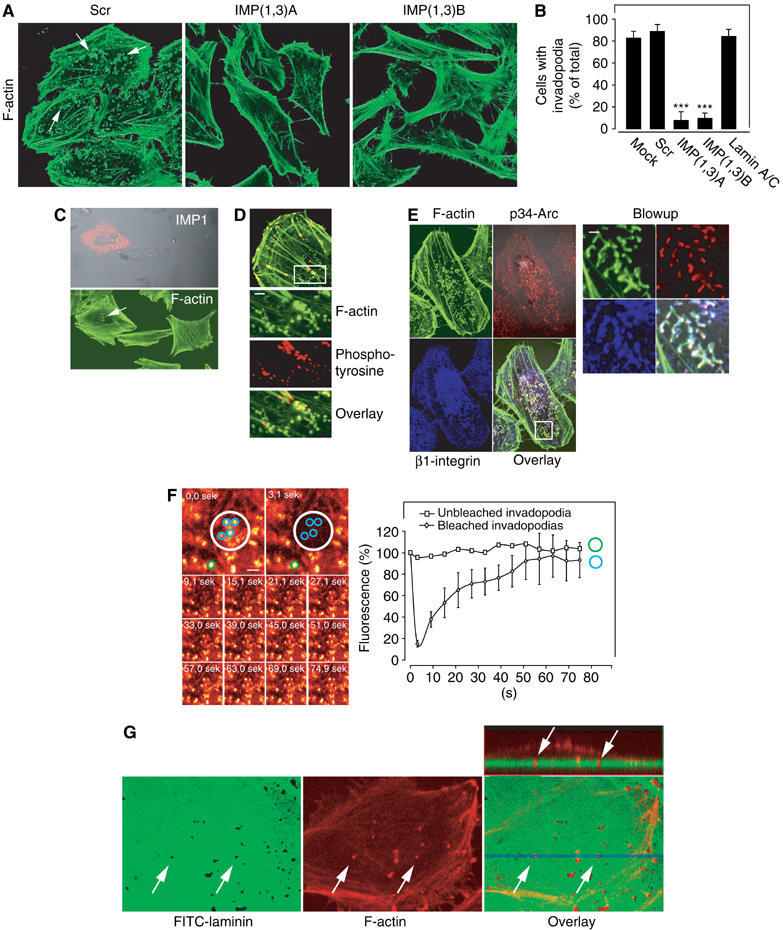
Invadopodia in IMP-depleted cells. (A) Loss of actin-rich structures located at the cell–substratum interface. HeLa cells were transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively, and stained with phalloidin to reveal F-actin-rich dot-like structures (arrows) resembling podosomes or invadopodia. (B) The percentile of cells containing podosomes or invadopodia after transfection with Scr, IMP(1,3)A, IMP(1,3)B or lamin A/C siRNAs was determined. Data are presented as the mean value±s.d. of three independent experiments. Asterisks depict statistically significant differences between the Scr siRNA-treated cells and the IMP(1,3)A and IMP(1,3)B siRNA-treated cells, respectively, by an unpaired, unequal t-test (***P<0.001). (C) HeLa cells were depleted for IMP and transfected with a mIMP1 expression plasmid—revealed by staining with anti-IMP1 (red)—which rescued podosome/invadopodia-like structures revealed by phalloidin staining (green). (D) Immunocytochemical analysis of tyrosine-phosphorylated proteins in the podosomes/invadopodia-like structures. HeLa cells were stained with Alexa Flour 488 conjugated phalloidin (green) and mouse anti-phosphotyrosine stained with Texas-Red conjugated anti-mouse antibody (red). The white box depicts the position of the blowups. Scale bar, 2 μm. (E) Immunocytochemical analysis of Arp2/3 complex protein p34-Arc and β1 integrin in the podosomes/invadopodia-like structures. HeLa cells were stained with phalloidin as above (green), in combination with rabbit anti-p34-Arc (red) and mouse anti-β1 integrin (blue) visualized with Texas-Red conjugated anti-rabbit antibody and Alexa Fluor 660 conjugated anti-mouse antibody, respectively (left panel). The white box depicts the position of the blowups (right panel). Scale bar, 2 μm. (F) Dynamics of podosome/invadopodia-like structures. HeLa cells were transfected with pEYFP-actin-expressing plasmid and actin turnover was examined by a FRAP analysis. The two upper pictures in the left panel show the area that was bleached before and after activation of the laser (white circle). The lower pictures show the recovery of EYFP-actin in the bleached area. The right panel shows the quantification of the recovery of EYFP-actin in the bleached area. The light intensity was measured in circular areas surrounding each of the four actin-rich structures present in the bleached area (blue circles). The green circle indicates the spot outside the bleached area that was used as reference. The results are stated as mean value±s.d. of the four blue circles in percentage of light emitted before bleaching. (G) ECM degradation assay. HeLa were seeded on an FITC-laminin-1-containing matrix, the cells were fixed and stained with Alexa Flour 660 conjugated phalloidin to depict the position of podosome/invadopodia-like structures. The dish was examined by confocal z-stacking, and the three lower pictures depict one of these slices, which show the colocalization between the matrix degradation spots and invadopodia structures. The upper right panel shows a vertical cross-section (lower right panel, blue line) of the cell and the underlying matrix with the projected invadopodia (arrows).
IMP1 and IMP3 knockdown gene expression profiling
To identify factors and pathways regulated by IMP proteins and obtain leads to the mechanism behind the phenotypic changes, we compared the gene expression profiles of IMP siRNA-treated cells and mock-treated cells. Expression profiles were generated from both the IMP(1,3)A and IMP(1,3)B siRNA sets. cRNA was hybridized to Affymetrix human U133A arrays, which contain 22 283 different probe sets. Totally, the signals of 32 probe sets corresponding to 27 different transcripts changed significantly. Seven transcripts were upregulated and 20 became downregulated. Five represented transcripts of unknown function. Among the 17 annotated transcripts that were downregulated, 11 encoded proteins involved in cell adhesion, motility, invasion and ECM (Table I). The five upregulated genes were DICER1 (BG109746), dimethylarginine dimethylaminohydrolase 1 (AL078459), kynureninase (BC000879), solute carrier family 31, member 1 (NM_001859) and a throphoblast-derived noncoding RNA (AU134977). The complete microarray results are available on www.rh-microarray.com and www.ebi.ac.uk/arrayexpress (accession number E-M-EXP-548). To verify the microarray findings, the expression of selected genes was also analysed by real-time reverse transcription–PCR (RT–PCR) (Table I). In all cases the results were in agreement with the microarray data.
Table 1.
