Abstract
Enteroviruses (EVs) overcome their host cells by usurping the translation machinery to benefit viral gene expression. This is accomplished through alternative translation initiation in a cap-independent manner at the viral internal ribosomal entry site (IRES). We have investigated the role of cis- and trans-acting viral factors in EV IRES translation in living cells. We observed that considerable portions of the viral genome, including the 5′-proximal open reading frame and the 3′ untranslated region, contribute to stimulation of IRES-mediated translation. With the IRES in proper context, translation via internal initiation in uninfected cells is as efficient as at capped messages with short, unstructured 5′ untranslated regions. IRES function is enhanced in cells infected with the EV coxsackievirus B3, but the related poliovirus has no significant stimulatory activity. This differential is due to the inherent properties of their 2A protease and is not coupled to 2A-mediated proteolytic degradation of the eukaryotic initiation factor 4G. Our results suggest that the efficiency of alternative translation initiation at EV IRESs depends on a properly configured template rather than on targeted alterations of the host cell translation machinery.
Plus-strand RNA viruses are among the most common human pathogens, responsible for devastating pandemics in the past (poliomyelitis) and present (hepatitis C virus infection and dengue hemorrhagic fever). Upon entering susceptible host cells, positive-strand RNA viruses utilize their genome as a template for viral gene expression in competition with host cell mRNAs for the translation machinery. To accomplish this, many positive-strand RNA viruses alter the host cell protein synthesis apparatus and employ unorthodox translation strategies involving 5′ untranslated region (5′UTR) and 3′UTR with uncommon structural features. This is exemplified by the Enterovirus genus of Picornaviridae.
Conventional translation of eukaryotic mRNAs occurs upon formation of a ribonucleoprotein network that engages the 43S preinitiation complex (53). This network bridges the 5′ terminus of eukaryotic mRNAs, universally modified by an m7Gppp cap, and a poly(A) tail. Components of the eukaryotic initiation factor 4F (eIF4F) complex mediate template closure: the cap-binding eIF4E binds to eIF4G, which interacts with the poly(A)-binding protein (PABP) (16, 43, 50). Since it also interacts with the RNA helicase eIF4A and via eIF3 with the 40S ribosomal subunit, eIF4G assumes a central scaffolding function for preinitiation complex assembly (20).
Despite their extreme genetic austerity, enteroviruses (EVs) devote a considerable portion of their genomes to elaborate, highly structured 5′- and 3′UTRs and encode a poly(A) tail of ∼50 nucleotides (nt) in length. The viral 5′UTR contains an internal ribosomal entry site (IRES) mediating 5′-end, cap-independent translation initiation of the uncapped genome (26, 40). The onset of viral gene expression coincides with severe disruptions of the intracellular milieu, including modification of canonical translation factors. EV infection produces proteolytic degradation of eIF4GI (14, 33, 55) and eIF4GII (17) yielding amino- and carboxy-terminal fragments. Physical separation of the eIF4E- and PABP-binding sites in the former from the eIF4A- and eIF3-binding sites in the latter disturbs the scaffolding function of eIF4G (30). In addition, PABP is cleaved during infection (27, 28). It is reasonable to assume that virus-induced rearrangements of the translation apparatus convey competitive advantages to IRES-driven viral RNAs with different requirements for translation initiation over cellular mRNAs. This competitive edge likely entails disrupting cap-dependent translation (host translation shutoff), while maintaining favorable conditions for IRES-driven translation.
Investigations of the molecular events determining translation rate at cellular and viral templates in EV-infected cells focused on the role of EV-rhinovirus 2Apro and aphthovirus L proteases. The 2Apro gene product participates in proteolytic processing of the viral polyprotein (52) and is involved in host translation shutoff (6) via eIF4G cleavage in EV-infected cells (14, 55). Complete shutoff does not occur upon eIF4GI cleavage alone (7, 41) and may involve a kinetic differential of eIF4GI versus eIF4GII cleavage (18) and/or concomitant degradation of PABP (28, 29). Similarly, 2Apro has been implicated in selective translation of viral genomic RNAs. This activity is mediated at least in part by IRES trans activation by 2Apro or indirectly by cellular trans activity induced by 2Apro (22). In addition, 2Apro-mediated eIF4G cleavage stimulated poliovirus (PV) (10) and rhinovirus (32) IRES-driven translation in vitro, albeit only ∼2 fold.
We report here experiments with subgenomic monocistronic reporter RNAs that recapitulate the conditions for translation initiation at the EV genome in vivo. Our experiments reveal that proximal (the 5′-proximal open reading frame [ORF]) and distal [the 3′UTR and poly(A) tail] sequences potently stimulate IRES function. Including all stimulatory sequences, IRES-driven translation in uninfected cells is as efficient as gene expression from mRNAs containing short unencumbered 5′ and 3′ termini.
We observed further enhancement of IRES function in cells infected with coxsackievirus B3 (CBV3) but not after infection with PV. This differential is attributable to inherent activity of their respective 2Apro gene products and coincides with the kinetics of proteolytic degradation of eIF4GI and -II. Experiments in cells expressing proteolytically inactive CBV3 2Apro revealed residual stimulatory activity in the absence of eIF4G cleavage, suggesting IRES trans activation by the 2A gene product. Expression of 2Apro cleavage-resistant eIF4GI did not prevent IRES stimulation, and exogenous eIF4GI proteolytic cleavage products had no stimulatory activity on their own in vivo. Our observations indicate that IRES-driven viral RNAs compete with cellular mRNAs, not via targeted disruption of host cell translation factors but rather through a highly competitive translation rate.
MATERIALS AND METHODS
Luciferase reporter constructs, chimeric viruses, and eIF4GI expression vectors.
Renilla luciferase (rLuc) expression vectors containing the CBV3 5′UTR and divergent 3′UTRs were constructed as follows. Plasmid pCBV3-0 (kindly provided by N. Chapman) was digested with SacI and ClaI, and the vector containing the CBV3 5′UTR was ligated with PCR-generated rLuc ORF (primers 1 and 2) and the desired 3′UTR (primer sequences are specified in Table 1). A12 and A50 [12- and 50-mer poly(A)] 3′UTRs were generated from complementary oligonucleotide pairs 3 and 4 and 5 and 6, respectively, and the CBV3+A12 3′UTR was PCR amplified with primers 7 and 8. The CBV3 3′UTR+A50 reporter was generated by digestion of the A50-containing construct with XbaI and ligation with the CBV3 3′UTR fragment lacking poly(A) (primers 7 and 9). To generate rLuc expression constructs with unrelated 3′UTR sequences, the A50-containing vector was digested with XbaI, treated with Klenow fragment, and ligated with a 100-bp SacI-ClaI segment of a modified pBlueScript vector (8), yielding a multiple cloning site (MCS) plus A50, or MCS+A50. To produce reporters harboring 22+A50, 41+A50, or 63+A50 3′UTRs, the MCS+A50 expression vector was digested with SacII-EcoRV, XbaI-EcoRV, or BamHI-PstI, respectively, followed by fill in with Klenow fragment and religation.
TABLE 1.