Downregulated transcripts in IMP-depleted cells
| Gene | Accession | Fold change | P-value |
|---|---|---|---|
| Cell adhesion, invasion and ECM | |||
| ALCAM | NM_001627 | −1.8 | 0.0001* |
| AMIGO2 | NM_181847 | −2.4 | 0.0005* |
| CD24 antigen | NM_013230 | −1.8 | 0.0023 |
| CD44 antigen | NM_000610 | −1.8 | 0.0094* |
| Collagen V, α1 | NM_000093 | −2.0 | 0.0000* |
| Dysadherin | NM_144779 | −2.2 | 0.0002 |
| Keratin 19 | NM_002276 | −2.7 | 0.0023 |
| Lumican | NM_002345 | −2.4 | 0.0122* |
| Matrix metalloproteinase 1 | NM_002421 | −2.2 | 0.0367 |
| MCAM | NM_006500 | −2.2 | 0.0001 |
| SynCAM | NM_014333 | −1.8 | 0.0019* |
| Other | |||
| ATPase Na+/K+ transporting β1 | NM_001677 | −1.9 | 0.0001 |
| Chloride intracellular channel 3 | NM_004669 | −2.4 | 0.0051 |
| eIF2 subunit 1α | NM_004094 | −2.2 | 0.0012 |
| IL-7 receptor | NM_002185 | −1.9 | 0.0045 |
| IMP3 | NM_006547 | −2.1 | 0.0010* |
| NIMA-related kinase 7 | NM_133493 | −2.2 | 0.0002 |
| The gene name, GenBank accession number, fold change and P-value of the analysis are shown. The asterisks indicate transcripts that were confirmed by RT–PCR and/or Western blot analysis (IMP3). IMP1 probe sets are not present on the U133A array. | |||
We also examined the expression and distribution of β-actin protein and mRNA in the cells. β-Actin protein levels were unchanged in the IMP-depleted cells and β-actin mRNA was found to localize to the leading lamella in only 4% of the control cells as well as the IMP-deficient cells (see Supplementary Figure S1). Finally, no expression of IGF2 and H19 was observed in HeLa cells and c-myc protein levels were unchanged after IMP depletion (data not shown).
CD44 knockdown results in loss of invadopodia formation
CD44 has previously been implicated in formation of invadopodia (Bourguignon et al, 1998); so we reasoned that downregulation of CD44 could play a role in the loss of invadopodia in IMP-depleted cells. CD44 was abundantly expressed at the surface of HeLa cells, especially at sites of membrane protrusions and around invadopodia (Figure 5D).
Figure 5.
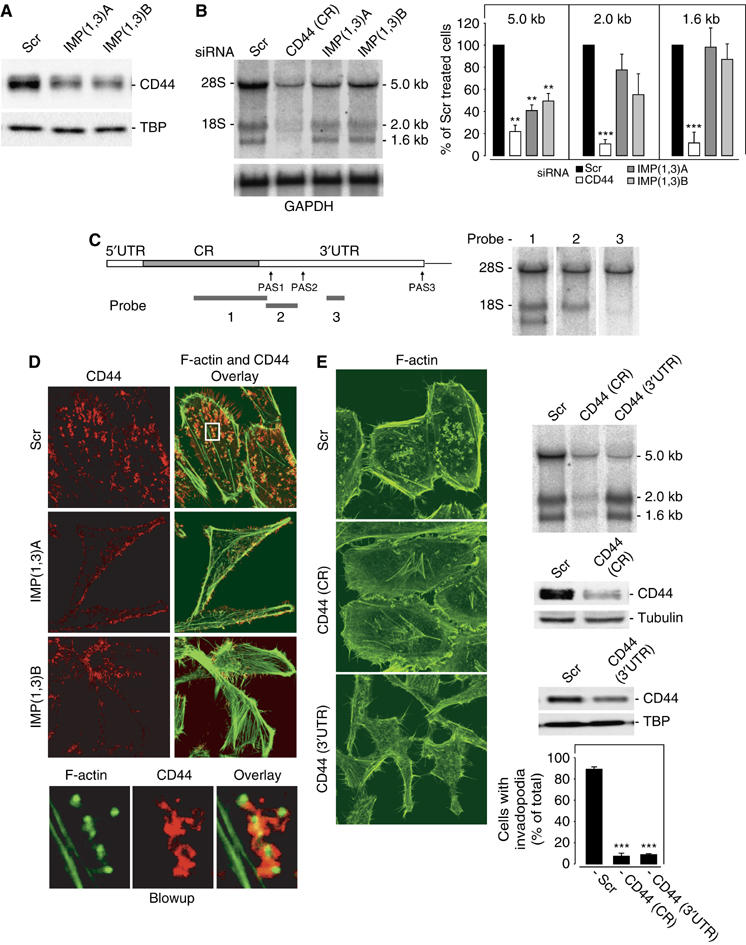
CD44 in invadopodia formation. CD44 protein and mRNA levels in IMP-depleted cells. (A) Western blot analysis of CD44 in HeLa cells transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs, respectively. TBP was used as loading control (B) Northern blot analysis of CD44 mRNA under the same conditions (left panel). The right panel shows the quantification of the 5.0, 2.0 and 1.6 kb CD44 transcripts during knockdown. GAPDH was used as loading control. (C) Schematic representation of the full-length CD44 transcript outlining the position of the different polyadenylation signals (PAS) and probes designated 1, 2 and 3, that were used in the Northern blot analysis shown to the right. 5′UTR—5′untranslated region; CR—coding region; 3′UTR—3′untranslated region. (D) CD44 is associated with invadopodia formation. HeLa cells were stained with mouse anti-CD44 antibody and phalloidin to depict F-actin. The blowup below shows that the CD44 protein is located in close proximity to invadopodia. (E) Loss of invadopodia in CD44-depleted cells. HeLa cells were treated with Scr siRNA or siRNA targeting the CD44 mRNA coding region (CR) or the 3′UTR (left panel). The upper right panel shows a Northern blot analysis of the siRNA-treated cells. At 72 h after transfection, the cells were harvested and the level of CD44 protein was determined by Western blot analysis (middle right panel). β-Tubulin or TBP was included as loading control. Finally, the cells were stained for F-actin and the number of invadopodia were counted (lower right panel). Data are presented as the mean value±s.d. of three independent experiments (lower right panel). Asterisks depict statistically significant differences between the Scr and CD44 siRNA-treated cells, by an unpaired, unequal t-test (***P<0.001).
HeLa cells were found to express three CD44 transcripts of 1.6, 2.0 and 5.0 kb (Figure 5B). A search among ESTs in the databases combined with Northern blot analysis with probes corresponding to positions 389–2327 in the coding region, and 2303–2886 and 3667–3952 in the 3′UTR (numbering from the AUG of the NM_000610 transcript), revealed that the three mRNAs exhibited identical 5′UTRs and coding regions but different 3′UTRs, due to alternative polyadenylation signals (PAS) starting at positions 2347, 2811 and/or 2895, and 5262, respectively (Figure 5C). Following IMP1 and IMP3 knockdown, the 5.0 kb transcript exhibiting the extended trailer became selectively downregulated (Figure 5B). The 5.0 kb CD44 mRNA and protein expression were reduced ∼2–3-fold (Figure 5B and A) and, obviously, no association between CD44 and invadopodias was found in the IMP-depleted cells. In all, 95% of the CD44 protein corresponded to the 90 kDa isoform without variable exons. The remaining CD44 consisted of 120 and 140 kDa isoforms containing combinations of variant exons 3 and 6–10 (data not shown).
To establish if CD44 was necessary for formation of invadopodia, we first performed a knockdown of all CD44 transcripts using a coding region siRNA (CD44(CR)). The amount of CD44 protein was reduced to approximately 15% and phalloidin staining of the CD44-depleted cells showed that invadopodia structures were present in 7±3% in CD44 siRNA-treated cells compared to 89±2% of Scr siRNA-treated cells (P<0.001) (Figure 5E). Subsequently, a selective knockdown of the 5.0 kb CD44 mRNA with a trailer-specific siRNA (CD44(3′UTR)) recapitulated the effect and reduced the number of cells with invadopodia to ∼10% of nontreated cells. In contrast to cells with complete CD44 knockdown, the spindle-shaped appearance that is characteristic of IMP-depleted cells was moreover apparent in the cells treated with the CD44 3′UTR siRNA (Figure 5E).
Taken together, the results demonstrate that the 5.0 kb CD44 mRNA is essential for formation of invadopodia.
IMP binds CD44 mRNA and stabilizes the transcript
To establish a causal relation between IMPs and CD44, as well as some of the other differentially regulated transcripts, we examined if ALCAM, CD24, CD44, MCAM and SynCAM mRNAs were associated with IMP RNP granules. Flag-tagged-IMP1 or Flag-tagged-ERG (negative control) proteins were transiently expressed in HeLa cells, and to ensure that Flag-IMP1 was bioactive we initially examined if Flag-IMP1 was able to associate with H19 RNA, which is a known IMP1 target (Runge et al, 2000) (Figure 6A). The IMP-containing RNP particles were pulled down by incubation with anti-Flag-coated beads and the mRNAs were detected by RT–PCR (Figure 6B). Whereas GAPDH mRNA was only detected in the cell lysates, CD44 (5.0 kb), CD24, MCAM, SynCAM and, to a smaller extent, ALCAM mRNAs were detected with pelleted Flag-IMP1. In contrast, none of the transcripts were present in the pelleted Flag-ERG beads, indicating that the mRNAs are associated with IMP RNP granules in vivo. Moreover, the subcellular distribution of CD44 mRNA and IMP proteins was characterized by combined in situ hybridization and immunocytochemical staining of Flag-IMP1, respectively (Figure 6C and D). CD44 mRNA and Flag-IMP1 proteins colocalized in about 70% of the characteristic 200–700 nm (optical diameter) Flag-IMP1-positive RNP granules distributed along the cytoskeletal components. There was no indication of nuclear retention in the absence of IMPs (data not shown). In contrast, GAPDH mRNA was mainly observed in smaller speckles randomly distributed in the cytoplasm, without particular colocalization (<15%) with Flag-IMP1.
Figure 6.
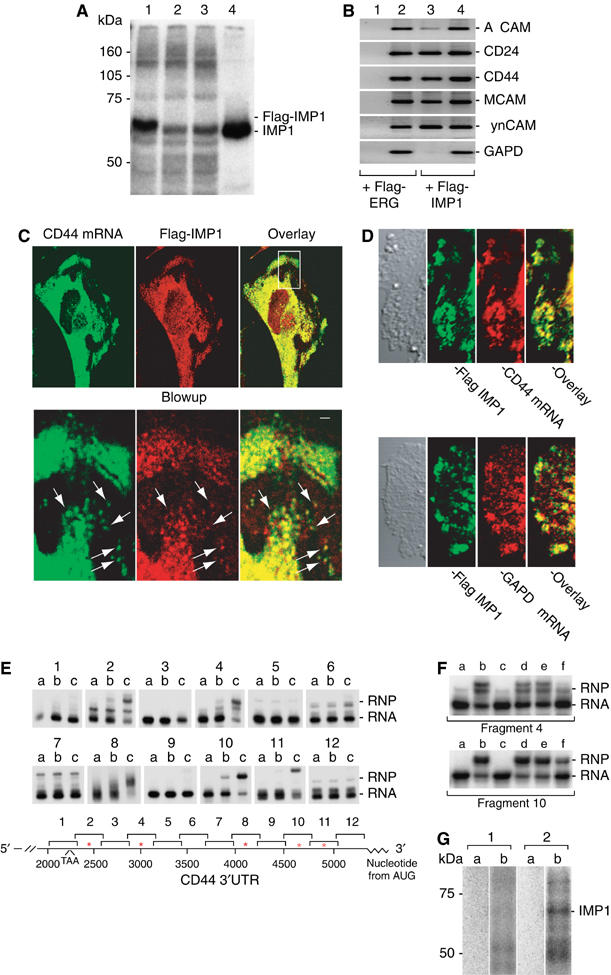
IMP binding to and colocalization with CD44 mRNA. (A) Binding of Flag-tagged IMP1 to H19 RNA target. UV crosslinking of [α-32P]UTP-labelled H19 RNA was performed with extracts from HeLa cells expressing Flag-IMP1 (lane 1), Flag-ERG (lane 2), nontransfected HeLa cells (lane 3) or with 5 nM recombinant IMP1 (lane 4). The position of IMP1 and Flag-IMP1 RNP complexes is indicated to the right. (B) Isolation of IMP1 RNP particles. Flag-tagged-ERG or Flag-tagged IMP1 proteins, respectively, were transiently expressed in HeLa cells. After lysis, the tagged proteins were pelleted by incubation with anti-Flag-coated beads. ALCAM, CD24, CD44(5.0 kb), MCAM, SynCAM and GAPDH transcripts were detected by RT–PCR in the pelleted beads (lanes 1 and 3) or in the lysates (lanes 2 and 4). (C) Colocalization of CD44 mRNA and Flag-IMP1 in lamellipodia and cellular protrusions and perinuclear regions. Flag-IMP1 was stained with Texas-Red conjugated anti-mouse IgG (red) and CD44 mRNA was detected with a digoxigenin-labelled probe followed by staining with HRP-conjugated sheep anti-digoxigenin antibody and Alexa Fluor 488 tyramide (green). The white box depicts the position of the blowups (lower panel) and the arrows indicate Flag-IMP1 and CD44 mRNA colocalized in RNP granules. Scale bar, 1500 nm. (D) Colocalization of CD44 mRNA and Flag-IMP1. HeLa cells were transfected with Flag-IMP1 expression vector and stained with mouse anti-Flag and visualized with FITC-conjugated anti-mouse IgG antibody (green). The localization of CD44 mRNA (red) was subsequently determined by in situ hybridization with a digoxigenin-labelled CD44 antisense probe detected with