List of oligonucleotides used in this work
| Number | Name | Sequence (5′-3′)a |
|---|---|---|
| 1 | rLuc-IRES(+) | CCGAGCTCAGACTTCGAAAGTTTATGATCC* |
| 2 | rLuc (−) | GGTCTAGAATTGTTCATTTTTGAGAACTC |
| 3 | poly(A)12 (+) | CTAG(A)12 |
| 4 | poly(A)12 (−) | CGA(T)12 |
| 5 | poly(A)50 (+) | CTAG(A)50 |
| 6 | poly(A)50 (−) | CGA(T)50 |
| 7 | CBV3 3′UTR (+) | CCTCTAGATTAGAGACAATTTG |
| 8 | CBV3+A12 3′UTR (−) | GGATCGA(T)12CCGCACCGAATGC |
| 9 | CBV3 3′UTR (−) | CCATCGATCCGCACCGAATGCGGAGA |
| 10 | T7/HRV2 5′UTR (+) | CGGCGGCCGCTAATACGACTCACTATAGGT TAAAACTGGA |
| 11 | HRV2 5′UTR (−) | CTGAGCTCCCATGGTGCCAATATATATATTG |
| 12 | HRV2 3′UTR (+) | CTAGATATAGAAATAGTAAACTGATAGTT TATTAGTTTTAT |
| 13 | HRV2 3′UTR (−) | CTAGATAAAACTAATAAACTATCAGTTT ACTATTTCTATAT |
| 14 | T7/PV1 5′UTR (+) | CGGGATCCTAATACGACTCACTATAGGT TAAAACAGCTCTGG |
| 15 | PV1 5′UTR (−) | CTGAGCTCCCATTATGATACAATTGTCTG |
| 16 | PV1 3′UTR (+) | GGTCTAGAAACCCTACCTCAGTCG |
| 17 | PV1 3′UTR (−) | GGGTCGAC(T)50CTCCGAATTA |
| 18 | CBV3 ORF (+) | GGGAGCTCAAGTATCAACGCAAAAG |
| 19 | CBV3 1/2 VP4 (−) | CTTTCGAAGTTGCGGCATCCTTGTAATAA TTAAC |
| 20 | CBV3 VP4 (−) | CTTTCGAAGTGTTGAGAGCTGGTAGTGATT |
| 21 | CBV3 1/3 VP0 (−) | CTTTCGAAGTGTCTGGTTGGGTCGGTTGG |
| 22 | HRV2 ORF (+) | GGGAGCTCAGGTTTCAAGACAAATGTTGG |
| 23 | HRV2 1/2 VP4 (−) | CTTTCGAAGTTGAAGCAGCATCTTTGAAAT |
| 24 | HRV2 VP4 (−) | CTTTCGAAGTCTGTAGTGTTGGTATTCCC |
| 25 | HRV2 1/3 VP0 (−) | CTTTCGAAGTGTTTTCACCAAAAATACCC |
| 26 | PV1 ORF (+) | GGGAGCTCAGGTTTCATCACAGAA |
| 27 | PV1 1/2 VP4 (−) | GTTTCGAAGCCGCGTTACTAGC |
| 28 | PV1 VP4 (−) | CTTTCGAAGTGTTTAGCATTGGGGCTGTT |
| 29 | PV1 1/3 VP0 (−) | CTTTCGAAGTTTGCCCAAAGAGTCCCATG |
| 30 | PV1-IRES (+) | CAACGCGTAAAACCAAGTTCAATAGAAGG |
| 31 | PV1-IRES (−) | CTGAGCTCCCATTATGATACAATTGTCTG |
| 32 | PV1 5′ junction (+) | CCAAGGTCACCTCCAAAATCAG |
| 33 | PV1 5′ junction (−) | CAAATGCGCCATATGTGGTCAGATCCTTGG |
| 34 | CBV3-2A (+) | GACCACATATGGCGCATTTGGACAACAATC |
| 35 | CBV3-2A (−) | CAATGGAACAGGGCATCACCAATTACA TAGAG |
| 36 | PV1 3′ junction (+) | TGGTGATGCCCTGTTCCATTGCATCATCTTC |
| 37 | PV1 3′ junction (−) | GGTACTGGTCACCATATTAGTC |
| 38 | rLuc-TNT(+) | CCCTCGAGACCATGACTTCGAAAGTTTATG ATCC |
| 39 | CBV3 2A(+) | CCCTCGAGACCATGACTTCGAAAGTTTATG ATCC |
| 40 | CBV3 2A(−) | TCTCTAGAACTGTTCCATTGCATCATCTTCC |
| 41 | PV1 2A(+) | CACTCGAGACCATGGGATTCGGACACCAAA ACAAAGC |
| 42 | PV1 2A(−) | CAGGTCGACCTATTGTTCCATGGCTTCTT CTTC |
| 43 | CBV3 2A Cys-Ala(−) | ACCGCCGGCGTCACCTGGTTCGGAAAATCC |
| 44 | CBV3 2A Cys-Ala(+) | GGTGACGCCGGCGGTATCCTAAGGTGTG |
| 45 | c-myc (+) | CTAGCACCATGGAGCAGAAACTCATCTCTG AAGAGGATCTGA |
| 46 | c-myc (−) | AGCTTCAGATCCTCTTCAGAGATGAGTTT CTGCTCCATGGTG |
| 47 | eIF4GI-b (+) | CCAAGCTTATGAACACGCCTTCTCAGCC |
| 48 | eIF4GI-e (+) | TTAAGCTTATGTCTGGGGCCCGCACTG |
| 49 | eIF4GI (−) | CAGCGGCCGCTCAGTTGTGGTCAGACTCC |
| 50 | cpN(−) | TTGCGGCCGCTCAACGGGTGCTAAGGGTT GTCC |
| 51 | cpC(+) | TTAAGCTTGGGCCCCCAAGGGGTGG |
| 52 | eIF4GI-R (−) | TGGCCCACCCCTTGGAGGCTCACGGGTGCTA AGGGTTGTGCGCGCAAGGTTGGCAAAGG ATGG |
| 53 | eIF4GI-Sbf (+) | CTCCTGCAGGCAGCAATCCAGGC |
Restriction sites used for cloning are underlined.
rLuc expression vectors with the human rhinovirus type 2 (HRV2) 5′- and 3′UTRs were constructed by substitution of corresponding CBV3 sequences. Briefly, the CBV3 A50-containing expression construct was digested with NotI and SacI and ligated with the T7/HRV2 5′UTR fragment generated by PCR with primers 10 and 11. The resulting construct was digested with XbaI and ligated with the HRV2 3′UTR generated from two complementary oligonucleotides (12 and 13), yielding HRV2+A50.
To generate PV+A50, pUC19 was digested with BamHI and SalI and ligated with three PCR-amplified fragments encompassing the T7 promoter and 5′ terminal segment (nt 1 to 754) of PV (primers 14 and 15), the rLuc ORF, and PV 3′UTR ending in A50 (primers 16 and 17). A reporter vector containing a heterologous IRES element was constructed by replacement of the PV IRES by its counterpart from HRV2 (21), yielding PV-RIPO+A50.
Reporters with the rLuc ORF fused to viral coding sequences (see Fig. 8A) were generated by insertion of the desired CBV3, HRV2, or PV fragment (primers 18 to 29) into the backbone CBV3+A50, HRV2+A50, PV+A50, or PV-RIPO+A50 rLuc vectors digested with SacI and SfuI.
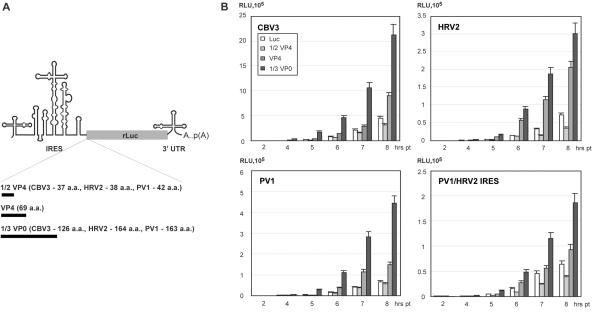
FIG. 8.
(A) Schematic representation of reporter constructs encoding rLuc-P1 fusion proteins. All reporters contained the intact 5′ and 3′UTRs (including A50) of the same species except the PV1/HRV2 chimera carrying an HRV2 IRES in a PV1 background. Various portions of the authentic P1 coding region (black bars) were inserted immediately downstream of the IRES in frame with the rLuc ORF. (B) Effect of P1 coding regions on EV IRES-dependent translation in HeLa cells. Reporter RNAs containing CBV3, PV1(M), or HRV2 sequences were evaluated, as well as a construct containing the HRV2 IRES in a PV1(M) background.
The chimeric virus carrying the PV type 1 (Mahoney) [PV1(M)] IRES in a CBV3 background was constructed as described previously (13), except that a PV IRES segment PCR generated with primers 30 and 31 was used instead of the HRV2 IRES. The PV1/CBV3 2A chimera was cloned with overlapping PCR (24); three PCR-generated fragments encompassing the PV 3′ P1 junction (primers 32 and 33), CBV3 2Apro ORF (primers 34 and 35) and PV 5′ 2B junction (primers 36 and 37) were fused in the second round of PCR performed with external primers 32 and 37. The resulting PCR fragment containing CBV3 2Apro flanked by PV1(M) sequences was digested with BstEII and ligated into a PV1(M) plasmid digested with the same endonuclease.
For in vitro synthesis of capped reporter mRNA, the rLuc ORF was PCR amplified with primers 2 and 38, digested with XhoI-XbaI, and cloned into the corresponding sites of the pTNT vector (Promega). To generate the reporter 3′UTR+A50, we replaced an XbaI-BamHI segment of pTNT by an XbaI-ClaI fragment from 22+A50. The pTNT-EGFP construct used to determine RNA transfection efficiency was generated by subcloning of a SalI-NotI fragment from pEGFP-N1 (Invitrogen) into the corresponding sites of the pTNT vector.
CBV3 or PV1(M) 2Apro ORFs generated with primer pairs 39 and 40 and 41 and 42, respectively, were cloned into the pTNT vector to produce capped 2Apro RNAs by in vitro transcription with T7 RNA polymerase. A mutation of Cys110 to Ala (Cys110→Ala) of CBV3 2Apro (23) was introduced by overlapping PCR using primer pairs 39 and 43 and 40 and 44. The fragment generated in the second round of PCR with primers 39 and 40 was cloned into pTNT.
eIF4GI expression vectors were constructed as described previously (12). Briefly, the c-myc epitope, generated with two complementary oligonucleotides, 45 and 46, was cloned into NheI-HindIII-digested pcDNA3.1, yielding pcDNA-myc. Fragments encoding eIF4GI-b and -e isoforms (9, 11) or N- and C-terminal cleavage products (31) were PCR amplified from full-length eIF4GI cDNA (pSPORT-eIF4GI, kindly provided by R. Lloyd) using primers 47 and 49 plus 48 and 49, or 47 and 50 plus 49 and 51, respectively, digested with HindIII and NotI, and inserted into pcDNA-myc. eIF4GI variants resistant to CBV3 2Apro cleavage (Gly675→Ala and Gly683→Glu) (56) were produced by PCR amplification of an ORF fragment with primers 52 and 53. The fragment was digested with SbfI and PflMI and used to replace a corresponding segment in eIF4GI-b or eIF4GI-e expression vectors, yielding eIF4GI-bR and eIF4GI-eR, respectively.