rhodamine-conjugated sheep anti-digoxigenin (upper panel). Control cells were incubated with a digoxigenin-labelled GAPDH probe (lower panel). (E) Binding of recombinant IMP1 to the 3′UTR of CD44 mRNA. An electrophoretic mobility shift analysis was performed (upper panel) with RNA fragments covering the entire CD44 3′UTR in the presence of buffer (lane a) or 1.5 (lane b) or 4.5 nM (lane c) recombinant IMP1, respectively. The migration of the RNP (RNP) complexes and the labelled RNA (RNA) is indicated to the right. The lower panel shows the positions of the individual RNA fragments (1–12), which are numbered according to their distances from the A in the translation start codon of CD44 mRNA (NM_000610). The positions of IMP1-binding sites are indicated by asterisks. (F) Competitive inhibition of IMP1 binding. An electrophoretic mobility shift assay was performed with CD44 3′UTR fragment 4 (upper panel) and CD44 3′UTR fragment 10 (lower panel) in the presence of buffer (lane a), 4.5 nM recombinant IMP1 (lane b), 4.5 nM IMP1 and 10 nM A11 SELEX RNA target (lane c), 4.5 nM IMP1 and 10 ng poly(A) (lane d), 4.5 nM IMP1 and 10 nM CD44 3′UTR fragment 7 (lane e), and 4.5 nM IMP1 and 10 ng total RNA from RD cells (lane f). The positions of the RNP complexes (RNP) and the labelled RNA (RNA) are indicated. (G) [α-32P]UTP-labelled CD44 RNA fragment 3133–3952 (1) and CD44 RNA fragment 3928–5040 (2) were subjected to UV light in the absence (a) or presence (b) of a cytoplasmic extract from Hela cells, followed by RNase A digestion and SDS polyacrylamide gel electrophoresis to resolve labelled proteins. The numbering of the fragments corresponds to the position numbering from the AUG codon of the CD44 transcript variant 1 (NM_000610). The position of the IMP1 RNP complex is indicated to the right.
To identify binding sites in the CD44 transcript, we performed a band-shift analysis with 21 in vitro synthesized RNA fragments covering the entire 3′UTR, and the nine variable exons (Figure 6E). All experiments were performed in the presence of a 20 000-fold molar excess of tRNA over labelled target RNA. Five high-affinity binding sites for recombinant IMP1 with KD's of 2–4 nM were detected in the 3′UTR at positions 2303–2627, 2862–3055, 3928–4236, 4567–4773 and 4749–5040, respectively (numbering from the A in the AUG start codon of the CD44 variant 1 transcript (NM_000610)). In contrast, no binding sites were observed in any of the variable exons (data not shown). The specificity of IMP binding to 32P-labelled CD44 RNA 2862–3055 and 4567–4773 was corroborated by competition with an IMP SELEX-target RNA (positive control) (Nielsen et al, 2004), poly(A) RNA, CD44 fragment 7 (3667–3952) RNA (negative control) and total RNA, respectively (Figure 6F). Whereas a complete displacement was observed in the presence of the SELEX-target RNA, total RNA caused only a minor reduction in binding, and CD44 fragment 7 (3667–3944) RNA or poly(A) RNA had no effect on binding. Finally, we performed a crosslinking experiment with 32P-labelled CD44 RNA corresponding to band-shift fragments 5, 6 and 7 (3133–3952), and 8, 9, 10 and 11 (3928–5040) comprising no and three IMP-binding sites, respectively (Figure 6G). RNAs were subjected to UV light in the presence of a cytoplasmic extract from Hela cells, followed by RNase A digestion and SDS polyacrylamide gel electrophoresis to resolve labelled proteins. Whereas CD44 3133–3952 RNA exhibited no crosslinked species, CD44 RNA fragment 3928–5040 exhibited a crosslink migrating with an apparent mass of 69 kDa, corresponding to IMP.
To define the function of IMP in the post-transcriptional control of 5.0 kb CD44 mRNA, we characterized the translation and decay of the transcript. The relative fraction of polysomal CD44 mRNA did not change significantly after IMP depletion (data not shown). The mRNA turnover was estimated by incubation with actinomycin D and quantification of the 5.0 kb transcript following Northern blot analysis. Linear regression of the decay curves demonstrated that the half-life of the 5.0 kb CD44 transcript was 14.1 h in cells treated with Scr siRNA and 4.5 and 5.5 h in IMP(1,3)A and IMP(1,3)B siRNA-treated cells (P<0.01 and P<0.001). GAPDH mRNA, that was included as a control, exhibited the same half-lives of 12.3, 12.1 and 13.3 h in Scr, IMP(1,3)A and (MPL1,3)B siRNA-treated cells, respectively (Figure 7A and B). Taken together, we infer that IMP stabilizes the 5.0 kb CD44 mRNA.
Figure 7.
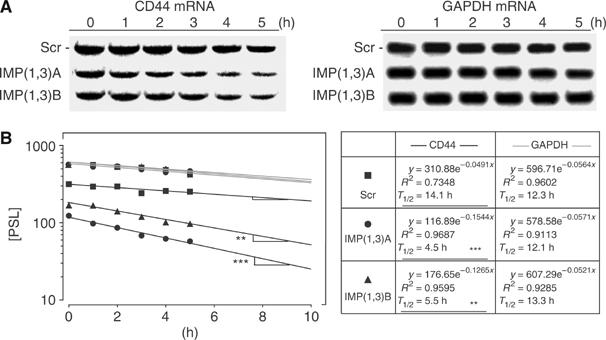
CD44 mRNA half-life in IMP-depleted cells. (A) HeLa cells were transfected with Scr, IMP(1,3)A or IMP(1,3)B siRNAs as indicated. After 72 h, they were incubated with 5 μM actinomycin D for the indicated times before the levels of CD44 and GAPDH mRNA were determined by Northern blot analysis. (B) The transcript levels were determined by phosphoimager counting and the left panel shows the mRNA decay plotted in a logarithmic scale. The lines represent the linear regression of the data. The right box shows the equation of the linear regression and the calculated half-life of the mRNAs. Asterisks depict statistically significant differences between the slope of the CD44 mRNA decline from Scr siRNA-treated cells and the slope of the CD44 mRNA decline from IMP(1,3)A or IMP(1,3)B siRNA-treated cells, respectively, calculated by the GraphPad Prism 4 software (**P<0.01, ***P<0.001).
Discussion
Following synthesis, IMPs enter the nucleus (Nielsen et al, 2003; Oleynikov and Singer, 2003), where they conceivably are loaded with target mRNAs, before they are exported to the cytoplasm and packaged in large RNP granules to reach their final destinations. Initially, we considered whether the effects of IMP knockdown could be rationalized in terms of altered regulation of reported target mRNAs. The IMP1 chicken orthologue ZBP1 has been implicated in localization of β-actin mRNA to the leading edge of fibroblasts (Ross et al, 1997), and local β-actin synthesis is important for cell-polarity and directional movement (Condeelis and Singer, 2005). Although this is an appealing model, which could rationalize—at least some of the observed phenotypic changes—the subcellular localization of the transcript was unaffected by loss of IMP1 and IMP3 (see Supplementary Figure S1). Moreover, HeLa cells do not express significant amounts of neither IGF2 nor H19 transcripts, and although c-myc is present in the cells, the protein level did not change following IMP depletion.
To obtain additional leads to the observed phenotype, we performed a global expression analysis. Similar to what we previously observed in IMP1 knockout mice, only few (27) transcripts changed. The majority were downregulated and only seven transcripts were upregulated after IMP depletion. Strikingly, 11 of the downregulated transcripts coded for proteins involved in cellular adhesion and/or invasion. ALCAM, CD24, CD44, MCAM and SynCAM transcripts were all associated with IMP-containing RNP granules, implying that IMPs mediate a coordinated regulation of the transcripts. In this way, the granules may be regarded as a post-transcriptional regulon. Downregulation of the adhesion proteins may collectively explain the loss of proper cell–cell contacts. Moreover, CD44, which originally was identified as a hyaluronan receptor, may be implicated in the reduced cell–matrix adhesion (Marhaba and Zoller, 2004). The reduced expression of lumican and collagen V, α1 is in fine agreement with previous observations in IMP1 null mice (Hansen et al, 2004), which exhibited reduced levels of the transcripts in gut, and further proves that IMPs control the composition of the ECM. Finally, it is striking that several of the downregulated factors are normally coexpressed with IMPs during development.
A distinct feature of the IMP-depleted cells was the loss of invadopodia. Invadopodia are composed of an actin-rich core and contain a variety proteins, such as the Arp2/3 complex, β1 integrin and multiple signalling molecules, and matrix metalloproteinases (MMP). They degrade and extend into the ECM (Buccione et al, 2004), and degradation of matrix around the invadopodia correlates with a cell's invasive potential (Bourguignon et al, 1998; Coopman et al, 1998; Kelly et al, 1998; Chuang et al, 2004; Hashimoto et al, 2004). To characterize the mechanism, whereby IMPs promote the formation of invadopodia, we focused on CD44. Overexpression of CD44 correlates with the invasive capacity of cancer cells (Jothy, 2003), and CD44 has previously been implicated in formation of invadopodia (Bourguignon et al, 1998). Furthermore, it has been shown that CD44 mediates collagen IV degradation and tumour cell invasion by anchoring MMPs (Yu and Stamenkovic, 1999). HeLa cells were found to express three CD44 transcripts of 1.6, 2.0 and 5.0 kb. Based on ESTs and Northern blot analysis, we show that the three mRNAs exhibit different 3′UTRs by employing alternative PAS. To establish if CD44 was necessary for formation of invadopodia, we first performed a knockdown of all CD44 transcripts using a coding region siRNA. CD44-depleted cells were essentially devoid of invadopodia, and, strikingly, a selective knockdown of the 5.0 kb CD44 mRNA completely recapitulated the effect indicating that only the 5.0 kb CD44 transcript is essential for formation of invadopodia. The 5.0 kb transcript was associated with IMP granules and the 3′UTR contained at least five IMP-binding sites with KD's of 2–4 nM. Following IMP1 and IMP3 depletion, it became selectively downregulated. The reduction in transcript level was ∼2.5-fold and this correlated with the observed decrease in the half-life of the transcript. The binding elements exhibited no obvious similarity to other known high-affinity binding sites, but, as described for other IMP-binding sites (Runge et al, 2000; Nielsen et al, 2004), they were characterized by few guanosines and stretches of pyrimidines yielding a low secondary structure potential. Not all 5.0 kb CD44 mRNA was associated with the RNP, and the effect on the stability of the subpopulation that provides new CD44 protein may be more pronounced than the global estimate. The fact that we find an almost complete loss of invadopodia only after an ∼2-fold reduction in total CD44 level may be rationalized in this way. CD44 expression is subject to both alternative splicing and post-translational modifications (Marhaba and Zoller, 2004), so the selective regulation of the CD44 transcripts by IMPs provides an additional level of complexity to the control of CD44 expression, in the sense that CD44 transcripts may be divided into an IMP-sensitive and an IMP-insensitive population.
IMP granules are transported towards the leading edge and a significant portion is located in cytoplasmic protrusions. The biological significance of the cytotopical localization in the context of CD44 or some of the other RNP-associated mRNAs is not resolved, but their synthesis may be regulated by local signalling events to ensure that the production of the factors is coordinated to adhesion, matrix degradation and membrane dynamics. RNA-binding proteins, including IMP2, were recently found in the so-called ‘spreading initiation centres' (SIC), which are formed during cell spreading (de Hoog et al, 2004). Our observations may provide a putative model for the function of SICs or similar structures in HeLa cells.
IMPs are expressed in various cancers, and overexpression of the IMP mouse orthologue CRD-BP in mammary epithelial cells leads to metastasizing mammary adenocarcinomas (Tessier et al, 2004). Although several studies have indicated that IMPs are dominant tumour promoters (Ioannidis et al, 2001; Gu et al, 2004; Tessier et al, 2004), it has been suggested that ZPB1 functions as a tumour suppressor by re-establishing normal patterns of β-actin mRNA targeting and maintaining a proper cell polarity (Condeelis and Singer, 2005). The present data reinforce that IMPs propagate cancer spreading, since formation of invadopodia increases the invasive capacity of the transformed cells (Bourguignon et al, 1998; Coopman et al, 1998; Kelly et al, 1998; Chuang et al, 2004; Hashimoto et al, 2004; Tessier et al, 2004). Moreover, upregulation of ALCAM, AMIGO2, MCAM, CD24, CD44, dysadherin and MMP1 has previously been associated with cancer, increased invasiveness and metastasis (Ino et al, 2002; Kristiansen et al, 2004; Rabenau et al, 2004; Weichert et al, 2004; Wu et al, 2004; Jiang et al, 2005; Miletti-Gonzalez et al, 2005; Poola et al, 2005).