Luciferase expression assays and in vivo RNA stability.
rLuc expression vectors linearized with XbaI (for reporters lacking 3′ terminal sequences), ClaI (CBV3/HRV2-based vectors), or SalI (PV/PV-RIPO-based vectors) were used as templates for in vitro transcription with T7 RNA polymerase (New England Biolabs). Capped rLuc and CBV3 or PV1(M) 2Apro RNAs were transcribed in the presence of an m7G(5′)ppp(5′)G cap analog (New England Biolabs). After a 1-h incubation at 37°C, in vitro transcription reaction mixtures were treated with RQ1 RNase-free DNase (Promega), and transcripts were purified using the RNeasy Kit (QIAGEN) and quantified by UV spectrophotometry.
To standardize the assay, preliminary tests were conducted to determine the optimal in vitro transcript concentration for transfection and subsequent rLuc activity measurement. For this, HeLa cell monolayers (corresponding to 106 cells) were transfected with serially diluted reporter transcript (twofold dilutions ranging from 0.1 μg to 4 μg). Standard transfection and luc assay conditions yielded a linear correlation of transcript concentration with luc activity.
All luc expression assays used equimolar amounts of in vitro transcript (0.5 μg per plate for truncated RNAs lacking 3′ terminal sequences) to transfect 106 HeLa cells with DMRIE-C reagent (Invitrogen). To assess the effect of 2Apro on IRES-dependent translation, capped CBV3 or PV1(M) 2Apro expression RNA (1 μg per plate) was cotransfected along with reporter RNA. After a 2-h incubation, the transfection mixture was replaced by growth medium containing 10% fetal bovine serum. At different intervals after transfection, the cells were lysed with 0.2 ml of Luc Assay Lysis buffer (Promega). The RNA transfection efficiency in HeLa cells was determined to fall within a range of 60 to 70% by cotransfection of a capped in vitro transcript derived from pTNT-EGFP. rLuc activity assays were performed according to the manufacturer's instructions with a Berthold LB9507 luminometer. Three independent transfection experiments were carried out for each construct, and the data shown represent the average values and standard deviation. RNA stability assays were performed as described previously (13).
One-step EV growth kinetics, transfection of EV-infected cells, and Western blot.
Synchronized infections of HeLa cells were performed as follows. Cell monolayers were incubated with virus at a multiplicity of infection (MOI) of 10 for 30 min while being gently rocked at room temperature. Afterwards, the cells were rinsed three times to remove unbound virus and overlaid with medium containing 2% fetal bovine serum (for growth kinetics) or transfected with reporter RNAs as described above. To analyze virus growth properties, infected cells were incubated at 37°C for specified periods until the cultures were frozen and treated for further testing by plaque assay. A 0-h sample, reflecting the portion of virus bound to the cells prior to particle internalization, was frozen immediately upon removal of unbound particles.
Cell lysates collected at different time points after reporter RNA transfection were subjected to luc assay and Western blotting. For the latter, lysates were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in precast 3 to 8% Tris-acetate or 4 to 12% Bis-Tris NuPAGE gels (Invitrogen) to detect eIF4GI-eIF4GII or PABP, respectively, and transferred to PROTRAN nitrocellulose membrane (Schleicher & Schuell). After being blocked overnight in 5 to 7% nonfat milk in TBST (100 mM Tris, 150 mM NaCl, 0.05% Tween-20, pH 8.0), membranes were incubated with α-eIF4GI directed against N-terminal epitopes, α-eIF4GI directed against C-terminal epitopes, or α-PABP (kindly provided by V. Pain, N. Sonenberg, and J. Keene, respectively). Following three washes with TBST, the membranes were treated with biotinylated α-rabbit immunoglobulin G (Vector Labs), rinsed, developed with streptavidin-peroxidase conjugate (Roche), and visualized with ECL Western blotting detection reagents (Amersham). Polyclonal rabbit α-eIF4GII antibody was raised against the N-terminal epitope CEENGEEAEPVRNGAESVSEGEGIDA (amino acids [aa] 569 to 593) (17) by standard protocols.
Expression of exogenous eIF4GI.
To assess the effect of eIF4GI overexpression on IRES-mediated translation, HeLa cells were transfected with the c-myc-tagged eIF4GI-b, eIF4GI-e, eIF4GI-bR, eIF4GI-eR, cpN, and/or cpC expression constructs (cpN and cpC are 2Apro-induced N-terminal and C-terminal cleavage products) or with pcDNA3.1 vector (1 μg per plate) using Lipofectamine 2000 (Invitrogen). Twenty-four hours after DNA transfection, cells were washed with serum-free medium, infected with CBV3 at an MOI of 10 or mock infected, transfected with reporter RNA, and subjected to the luc assay and Western blotting as described above. α-c-myc antibody 9E10 (Sigma) and biotinylated α-mouse immunoglobulin G (Vector Laboratories) were used to detect exogenous eIF4GI.
Ribosomal profiles.
HeLa S3 in vitro translation extracts (5) were incubated at 37°C with radiolabeled reporter RNAs (3 ng/μl) in buffer containing 20 mM creatine phosphate, 0.1-mg/ml creatine kinase, 0.1 mM spermidine, 100 μM amino acids, 90 mM potassium acetate, 3 mM Mg diacetate, and 16 mM HEPES (pH 7.4) in the presence of 0.5 mM cycloheximide. At the indicated time points, reactions were diluted in ice-cold gradient buffer (39), applied onto 5 to 20% sucrose gradients, and centrifuged at 37,000 rpm at 4°C for 3 h 45 min in an SW41 rotor. Absorbance spectra and fractions (each, 0.52 ml) were obtained for each gradient with an ISCO density gradient fractionation system. Collected fractions were subjected to liquid scintillation counting (Beckman). RNA distribution throughout the gradient is presented as a percentage of he total amount of radiolabeled RNA (Fig. 1D).
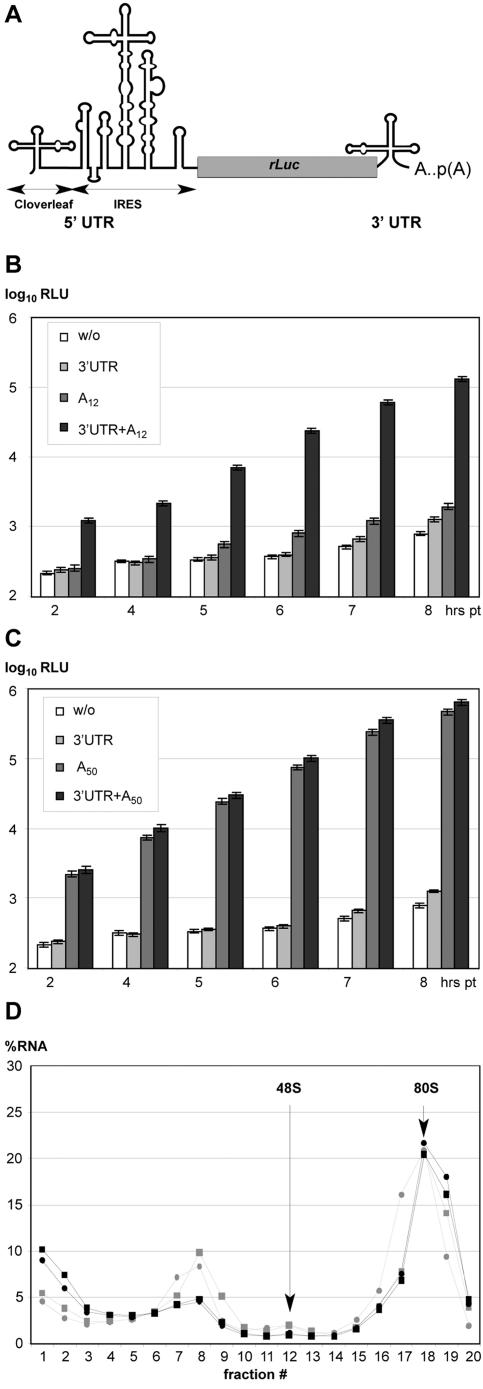
FIG. 1.
Effect of 3′ terminus on EV IRES-dependent translation. (A) Genetic structure of IRES reporter constructs. (B and C) Efficiency of rLuc expression from reporters with diverse 3′ termini. w/o, termination codon of rLuc ORF; 3′UTR, CBV3 3′UTR; A12/A50, 12/50-mer poly(A); 3′UTR+A12/A50, CBV3 3′UTR and 12/50-mer poly(A). rLuc activity was measured in HeLa cell lysates collected at designated intervals after transfection with reporter RNAs carrying the specified 3′ terminus. rLuc expression of w/o, 3′UTR, and A12 was marginal (B), reaching <0.5% of 3′UTR+A50 at 8 h PT (C). The data for all reporter RNAs were generated in the same experiment. (D) Ribosomal profile of A50 (circles) and 3′UTR+A50 (squares) RNAs after 5-min (grey circles and squares) and 10-min (black circles and squares) incubation in HeLa cell in vitro translation extracts.
RESULTS
The 3′UTR and poly(A) tail synergistically stimulate EV IRES-mediated translation in vivo.
We have shown previously that EV 3′UTRs can modulate particle propagation in a cell-type-specific manner (13). To examine the effects of the 3′UTR in conjunction with poly(A) on IRES-mediated translation in vivo, we generated a series of rLuc reporter gene constructs under translation control of the CBV3 IRES (Fig. 1A). The 5′ terminus of all reporter RNAs was formed by the viral cloverleaf structure lacking an m7GpppN-cap (Fig. 1A).
Since the CBV3 genome 3′ terminus consists of the 3′UTR and a genetically encoded poly(A) tail, we analyzed their effect on IRES-driven translation separately. We generated reporters with the following 3′ termini: a construct where the rLuc termination codon is the 3′ terminus, the CBV3 3′UTR alone, a poly(A) tail alone, or the CBV3 3′UTR and poly(A) tail combined (Fig. 1). The average length of picornavirus poly(A) tails has been determined to be 35 to 60 nt (1); we confirmed this range for CBV3 and PV inith infected HeLa cells (data not shown). To test the effects of poly(A) length on translation, we used reporters ending with A12 (Fig. 1B) or A50 (Fig. 1C). In vitro transcript RNAs were purified, quantified, and prepared for transfection into HeLa cells; measurement of rLuc activity in cell lysates at various intervals posttransfection (PT) revealed the effect of a 3′-terminal structure on translation efficiency (Fig. 1B and C).
A reporter lacking all 3′UTR sequences yielded near-background rLuc activity (Fig. 1B). Addition of the CBV3 3′UTR or A12 alone barely increased rLuc levels, suggesting that low levels of rLuc expression from the latter were not due to 3′-5′ exonucleolytic degradation at its unprotected 3′ end (Fig. 1B). However, combination of the CBV3 3′UTR and A12 induced CBV3 IRES-dependent translation by up to 150 fold (Fig. 1B). This stimulatory effect was evident only when CBV3 3′UTR+A12 was supplied in cis (data not shown), indicating that intramolecular interactions with the 3′ terminus were required for IRES stimulation. Unlike A12, the presence of A50 alone strongly stimulated IRES translation (Fig. 1C). Combining the CBV3 3′UTR and A50 resulted in a further enhancement of CBV3 IRES translation (Fig. 1C), although the stimulatory effect of the CBV3 3′UTR on A50 was less pronounced than that on A12.
To determine if the stimulatory effect of the 3′UTR on IRES-mediated translation is due to enhanced initiation, we analyzed formation of ribosomal complexes on polyadenylated (A50) reporter RNAs with and without 3′UTR in vitro (Fig. 1D). We obtained very similar patterns of RNA distribution throughout the ribosomal profiles for both RNAs, regardless of the presence of a 3′UTR (Fig. 1D). This suggests that the minor stimulatory role of the 3′UTR to RNAs with 50-residue poly(A) tails is due to postinitiation events, possibly termination and/or release of the completed rLuc polypeptide.
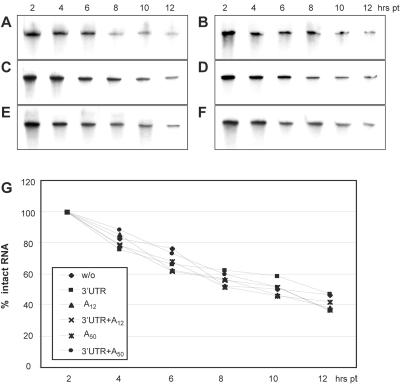
The 3′ terminal sequences have no effect on reporter RNA stability.
Poly(A)'s role as a guardian of mRNA integrity (54) suggests that the influence of poly(A) tail length on translation could reflect differential rates of transcript decay. Therefore, we analyzed the in vivo decay of [α32P]UTP-labeled reporters with variable 3′ termini tested by translation assays (Fig. 2). Total RNA preparations from lysed cells were subjected to denaturing gel electrophoresis, and the proportion of intact reporter RNA associated with the transfected cells was calculated at various time points PT. No significant discrepancy in the stability of different RNA species was observed (Fig. 2), suggesting that stimulatory effects of 3′UTRs and/or poly(A) tails on CBV3 IRES-driven translation were not due to enhanced RNA stability.
FIG. 2.
Stability of reporter RNAs with different 3′ termini in HeLa cells. w/o (A); 3′UTR (B); A12 (C); 3′UTR+A12 (D); A50 (E); 3′UTR+A50 (F). Total RNA from transfected cells was separated by gel electrophoresis in 4% polyacrylamide gel-8 M urea, and radiolabeled reporter RNA was detected and quantified by phosphorimaging. (G) Percentage of remaining intact reporter RNA present in cells at the indicated intervals PT.
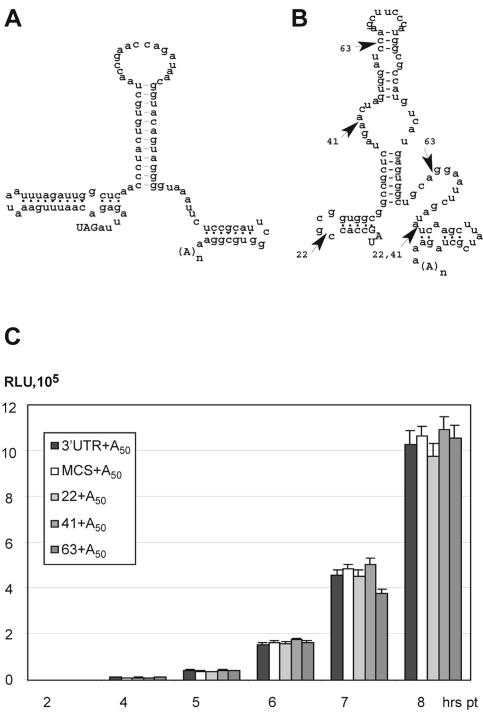
Stimulation of IRES-dependent translation by 3′UTRs occurs independently of primary sequence and structure.
We investigated whether translation stimulation by the CBV3 3′UTR depends on its primary sequence or structure. We constructed a set of IRES-rLuc expression vectors featuring A50 downstream of unrelated 3′UTRs of various length (Fig. 3). A fragment of the pBlueScript MCS with size (100 nt) and predicted secondary structure arrangement reminiscent of the CBV3 3′UTR (Fig. 3B) served as 3′UTR with irrelevant primary sequence. In addition, several internal deletions within MCS yielded 3′UTRs of 22, 41, or 63 nt in length (Fig. 3B). Reporter constructs containing the artificial MCS 3′UTR or the authentic CBV3 counterpart exhibited indistinguishable rLuc expression levels (Fig. 3C). Similarly, none of the deletions introduced in the MCS sequence had significant effects on CBV3 IRES-dependent translation (Fig. 3C), suggesting that neither the size beyond 22 nt nor the origin of the sequences preceding the poly(A) tail determines stimulation of IRES-driven translation.
FIG. 3.
Effect of 3′UTR structure and size on IRES translation. (A) Predicted secondary structure of the CBV3 3′UTR (42). (B) A fragment encompassing the pBlueScript MCS or its internal deletion variants (indicated as 22, 41, and 63 by arrowheads) were used to replace the native CBV3 3′UTR. The secondary structure of the MCS fragment was derived from sequence analysis with Mfold 3.1 (57). All reporters carry an A50 tail. (C) rLuc expression from reporter RNAs containing the specified 3′UTR.
CBV3 infection stimulates EV IRES-mediated translation.
Poly(A)-dependent IRES-mediated translation may reflect recruitment of PABP, which has been proposed to stimulate picornavirus IRES activity via eIF4G interaction (48). Since eIF4GI, eIF4GII, and PABP, components of the canonical template-closing RNP network, are degraded during EV infection, conditions within EV-infected cells may profoundly affect the IRES translation rate. To test this hypothesis, we assayed CBV3 IRES-driven translation of reporter RNAs containing the cognate 3′UTR and A50 tail in EV-infected cells. To achieve the authentic pattern of eIF4G and PABP cleavage in living cells, HeLa cell monolayers were infected with either CBV3 or PV, followed by reporter RNA transfection (see Materials and Methods). One-step growth kinetics analysis of HeLa cells revealed parallel propagation rates for both viruses (Fig. 4A). Virus-infected cells were transfected with reporter RNA, and rLuc expression was assayed over a course of 6 h (Fig. 4B). We did not extend the assay period beyond 6 h (unlike all other translation assays) because translation levels dropped precipitously with the onset of cytopathic effects (data not shown). Surprisingly, CBV3 infection enhanced IRES translation ∼3 fold, while PV had a negligible effect (Fig. 4B). Fading IRES-mediated translation in PV infected cells at 6 h PT (Fig. 4B) likely was due to the onset of cytopathic effects and generalized disruption of translation activity. Since both CBV3 and PV are known to induce identical alterations of the host cell translation machinery, this result was unexpected.
FIG. 4.