Taken together, our results show that IMPs are involved in cell adhesion and invadopodia formation, and we infer that overexpression of IMPs in cancer cells may promote their invasive capacity.
Materials and methods
Reverse transcription PCR and Northern blot analysis
Total RNA was isolated with Trizol reagent (Invitrogen). For RT–PCR, cDNA was synthesized with the avian myeloblastosis virus reverse transcriptase. The cDNAs were amplified using the primers listed in Supplementary data S3.
Northern blot analysis was performed as described previously (Nielsen et al, 1999) using 10 μg of total RNA hybridized with [α-32P]dCTP-labelled CD44 probes (bp 389–2327, 2303–2886 and 3667–3952 of the CD44 variant 1 transcript (NM_000610) numbering from the A in the AUG start codon) or with [α-32P]dCTP-labelled GAPDH probe (coding region). For mRNA decay analysis, cells were treated with 5 μg/ml actinomycin D for the indicated times before harvesting. The intensities of CD44 and GAPDH mRNA were measured with a BAS 2000 phosphoimager (Fuji). The decay rate was determined by a linear regression of the log-transformed data using GraphPad Prism 4 software.
Cell adhesion and cytoplasmic spreading
Adhesion assays were performed on siRNA-treated cells 72 h after transfection. HeLa cells were detached by trypsinization and counted before the cells were reseeded on uncoated or laminin-1-coated (2 μg/cm2) glass bottom dishes. Cells were allowed to re-adhere for 1 h before they were gently washed with phosphate-buffered saline (PBS) and fixed in 4% formaldehyde for 15 min. After staining with 0.1% crystal violet, the cells were examined by microscopy. Five randomly chosen visual fields, representing approximately 15% of the dish surface area, were photographed and the number of attached cells was counted (at least 1000 cells). The cell-spreading assay was performed the same way. The cells were scored as either nonspreading or spreading, depending on whether the cell had obtained cytoplasm extensions.
The cells' 2D area and circumference were measured using the Zeiss Image browser software. Furthermore, their protrusive circumference was also measured. The edges were scored as protrusive if they were convex shaped and exhibited closely lying FXs to support them. To describe the 2D shape, the value (length of circumference)2/(4π*cell area) was calculated for each cell. The score is 1 for a perfect circle, since (2πr)2/(4π*πr2)=1; so the more a cell differs from a circular shape, the higher the score.
Immunocytochemistry and FRAP analysis
Cells were fixed with 4% formaldehyde for 15 min at room temperature and permeabilized with 0.1% Triton X-100 in PBS for 3 min. After blocking with 1% bovine serum albumin (BSA) in PBS for 1 h at room temperature, they were incubated with primary antibody for 2 h or overnight at 4°C, washed in PBS and stained with Texas-Red anti-mouse, Texas-Red anti-rabbit or Alexa Flour 660 anti-mouse antibodies for 1.5 h at room temperature. F-actin staining was performed with Alexa Flour 488 phalloidin and Alexa Flour 660 phalloidin in 1% BSA in PBS for 1 h. FRAP analysis of actin turnover in invadopodia was performed on pEYFP-actin-transfected cells 24 h after transfection. The fluorescent recovery of YFP-actin in the bleached areas was measured every 6 s in areas surrounding each invadopodium.
ECM degradation assay
A fluorescent ECM was prepared by conjugating fluorescein-5-isothiocyanate (FITC) (Molecular probes) to laminin-1 as described by the manufacturer. Briefly, laminin-1 was chromatographed, dissolved in 0.1 M NaHCO3 (pH 9.0) and mixed with 10 mg/ml FITC in diformaldehyde at a ratio of 10:1. Conjugated FITC-lamin-1 was separated from unreacted FITC, and FITC-laminin-1-coated glass-bottom dishes were prepared by covering dishes with FITC-laminin-1 in PBS and allowing it to dry out. HeLa cells were seeded on the coated dishes and examined 24 h after seeding.
In situ hybridization
In situ hybridization was performed 72 h after transfection. In the case of β-actin, the procedure was also performed on cells that had been trypsin detached and allowed to re-adhere for 1 h on a laminin-1-coated surface as described above. Briefly, the cells were fixed for 30 min in freshly prepared 4% paraformaldehyde and 0.9% NaCl (pH 7.4) at room temperature. After washing and permeabilization in 0.5% Triton X-100, the cells were prehybridized in 50% formamide, 750 mM NaCl, 5 mM EDTA, 50 mM sodium phosphate (pH 7.0), 5 × Denhardt's solution, 0.5% SDS and 100 ng/ml salmon sperm DNA for 30 min. The cells were then hybridized with CD44 digoxigenin-labelled (DIG) antisense probe (bp 2027–2327 numbering from the A in the AUG start codon of the CD44 variant 1 transcript), DIG-labelled GAPDH antisense coding region probe, DIG-labelled β-actin antisense probe (bp 1386–1481) or DIG-labelled YFP antisense probe for 4–16 h at 37°C, before the cells were washed in 60% formamide, 300 mM NaCl and 30 mM sodium citrate. Bound DIG-labelled RNA probe was detected either by incubation with alkaline phosphatase-coupled sheep anti-DIG (Roche) for 90 min at room temperature and developed with nitro-blue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate for 5–10 min, or by incubation with HRP-coupled sheep anti-DIG (Roche) for 90 min at room temperature followed by development using a tyramide signal amplification kit (Molecular Probes) with the Alexa Flour 488 fluorophor according to the manufacturer's instructions. Finally, in order to obtain better spatial resolution, we stained the DIG-labelled probes with rhodamine-conjugated sheep anti-DIG (Roche). The percentage of IMP1 granules containing CD44 mRNA was determined with the LSM IMAGE examiner software from Zeiss. IMP1 granules were marked while blinded for CD44 mRNA staining. Subsequently, the IMP1 staining was blinded and CD44 mRNA was visualized and overlapping granules were counted. In addition, the colocalization software was used to determine the percentage of overlapping Flag-IMP1-stained pixels and CD44 mRNA-stained pixels.
Isolation of IMP1 ribonucleoprotein particles
HeLa cells were transfected with the 3XFlag-IMP1 plasmid or with Flag-ERG plasmid (a kind gift from Bo Porse, Rigshospitalet). After 24 h the cells were harvested and lysed in 2 ml lysis buffer (50 mM Tris–HCl (pH 7.4), 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.5 mM PMSF and 0.5% protease inhibitor cocktail (Sigma)), followed by sonication on ice. A volume of 200 μl of lysate was recovered to measure the total amount of RNA. The 3XFlag-IMP1 fusion protein and Flag-ERG were purified by immunoprecipitation using agarose anti-Flag (M2) beads (Sigma) at 4°C overnight. After precipitation, the nonbound protein fraction was removed by centrifugation. The beads were washed four times in TBS (50 mM Tris–HCl (pH 7.4), 150 mM NaCl), and the bound RNA was isolated by addition of 5 μg yeast ribosomal RNA as carrier and 1 ml Trizol.
Electrophoretic mobility-shift analysis and UV crosslinking
The primers used are listed in Supplementary data S3. In vitro transcripts were purified by denaturing gel electrophoresis. Recombinant proteins were expressed and purified as described previously (Nielsen et al, 1999). Radiolabelled RNA (200 amol) and 100 ng Escherichia coli tRNA were incubated with recombinant IMP1 at concentrations of 1.5 and 4.5 nM for 20 min at 30°C in 10 μl 20 mM Tris–HCl (pH 7.8), 140 mM KCl, 2 mM MgCl2 and 0.1% Triton X-100. After addition of Ficoll (2%) and bromophenol blue at 30°C, samples were applied directly to a cold 1 mm 5% polyacrylamide gel (19:1) in 90 mM Tris-borate (pH 8.3), and run at 80 V for 5 h. UV crosslinking was performed with H19 RNA segment H and CD44 RNA fragments as described (Runge et al, 2000), except that 10 ng/μl poly(U) RNA was added during the incubation with the HeLa cell extracts.
Supplementary Material
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Acknowledgments
We thank Joan Christiansen, Lis Nielsen, Pernille Ekstroem and Lena Bjoern Johansson for technical assistance. This work was supported by the Danish Natural Science and Medical Research Councils, the Danish Cancer Society, the Novo Nordisk Foundation, the University of Copenhagen and the Toyota Foundation.
References
- Atlas R, Behar L, Elliott E, Ginzburg I (2004) The insulin-like growth factor mRNA binding-protein IMP-1 and the Ras-regulatory protein G3BP associate with tau mRNA and HuD protein in differentiated P19 neuronal cells. J Neurochem 89: 613–626 [DOI] [PubMed] [Google Scholar]
- Bourguignon LY, Gunja-Smith Z, Iida N, Zhu HB, Young LJ, Muller WJ, Cardiff RD (1998) CD44v(3,8–10) is involved in cytoskeleton-mediated tumor cell migration and matrix metalloproteinase (MMP-9) association in metastatic breast cancer cells. J Cell Physiol 176: 206–215 [DOI] [PubMed] [Google Scholar]
- Buccione R, Orth JD, McNiven MA (2004) Foot and mouth: podosomes, invadopodia and circular dorsal ruffles. Nat Rev Mol Cell Biol 5: 647–657 [DOI] [PubMed] [Google Scholar]
- Chuang YY, Tran NL, Rusk N, Nakada M, Berens ME, Symons M (2004) Role of synaptojanin 2 in glioma cell migration and invasion. Cancer Res 64: 8271–8275 [DOI] [PubMed] [Google Scholar]
- Condeelis J, Singer RH (2005) How and why does beta-actin mRNA target? Biol Cell 97: 97–110 [DOI] [PubMed] [Google Scholar]
- Coopman PJ, Do MT, Thompson EW, Mueller SC (1998) Phagocytosis of cross-linked gelatin matrix by human breast carcinoma cells correlates with their invasive capacity. Clin Cancer Res 4: 507–515 [PubMed] [Google Scholar]
- de Hoog CL, Foster LJ, Mann M (2004) RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell 117: 649–662 [DOI] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Abramson T, Schnapp BJ (1998) A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol 8: 489–496 [DOI] [PubMed] [Google Scholar]
- Doyle GA, Betz NA, Leeds PF, Fleisig AJ, Prokipcak RD, Ross J (1998) The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic Acids Res 26: 5036–5044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle GA, Bourdeau-Heller JM, Coulthard S, Meisner LF, Ross J (2000) Amplification in human breast cancer of a gene encoding a c-myc mRNA-binding protein. Cancer Res 60: 2756–2759 [PubMed] [Google Scholar]
- Farina KL, Huttelmaier S, Musunuru K, Darnell R, Singer RH (2003) Two ZBP1 KH domains facilitate beta-actin mRNA localization, granule formation, and cytoskeletal attachment. J Cell Biol 160: 77–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu L, Shigemasa K, Ohama K (2004) Increased expression of IGF II mRNA-binding protein 1 mRNA is associated with an advanced clinical stage and poor prognosis in patients with ovarian cancer. Int J Oncol 24: 671–678 [PubMed] [Google Scholar]
- Hansen TV, Hammer NA, Nielsen J, Madsen M, Dalbaeck C, Wewer UM, Christiansen J, Nielsen FC (2004) Dwarfism and impaired gut development in insulin-like growth factor II mRNA-binding protein 1-deficient mice. Mol Cell Biol 24: 4448–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto S, Onodera Y, Hashimoto A, Tanaka M, Hamaguchi M, Yamada A, Sabe H (2004) Requirement for Arf6 in breast cancer invasive activities. Proc Natl Acad Sci USA 101: 6647–6652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK (1998) RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev 12: 1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ino Y, Gotoh M, Sakamoto M, Tsukagoshi K, Hirohashi S (2002) Dysadherin, a cancer-associated cell membrane glycoprotein, down-regulates E-cadherin and promotes metastasis. Proc Natl Acad Sci USA 99: 365–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, Trangas T, Dimitriadis E, Samiotaki M, Kyriazoglou I, Tsiapalis CM, Kittas C, Agnantis N, Nielsen FC, Nielsen J, Christiansen J, Pandis N (2001) C-MYC and IGF-II mRNA-binding protein (CRD-BP/IMP-1) in benign and malignant mesenchymal tumors. Int J Cancer 94: 480–484 [DOI] [PubMed] [Google Scholar]
- Jiang X, Dutton CM, Qi WN, Block JA, Garamszegi N, Scully SP (2005) siRNA mediated inhibition of MMP-1 reduces invasive potential of a human chondrosarcoma cell line. J Cell Physiol 202: 723–730 [DOI] [PubMed] [Google Scholar]