Effect of concomitant EV infection on IRES-driven translation. (A) One-step growth curves of CBV3 and PV1(M) in HeLa cells. (B) rLuc activity in mock-infected cells transfected with reporter RNA carrying the CBV3 IRES and CBV3 3′UTR+A50 and in cells infected with CBV3 or PV1(M). (C and D) Kinetics of PABP (C) and eIF4GI and -II (D) degradation in HeLa cells at specified intervals after infection with CBV3 or PV1. Arrowheads indicate intact proteins or their cleavage products (cp). A C-terminal specific (cpC) α-eIF4GI was used for Western blotting (48). (E) rLuc activity in mock-infected cells transfected with reporter RNA carrying the PV1(M) IRES and 3′UTR+A50 and in cells infected with CBV3 or PV1(M).
IRES stimulation by EV infection coincides with proteolytic activity of 2Apro.
To investigate the molecular mechanism of differential IRES stimulation by CBV3 and PV infection, we analyzed the kinetics of viral cleavage of PABP, eIF4GI, and eIF4GII (Fig. 4C and D). Western blot analysis demonstrated similar kinetics of PABP degradation with both viruses (Fig. 4C). Although PABP cleavage products could be detected at 4 h postinfection (hpi), intact protein was still abundant in cells at 10 hpi with CBV3 and PV (Fig. 4C). PV infection produced a unique proteolytic cleavage product not observed with CBV3-infected cells (Fig. 4C). In contrast, the kinetics of eIF4GI and eIF4GII cleavage differed markedly in CBV3- and PV-infected cells (Fig. 4D); eIF4GI-eIF4GII degradation and the appearance of cleavage products occurred much faster in the former. At 3 hpi with CBV3, intact eIF4GI and eIF4GII were already undetectable, whereas in PV-infected cells complete cleavage occurred with a 4- to 5-h delay (Fig. 4D).
Enhanced IRES stimulation in CBV3-infected cells might be due to (i) species-specific effects, preventing CBV3 IRES stimulation by PV; (ii) inherent higher CBV3 IRES efficiency, resulting in elevated levels of viral gene products (e.g., 2Apro); or (iii) intrinsically higher CBV3 2Apro activity, leading to accelerated eIF4GI-eIF4GII degradation. We tested these hypotheses separately. First, we tested the effect of CBV3-PV infection on translation at the PV IRES (Fig. 4E). For this purpose, we replaced the CBV3 5′ and 3′UTRs in the rLuc reporter construct with the equivalent PV structures. Transfection of PV reporter RNA into HeLa cells previously infected with CBV3 and PV produced an identical stimulation profile, as observed with the CBV3 construct (Fig. 4E), ruling out possible species-specific effects. Next, to test whether intrinsic IRES efficiency determines viral gene expression levels and eIF4G degradation, we constructed a CBV3 chimera replicating under control of the PV IRES. IRES exchange had no effect on the kinetics of CBV3 gene expression, growth kinetics, or eIF4GI cleavage; the chimera stimulated reporter gene expression at the CBV3 and PV IRESs to the same extent as wild-type (wt) CBV3 (data not shown).
Last, we tested IRES stimulation and eIF4GI cleavage by CBV3 2Apro in a PV background to compare the CBV3 and PV enzymes side by side (Fig. 5). Insertion of the CBV3 2Apro coding sequence into the PV genome altered the P1-P2 junction, potentially impeding 2Apro cis cleavage (Fig. 5A); indeed, growth of the 2Apro chimera was slightly protracted compared to that of wt CBV3 (Fig. 5B). Nevertheless, CBV3 2Apro in a PV context stimulated CBV3 IRES translation and cleaved eIF4GI almost as efficiently as wt CBV3 (Fig. 5C and D). Slightly slower eIF4GI degradation by the 2Apro chimera probably reflected impaired 2Apro P1-P2 cis cleavage. Since the cleavage kinetics of eIF4GI and eIF4GII in CBV3- and PV-infected cells progressed in parallel (Fig. 4D), we only analyzed eIF4GI.
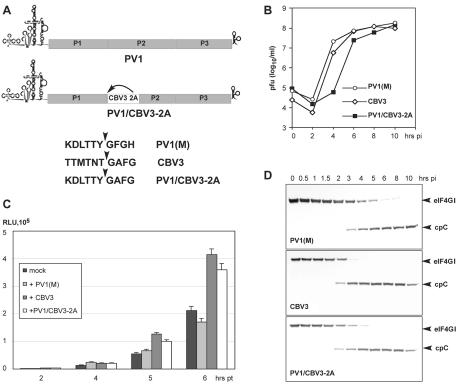
FIG. 5.
Role of CBV3 2Apro in modulation of IRES-dependent translation. (A) Genetic structure of wt PV1 (top) and a chimeric PV1/CBV3 2A virus (bottom) encoding CBV3 2Apro in a PV1(M) background. The autoproteolytic cleavage site for CBV3 2Apro (P1-P2) is indicated (arrow). The amino acid sequence of the P1-P2 junction of PV1(M), CBV3, and the chimera are shown. (B) One-step growth kinetics of parental PV1(M), CBV3, and the PV1/CBV3 2A chimera. (C) Effect of infection with specified viruses on rLuc expression levels. (D) Comparison of eIF4GI degradation rates in HeLa cells infected at an MOI of 10 with PV1(M), CBV3, or the PV1/CBV3 2A chimera.
IRES stimulation is mediated by 2Apro alone.
Differential IRES stimulation and eIF4G cleavage by CBV3 and PV correlates with their 2Apro activities. To exclude contributing effects of other viral proteins (e.g., 3Cpro), we analyzed IRES-driven reporter expression and eIF4GI cleavage in cells cotransfected with transcripts encoding CBV3 or PV 2Apro (Fig. 6A). We found that 2Apro was sufficient to achieve IRES stimulation at levels exceeding those in infected cells (Fig. 6A). Moreover, this assay confirmed intrinsically higher IRES stimulation and accelerated eIF4GI cleavage with CBV3 versus PV 2Apro (Fig. 6A and B). Stronger IRES stimulation by 2Apro coexpression relative to virus infection (compare Fig. 6A to Fig. 4B) may be due to higher 2Apro levels in transfected versus infected cells and/or to the lack of translation inhibition by 3Cpro in the former (J. A. Bonderoff, N. M. Kuyumcu-Martinez, and R. E. Lloyd, Abstr. XIIIth Meet. Eur. Study Group Mol. Biol. Picornaviruses, abstr. C06, 2005). The proportion of uncleaved eIF4GI in 2Apro RNA cotransfected cells likely reflects intact protein in untransfected cells (RNA transfection efficiency in HeLa cells was 60 to 70%; see Materials and Methods).
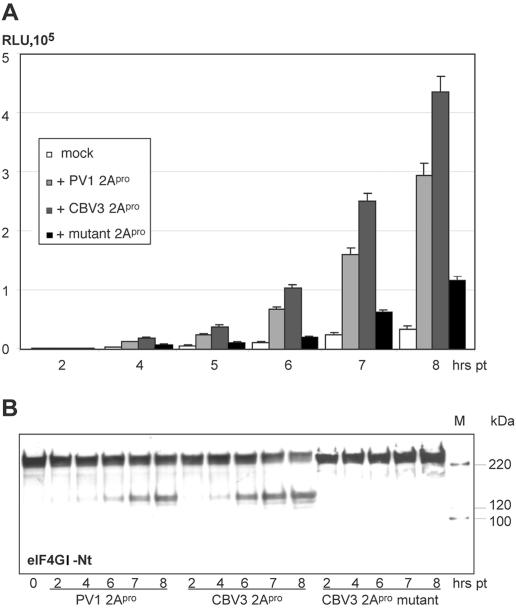
FIG. 6.
Stimulation of IRES-mediated translation by 2Apro coexpression. (A) HeLa cells were transfected with CBV3 IRES reporter RNA alone (mock) or cotransfected with capped PV, CBV3 2Apro RNAs, or a CBV3 2Apro active-site mutant. (B) Expression and proteolytic activity of 2Apro were detected by its ability to induce eIF4GI cleavage. Immunoblotting was performed with an α-eIF4GI antibody against an N-terminal epitope (eIF4GI-Nt) (12).
Covariation of IRES stimulation and eIF4GI cleavage suggests a causal relation. Therefore, we evaluated IRES stimulation by a CBV3 2Apro variant with an active-site mutation (23): substitution of amino acid residue 110 (Cys→Ala) abolished eIF4GI proteolysis (Fig. 6B). Although stimulation was reduced, proteolytically inactive CBV3 2Apro enhanced IRES-mediated translation ∼2.5 fold, suggesting that residual IRES stimulation is independent of proteolytic activity.
The effect of eIF4GI on EV IRES translation in vivo.
To address a role for 2Apro-mediated eIF4GI degradation in IRES stimulation, we followed a strategy first reported by Zhao et al. (56). First, we constructed expression vectors encoding eIF4GI isoforms via initiation at two of the five authentic initiating AUGs: b (41), and e (197) (numbers in paretheses correspond to the amino acid numbers of eIF4GI-a) (Fig. 7A). To enhance expression and track exogenous eIF4GI in transfected cells, we added an N-terminal c-myc epitope tag (Fig. 7A) (12). We evaluated eIF4GI-b and eIF4GI-e separately, since the latter lacks the PABP-binding domain (Fig. 7A). Furthermore, we constructed eIF4G-b and eIF4GI-e variants resistant to 2Apro cleavage (eIF4G-bR and eIF4GI-eR) by altering the primary (aa 682 to 683) and secondary (aa 674 to 675) eIF4GI cleavage sites (Fig. 7A) (56). HeLa cells were transfected with plasmids expressing eIF4GI-b-eIF4GI-bR or eIF4GI-e-eIF4GI-eR or with pcDNA3.1 vector DNA. DNA transfection efficiency was determined to be ∼80% by cotransfection of pEGFP-N1 expression vector DNA (data not shown). After incubation at 37°C for 24 h, the cells were either mock infected or infected with CBV3 and transfected with a CBV3 IRES reporter to evaluate translation efficiency (Fig. 7B).
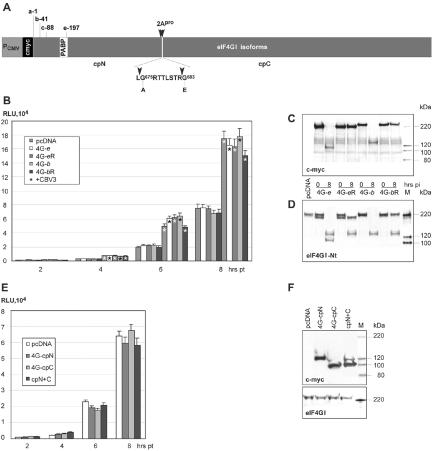
FIG. 7.
Effect of eIF4GI overexpression on EV IRES-mediated translation. (A) eIF4GI expression construct structure; an N-terminal c-myc tag (black box) is fused to the eIF4GI ORF at authentic initiation codons b and e (9, 11). PABP binding (white box) and EV 2Apro cleavage sites (arrowheads) are indicated. The 2Apro-induced N-terminal and C-terminal cleavage products are designated cpN and cpC, respectively. Two amino acids, Gly683 and Gly675 (56), were mutated to generate 2Apro-resistant eIF4GI variants. (B) rLuc translation in HeLa cells transiently expressing exogenous eIF4GI isoforms and their cleavage-resistant variants. (C) Expression of c-myc-tagged eIF4GI isoforms in HeLa cells at 0 and 8 hpi with CBV3. The integrity of exogenous eIF4GI was determined by Western blotting with α-c-myc. (D) Expression levels and integrity of total cellular eIF4GI were determined by Western blotting with α-eIF4GI-Nt. (E) CBV3 IRES-driven rLuc translation in HeLa cells expressing eIF4GI cleavage products. (F) Expression levels of eIF4GI proteolytic cleavage products and endogenous eIF4GI.
Comparison of cells expressing exogenous eIF4GI isoforms with those transfected with vector alone revealed that enhanced eIF4GI levels did not modify IRES translation in uninfected cells (Fig. 7B). CBV3 infection of HeLa cells expressing cleavage-susceptible eIF4GI-b and -e isoforms resulted in complete degradation of both exogenous (Fig. 7C) and endogenous (Fig. 7D) eIF4GI by 8 hpi. In contrast, in cells expressing eIF4GI-bR and -eR, intact exogenous product was detected throughout the assay interval (Fig. 7C). The expression levels of exogenous eIF4GI variants were equivalent to those of intrinsic protein in uninfected cells transfected with empty vector (Fig. 7D).
CBV3-mediated IRES stimulation in cells expressing cleavage-resistant forms of eIF4GI did not differ from those expressing cleavage-susceptible variants (Fig. 7B). Thus, the presence of intact eIF4GI in the former did not preclude IRES stimulation by CBV3 infection. This rules out a possible inhibitory effect of intact eIF4GI on EV IRES-mediated translation. We separately evaluated a possible stimulatory role of eIF4GI proteolytic cleavage products by analyzing IRES activity in cells expressing N- and C-terminal fragments (cpN and cpC) either individually or in combination (Fig. 7F). Expression of exogenous cpN, cpC, or both combined had no effect on IRES-dependent reporter translation (Fig. 7E).
IRES-driven translation is modulated by picornaviral coding sequences.
We extended our analyses of cis determinants of IRES function to the viral 5′ proximal ORF, a scenario suggested by precedence with the hepatitis C virus IRES (34, 44). We generated a series of rLuc constructs driven by the CBV3, HRV2, or PV IRES with authentic corresponding 3′UTRs terminating in A50. Various segments (ranging from 0 to 492 nt) of the viral coding region were fused to the rLuc ORF (Fig. 8). The insert sizes varied for different virus species (Fig. 8), because we considered predicted secondary structure arrangements in the design of our constructs, to avoid disruption of putative stem-loop structures (data not shown).
Similar effects of 5′ proximal ORF sequences were observed for all type 1 picornavirus IRESs (Fig. 8). While insertion of a portion of the VP4 coding region (111 to 126 nt) slightly reduced IRES performance, further addition of coding sequences dramatically stimulated IRES-dependent reporter expression (Fig. 8). Optimal stimulation of translation, outperforming constructs lacking viral coding sequences at least fivefold, was achieved with approximately one-third of VP0 coding sequences fused to rLuc (Fig. 8). Insertion of larger ORF fragments stimulated rLuc activity to a lesser degree (data not shown), but this was due to adverse effects on rLuc enzymatic activity by the fused sequences rather than reduced expression levels. This was determined by immunoprecipitation of [35S]Met-labeled rLuc from IRES-reporter transfected cells (data not shown). We found that all CBV3 IRES-driven constructs featuring variable portions of the VP0 coding sequence exhibited similar decay profiles in vivo (data not shown). This excluded effects on rLuc expression levels due to transcript stability covarying with ORF structure. To test if ORF sequences only stimulated homologous IRESs, we tested PV constructs in which the cognate IRES was replaced by the HRV2 IRES. The PV reporter with a heterologous HRV2 IRES exhibited rLuc expression and coding sequence stimulation patterns identical to those of the parental PV construct (Fig. 8).
EV genomes compete with capped mRNAs by highly efficient translation initiation.
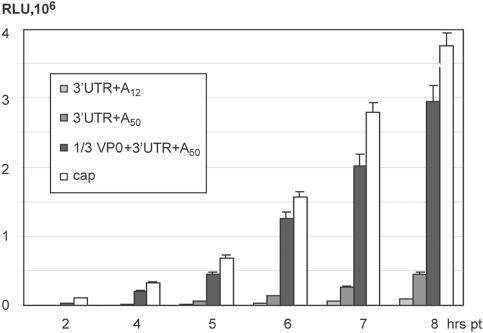
Our experiments have identified the key cis- and trans-acting factors influencing translation rate at picornaviral type 1 IRESs. Early after infection, few viral genomes need to be translated efficiently in the absence of viral gene products and their effect on the host's translation apparatus. We evaluated the relative translation efficiency of IRES-driven RNA reporters and capped messages containing the 5′ β-globin leader and poly(A50) (Fig. 9). IRES-driven reporter constructs containing all cis-acting stimulatory sequences were translated in vivo at levels equivalent to those of capped mRNAs (Fig. 9).
FIG. 9.
Cumulative effects of EV cis-acting sequence elements on EV IRES translation in vivo. Efficiency of rLuc expression from specified CBV3 IRES-containing reporter RNAs is shown in comparison with a capped reporter RNA containing the β-globin 5′ leader and poly(A) tail.
DISCUSSION
In contrast to host mRNAs (including those purported to contain IRESs) picornaviral RNAs are uncapped (37) and hence rely uniquely on their IRESs to attract ribosomes. Picornavirus translation is considered to depend on viral manipulation of the host's protein synthesis apparatus for competition with host mRNAs. We show here that, although IRES efficiency undoubtedly benefits from expression of viral gene products, viral IRES function in the proper context is surprisingly robust in uninfected cells. In the presence of authentic genomic termini and a 5′-proximal portion of the viral ORF, the IRES-driven gene expression rate equals conventional translation at capped templates. Efficient translation of the viral genome may be particularly important in early phases of the virus life cycle, with few viral genomes released into a hostile environment prior to accumulation of viral gene products.
In accordance with previous reports, we found that IRES function strictly depends on poly(A) tail length (5, 36, 48). In our system, A12 is unable to compensate for 3′UTR deletion, but A50 stimulation exceeds the level of 3′UTR+A12. Since A12 may accommodate only a single PABP molecule (46), an upsurge of translation with longer poly(A) tails suggests a role of PABP in the poly(A)-mediated stimulation of IRES translation (5). This stimulatory role has been proposed to rely, as with cap-dependent translation, on eIF4G-PABP bridging (36, 48). However, in CBV3-infected cells and in in vitro translation extracts, efficient poly(A)-dependent IRES function occurs in the absence of intact eIF4G. Similarly, eIF4G integrity was shown to be dispensable for cellular IRESs (51), and the eIF4GI middle apoptotic fragment harboring eIF3- and eIF4A-binding sites was sufficient for the functional replacement of the full-length protein (25). Moreover, elegant experiments with immunodepleted in vitro translation extracts have suggested PABP-independent stimulation of translation by poly(A) for cellular IRES elements (51). Although we have not provided direct evidence for a role of PABP in EV IRES translation, our observations support its involvement (see below).