- Jothy S (2003) CD44 and its partners in metastasis. Clin Exp Metastasis 20: 195–201 [DOI] [PubMed] [Google Scholar]
- Katsumi A, Milanini J, Kiosses WB, del Pozo MA, Kaunas R, Chien S, Hahn KM, Schwartz MA (2002) Effects of cell tension on the small GTPase Rac. J Cell Biol 158: 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly T, Yan Y, Osborne RL, Athota AB, Rozypal TL, Colclasure JC, Chu WS (1998) Proteolysis of extracellular matrix by invadopodia facilitates human breast cancer cell invasion and is mediated by matrix metalloproteinases. Clin Exp Metastasis 16: 501–512 [DOI] [PubMed] [Google Scholar]
- Kristiansen G, Sammar M, Altevogt P (2004) Tumour biological aspects of CD24, a mucin-like adhesion molecule. J Mol Histol 35: 255–262 [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH (2003) The analysis of gene expression data: methods and software. In DNA-Chip Analyser (dChip), Parmigiani G, Garret ES, Irizarry R, Zeger SL (eds), pp 120–141. New York: Springer [Google Scholar]
- Liao B, Hu Y, Herrick DJ, Brewer G (2005) The RNA-binding protein IMP-3 is a translational activator of insulin-like growth factor II leader-3 mRNA during proliferation of human K562 leukemia cells. J Biol Chem 280: 18517–18524 [DOI] [PubMed] [Google Scholar]
- Marhaba R, Zoller M (2004) CD44 in cancer progression: adhesion, migration and growth regulation. J Mol Histol 35: 211–231 [DOI] [PubMed] [Google Scholar]
- Miletti-Gonzalez KE, Chen S, Muthukumaran N, Saglimbeni GN, Wu X, Yang J, Apolito K, Shih WJ, Hait WN, Rodriguez-Rodriguez L (2005) The CD44 receptor interacts with P-glycoprotein to promote cell migration and invasion in cancer. Cancer Res 65: 6660–6667 [DOI] [PubMed] [Google Scholar]
- Nielsen FC, Nielsen J, Christiansen J (2001) A family of IGF-II mRNA binding proteins (IMP) involved in RNA trafficking. Scand J Clin Lab Invest Suppl 234: 93–99 [PubMed] [Google Scholar]
- Nielsen J, Adolph SK, Rajpert-De Meyts E, Lykke-Andersen J, Koch G, Christiansen J, Nielsen FC (2003) Nuclear transit of human zipcode-binding protein IMP1. Biochem J 376: 383–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Christiansen J, Lykke-Andersen J, Johnsen AH, Wewer UM, Nielsen FC (1999) A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Mol Cell Biol 19: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J (2004) Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic Acids Res 32: 4368–4376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleynikov Y, Singer RH (2003) Real-time visualization of ZBP1 association with beta-actin mRNA during transcription and localization. Curr Biol 13: 199–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poola I, DeWitty RL, Marshalleck JJ, Bhatnagar R, Abraham J, Leffall LD (2005) Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat Med 11: 481–483 [DOI] [PubMed] [Google Scholar]
- Rabenau KE, O'Toole JM, Bassi R, Kotanides H, Witte L, Ludwig DL, Pereira DS (2004) DEGA/AMIGO-2, a leucine-rich repeat family member, differentially expressed in human gastric adenocarcinoma: effects on ploidy, chromosomal stability, cell adhesion/migration and tumorigenicity. Oncogene 23: 5056–5067 [DOI] [PubMed] [Google Scholar]
- Ross AF, Oleynikov Y, Kislauskis EH, Taneja KL, Singer RH (1997) Characterization of a beta-actin mRNA zipcode-binding protein. Mol Cell Biol 17: 2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge S, Nielsen FC, Nielsen J, Lykke-Andersen J, Wewer UM, Christiansen J (2000) H19 RNA binds four molecules of insulin-like growth factor II mRNA-binding protein. J Biol Chem 275: 29562–29569 [DOI] [PubMed] [Google Scholar]
- Tessier CR, Doyle GA, Clark BA, Pitot HC, Ross J (2004) Mammary tumor induction in transgenic mice expressing an RNA-binding protein. Cancer Res 64: 209–214 [DOI] [PubMed] [Google Scholar]
- Tiruchinapalli DM, Oleynikov Y, Kelic S, Shenoy SM, Hartley A, Stanton PK, Singer RH, Bassell GJ (2003) Activity-dependent trafficking and dynamic localization of zipcode binding protein 1 and beta-actin mRNA in dendrites and spines of hippocampal neurons. J Neurosci 23: 3251–3261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji T, Ishizaki T, Okamoto M, Higashida C, Kimura K, Furuyashiki T, Arakawa Y, Birge RB, Nakamoto T, Hirai H, Narumiya S (2002) ROCK and mDia1 antagonize in Rho-dependent Rac activation in Swiss 3T3 fibroblasts. J Cell Biol 157: 819–830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M, Kunsch S, Duerschmied D, Beil M, Adler G, Mueller F, Gress TM (2003) Transgenic overexpression of the oncofetal RNA binding protein KOC leads to remodeling of the exocrine pancreas. Gastroenterology 124: 1901–1914 [DOI] [PubMed] [Google Scholar]
- Weichert W, Knosel T, Bellach J, Dietel M, Kristiansen G (2004) ALCAM/CD166 is overexpressed in colorectal carcinoma and correlates with shortened patient survival. J Clin Pathol 57: 1160–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GJ, Peng Q, Fu P, Wang SW, Chiang CF, Dillehay DL, Wu MW (2004) Ectopical expression of human MUC18 increases metastasis of human prostate cancer cells. Gene 327: 201–213 [DOI] [PubMed] [Google Scholar]
- Yaniv K, Fainsod A, Kalcheim C, Yisraeli JK (2003) The RNA-binding protein Vg1 RBP is required for cell migration during early neural development. Development 130: 5649–5661 [DOI] [PubMed] [Google Scholar]
- Yaniv K, Yisraeli JK (2002) The involvement of a conserved family of RNA binding proteins in embryonic development and carcinogenesis. Gene 287: 49–54 [DOI] [PubMed] [Google Scholar]
- Yisraeli JK (2005) VICKZ proteins: a multi-talented family of regulatory RNA-binding proteins. Biol Cell 97: 87–96 [DOI] [PubMed] [Google Scholar]
- Yu Q, Stamenkovic I (1999) Localization of matrix metalloproteinase 9 to the cell surface provides a mechanism for CD44-mediated tumor invasion. Genes Dev 13: 35–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3