In addition, we found the viral 3′UTR to enhance translation at the IRES synergistically with poly(A). 3′UTR length has been reported to influence translation rate of capped mRNAs; similar to our data with 3′UTRs of at least 22 nt, Tanguay et al. reported stimulation of cap-dependent translation by 3′UTRs with a minimum of 27 nt (49). As with capped mRNAs, 3′UTR primary sequence is irrelevant for IRES stimulation, and 3′UTRs beyond 22 nt do not produce incremental translation rate increases. The stimulatory effect of the viral 3′UTR is likely due to postinitiation events, because it has no influence on RNA incorporation into 80S monosomes in vitro. We favor an interpretation by Tanguay et al. that the 3′UTR may physically separate ribosomes approaching the termination codon from existing PABP-poly(A) complexes. A spacer function of the 3′UTR aiding in proper termination and ribosome release lends further indirect support to a role for PABP in poly(A)-mediated stimulation of IRES translation.
We observed strong translation enhancement at EV and rhinovirus IRESs by authentic 5′-proximal ORFs. A similar stimulatory role of downstream coding sequences has been described for the IRESs of hepatitis C virus (34, 44), classical swine fever virus (15), and the picornavirus hepatitis A virus, which is structurally related to type 2 IRESs (19). Since the 5′-proximal portion of the viral ORF is fused in frame to the luc ORF, we speculate that IRES stimulation is due to its influence on the structural arrangement of the template. Thus, cis-acting RNA elements alone potently stimulate IRES-dependent translation, and properly configured viral templates efficiently compete with capped mRNAs.
In addition to cis-acting viral sequences, we show IRES stimulation upon CBV3 infection. Surprisingly, although all EVs cause identical modifications of the translation apparatus, CBV3 infection stimulates both its own and the PV IRES to a far greater extent than PV. The IRES stimulatory effect of virus infection is recapitulated with viral 2Apro alone, and the CBV3-PV differential reflects the intrinsic stimulatory activity of their 2Apro gene products.
Stimulation of IRES-driven translation by 2Apro has been assigned to direct IRES trans activation (22) or its proteolytic activity targeting eIF4G (4). Although 2Apro proteolytic activity certainly plays a major role, IRES stimulation by a 2Apro active-site mutant incapable of eIF4G cleavage supports trans-acting functions independent of proteolytic activity. A possible stimulatory role of eIF4G degradation has been ascribed to (i) enhanced efficiency of the C-terminal eIF4G fragment in promoting internal initiation (10, 35, 38), (ii) IRES repression by intact eIF4G (56), or (iii) translation shutoff of capped messages conveying a competitive advantage to the IRES. Our results do not support a role for 2Apro-mediated eIF4G degradation in IRES stimulation: ectopic expression of 2Apro-generated eIF4GI cleavage products did not enhance IRES-driven gene expression in vivo. Similarly, Roberts et al. reported no effects of eIF4GI cleavage products on EV IRES translation in vivo (45); contrary results from in vitro translation extracts (10, 38) may reflect inherent differences in these assay systems. Expression of cleavage-resistant eIF4GI variants in CBV3-infected cells did not prevent stimulation of IRES-mediated translation, ruling out possible inhibitory effects of intact eIF4G. The failure to prevent stimulation by CBV3 infection in cells expressing cleavage-resistant eIF4G isoforms argues against IRES stimulation through competitive advantage in the absence of intact eIF4G.
Taken together, our results are inconsistent with a hypothetical switch from cap-dependent to IRES-mediated translation caused by EV 2Apro cleavage of eIF4G alone. Many other proteins modified during EV infection could modulate IRES activity. For example, IRES trans-acting factors such as the La autoantigen and polypyrimidine tract-binding protein are cleaved in infected cells and result in opposing effects on PV IRES function (2, 47). PV infection or transient 2Apro expression increases permeability of the nuclear envelope, resulting in translocation of nuclear proteins into cytoplasm (3). Proteomic analysis of HRV1A-infected cells also revealed a number of affected proteins including those engaged in transcription and translation (S. Amineva, R. Brockman-Schneider, and J. Gern, Abstr. XIIIth Meet. Eur. Study Group Mol. Biol. Picornaviruses, abstr. G10, 2005). The role of these events in modulation of IRES-dependent translation has yet to be determined.
Acknowledgments
We are grateful to N. Chapman (University of Nebraska, Omaha) for CBV3 cDNA, N. Sonenberg (McGill University, Montreal, Canada) and V. Pain (University of Sussex, Brighton, United Kingdom) for α-eIF4GI antibodies, J. Keene (Duke University, Durham, NC) for α-PABP antibodies, and R. Lloyd (Baylor College of Medicine, Houston, TX) for pSPORT-eIF4GI. We thank S. Bradrick for critical reading of the manuscript.
R.N.G. is a Howard Hughes Medical Institute Medical Research Training Fellow. This work was supported by Public Health Service grant CA87537 (M.G.). M.G. is the recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences.
REFERENCES
- 1.Ahlquist, P., and P. Kaesberg. 1979. Determination of the length distribution of poly(A) at the 3′ terminus of the virion RNAs of EMC virus, poliovirus, rhinovirus, RAV-61 and CPMV and of mouse globin mRNA. Nucleic Acids Res. 7:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Back, S. H., Y. K. Kim, W. J. Kim, S. Cho, H. R. Oh, J.-E. Kim, and S. K. Jang. 2002. Translation of polioviral mRNA is inhibited by cleavage of polypyrimidine tract-binding proteins executed by polioviral 3Cpro. J. Virol. 76:2529-2542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belov, G. A., P. V. Lidsky, O. V. Mikitas, D. Egger, K. A. Lukyanov, K. Bienz, and V. I. Agol. 2004. Bidirectional increase in permeability of nuclear envelope upon poliovirus infection and accompanying alterations of nuclear pores. J. Virol. 78:10166-10177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belsham, G. J., and R. J. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 5.Bergamini, G., T. Preiss, and M. W. Hentze. 2000. Picornavirus IRESes and the poly(A) tail jointly promote cap-independent translation in a mammalian cell-free system. RNA 6:1781-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, H. D., N. Sonenberg, and D. Baltimore. 1985. Poliovirus mutant that does not selectively inhibit host cell protein synthesis. Mol. Cell. Biol. 5:2913-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonneau, A. M., and N. Sonenberg. 1987. Proteolysis of the p220 component of the cap-binding protein complex is not sufficient for complete inhibition of host cell protein synthesis after poliovirus infection. J. Virol. 61:986-991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Borovkov, A. Y., and M. I. Rivkin. 1997. XcmI-containing vector for direct cloning of PCR products. BioTechniques 22:812-814. [DOI] [PubMed] [Google Scholar]
- 9.Bradley, C. A., J. C. Padovan, T. L. Thompson, C. A. Benoit, B. T. Chait, and R. E. Rhoads. 2002. Mass spectrometric analysis of the N terminus of translational initiation factor eIF4G-1 reveals novel isoforms. J. Biol. Chem. 277:12559-12571. [DOI] [PubMed] [Google Scholar]
- 10.Buckley, B., and E. Ehrenfeld. 1987. The cap-binding protein complex in uninfected and poliovirus-infected HeLa cells. J. Biol. Chem. 262:13599-13606. [PubMed] [Google Scholar]
- 11.Byrd, M. P., M. Zamora, and R. E. Lloyd. 2002. Generation of multiple isoforms of eukaryotic translation initiation factor 4GI by use of alternate translation initiation codons. Mol. Cell. Biol. 22:4499-4511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coldwell, M. J., L. Hashemzadeh-Bonehi, T. M. Hinton, S. J. Morley, and V. M. Pain. 2004. Expression of fragments of translation initiation factor eIF4GI reveals a nuclear localisation signal within the N-terminal apoptotic cleavage fragment N-FAG. J. Cell Sci. 117:2545-2555. [DOI] [PubMed] [Google Scholar]
- 13.Dobrikova, E., P. Florez, S. Bradrick, and M. Gromeier. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. USA 100:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 15.Fletcher, S. R., I. K. Ali, A. Kaminski, P. Digard, and R. J. Jackson. 2002. The influence of viral coding sequences on pestivirus IRES activity reveals further parallels with translation initiation in prokaryotes. RNA 8:1558-1571. [PMC free article] [PubMed] [Google Scholar]
- 16.Gallie, D. R. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev. 5:2108-2116. [DOI] [PubMed] [Google Scholar]
- 17.Gradi, A., H. Imataka, Y. V. Svitkin, E. Rom, B. Raught, S. Morino, and N. Sonenberg. 1998. A novel functional human eukaryotic translation initiation factor 4G. Mol. Cell. Biol. 18:334-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gradi, A., Y. V. Svitkin, H. Imataka, and N. Sonenberg. 1998. Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. USA 95:11089-11094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graff, J., and E. Ehrenfeld. 1998. Coding sequences enhance internal initiation of translation by hepatitis A virus RNA in vitro. J. Virol. 72:3571-3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grifo, J. A., S. M. Tahara, M. A. Morgan, A. J. Shatkin, and W. C. Merrick. 1983. New initiation factor activity required for globin mRNA translation. J. Biol. Chem. 258:5804-5810. [PubMed] [Google Scholar]
- 21.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hambidge, S. J., and P. Sarnow. 1992. Translational enhancement of the poliovirus 5′ noncoding region mediated by virus-encoded polypeptide 2A. Proc. Natl. Acad. Sci. USA 89:10272-10276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellen, C. U., M. Facke, H. G. Krausslich, C. K. Lee, and E. Wimmer. 1991. Characterization of poliovirus 2A proteinase by mutational analysis: residues required for autocatalytic activity are essential for induction of cleavage of eukaryotic initiation factor 4F polypeptide p220. J. Virol. 65:4226-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho, S. N., H. D. Hunt, R. M. Horton, J. K. Pullen, and L. R. Pease. 1989. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene 77:51-59. [DOI] [PubMed] [Google Scholar]
- 25.Hundsdoerfer, P., C. Thoma, and M. W. Hentze. 2005. Eukaryotic translation initiation factor 4GI and p97 promote cellular internal ribosome entry sequence-driven translation. Proc. Natl. Acad. Sci. USA 102:13421-13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerekatte, V., B. D. Keiper, C. Badorff, A. Cai, K. U. Knowlton, and R. E. Rhoads. 1999. Cleavage of poly(A)-binding protein by coxsackievirus 2A protease in vitro and in vivo: another mechanism for host protein synthesis shutoff? J. Virol. 73:709-717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuyumcu-Martinez, N. M., M. E. Van Eden, P. Younan, and R. E. Lloyd. 2004. Cleavage of poly(A)-binding protein by poliovirus 3C protease inhibits host cell translation: a novel mechanism for host translation shutoff. Mol. Cell. Biol. 24:1779-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lamphear, B. J., R. Kirchweger, T. Skern, and R. E. Rhoads. 1995. Mapping of functional domains in eukaryotic protein synthesis initiation factor 4G (eIF4G) with picornaviral proteases. Implications for cap-dependent and cap-independent translational initiation. J. Biol. Chem. 270:21975-21983. [DOI] [PubMed] [Google Scholar]
- 31.Lamphear, B. J., R. Yan, F. Yang, D. Waters, H. D. Liebig, H. Klump, E. Kuechler, T. Skern, and R. E. Rhoads. 1993. Mapping the cleavage site in protein synthesis initiation factor eIF-4 gamma of the 2A proteases from human coxsackievirus and rhinovirus. J. Biol. Chem. 268:19200-19203. [PubMed] [Google Scholar]
- 32.Liebig, H. D., E. Ziegler, R. Yan, K. Hartmuth, H. Klump, H. Kowalski, D. Blaas, W. Sommergruber, L. Frasel, B. Lamphear, et al. 1993. Purification of two picornaviral 2A proteinases: interaction with eIF-4 gamma and influence on in vitro translation. Biochemistry 32:7581-7588. [DOI] [PubMed] [Google Scholar]
- 33.Lloyd, R. E., M. J. Grubman, and E. Ehrenfeld. 1988. Relationship of p220 cleavage during picornavirus infection to 2A proteinase sequencing. J. Virol. 62:4216-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu, H. H., and E. Wimmer. 1996. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc. Natl. Acad. Sci. USA 93:1412-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Macadam, A. J., G. Ferguson, T. Fleming, D. M. Stone, J. W. Almond, and P. D. Minor. 1994. Role for poliovirus protease 2A in cap independent translation. EMBO J. 13:924-927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Michel, Y. M., D. Poncet, M. Piron, K. M. Kean, and A. M. Borman. 2000. Cap-poly(A) synergy in mammalian cell-free extracts. Investigation of the requirements for poly(A)-mediated stimulation of translation initiation. J. Biol. Chem. 275:32268-32276. [DOI] [PubMed] [Google Scholar]
- 37.Nomoto, A., Y. F. Lee, and E. Wimmer. 1976. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl. Acad. Sci. USA 73:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohlmann, T., M. Rau, V. M. Pain, and S. J. Morley. 1996. The C-terminal domain of eukaryotic protein synthesis initiation factor (eIF) 4G is sufficient to support cap-independent translation in the absence of eIF4E. EMBO J. 15:1371-1382. [PMC free article] [PubMed] [Google Scholar]
- 39.Otto, G. A., and J. D. Puglisi. 2004. The pathway of HCV IRES-mediated translation initiation. Cell 119:369-380. [DOI] [PubMed] [Google Scholar]
- 40.Pelletier, J., G. Kaplan, V. R. Racaniello, and N. Sonenberg. 1988. Cap-independent translation of poliovirus mRNA is conferred by sequence elements within the 5′ noncoding region. Mol. Cell. Biol. 8:1103-1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez, L., and L. Carrasco. 1992. Lack of direct correlation between p220 cleavage and the shut-off of host translation after poliovirus infection. Virology 189:178-186. [DOI] [PubMed] [Google Scholar]
- 42.Pilipenko, E. V., S. V. Maslova, A. N. Sinyakov, and V. I. Agol. 1992. Towards identification of cis-acting elements involved in the replication of enterovirus and rhinovirus RNAs: a proposal for the existence of tRNA-like terminal structures. Nucleic Acids Res. 20:1739-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Preiss, T., and M. W. Hentze. 1998. Dual function of the messenger RNA cap structure in poly(A)-tail-promoted translation in yeast. Nature 392:516-520. [DOI] [PubMed] [Google Scholar]
- 44.Reynolds, J. E., A. Kaminski, H. J. Kettinen, K. Grace, B. E. Clarke, A. R. Carroll, D. J. Rowlands, and R. J. Jackson. 1995. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 14:6010-6020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roberts, L. O., R. A. Seamons, and G. J. Belsham. 1998. Recognition of picornavirus internal ribosome entry sites within cells; influence of cellular and viral proteins. RNA 4:520-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sachs, A. B., R. W. Davis, and R. D. Kornberg. 1987. A single domain of yeast poly(A)-binding protein is necessary and sufficient for RNA binding and cell viability. Mol. Cell. Biol. 7:3268-3276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shiroki, K., T. Isoyama, S. Kuge, T. Ishii, S. Ohmi, S. Hata, K. Suzuki, Y. Takasaki, and A. Nomoto. 1999. Intracellular redistribution of truncated La protein produced by poliovirus 3Cpro-mediated cleavage. J. Virol. 73:2193-2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Svitkin, Y. V., H. Imataka, K. Khaleghpour, A. Kahvejian, H. D. Liebig, and N. Sonenberg. 2001. Poly(A)-binding protein interaction with elF4G stimulates picornavirus IRES-dependent translation. RNA 7:1743-1752. [PMC free article] [PubMed] [Google Scholar]
- 49.Tanguay, R. L., and D. R. Gallie. 1996. Translational efficiency is regulated by the length of the 3′ untranslated region. Mol. Cell. Biol. 16:146-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tarun, S. Z., Jr., and A. B. Sachs. 1995. A common function for mRNA 5′ and 3′ ends in translation initiation in yeast. Genes Dev. 9:2997-3007. [DOI] [PubMed] [Google Scholar]
- 51.Thoma, C., G. Bergamini, B. Galy, P. Hundsdoerfer, and M. W. Hentze. 2004. Enhancement of IRES-mediated translation of the c-myc and BiP mRNAs by the poly(A) tail is independent of intact eIF4G and PABP. Mol. Cell 15:925-935. [DOI] [PubMed] [Google Scholar]
- 52.Toyoda, H., M. J. Nicklin, M. G. Murray, C. W. Anderson, J. J. Dunn, F. W. Studier, and E. Wimmer. 1986. A second virus-encoded proteinase involved in proteolytic processing of poliovirus polyprotein. Cell 45:761-770. [DOI] [PubMed] [Google Scholar]
- 53.Wells, S. E., P. E. Hillner, R. D. Vale, and A. B. Sachs. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol. Cell 2:135-140. [DOI] [PubMed] [Google Scholar]
- 54.Wilusz, C. J., M. Wormington, and S. W. Peltz. 2001. The cap-to-tail guide to mRNA turnover. Nat. Rev. Mol. Cell Biol. 2:237-246. [DOI] [PubMed] [Google Scholar]
- 55.Wyckoff, E. E., J. W. Hershey, and E. Ehrenfeld. 1990. Eukaryotic initiation factor 3 is required for poliovirus 2A protease-induced cleavage of the p220 component of eukaryotic initiation factor 4F. Proc. Natl. Acad. Sci. USA 87:9529-9533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhao, X., B. J. Lamphear, D. Xiong, K. Knowlton, and R. E. Rhoads. 2003. Protection of cap-dependent protein synthesis in vivo and in vitro with an eIF4G-1 variant highly resistant to cleavage by coxsackievirus 2A protease. J. Biol. Chem. 278:4449-4457. [DOI] [PubMed] [Google Scholar]
- 57.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]