Abstract
We characterized human immunodeficiency virus type 1 (HIV-1) envelope glycoproteins (Env) isolated from two HIV-1-infected CCR5Δ32 homozygotes. Envs from both subjects used CCR5 and CXCR4 for entry into transfected cells. Most R5X4 Envs were lymphocyte-tropic and used CXCR4 exclusively for entry into peripheral blood mononuclear cells (PBMC), but a subset was dually lymphocyte- and macrophage-tropic and used either CCR5 or CXCR4 for entry into PBMC and monocyte-derived macrophages. The persistence of CCR5-using HIV-1 in two CCR5Δ32 homozygotes suggests the conserved CCR5 binding domain of Env is highly stable and provides new mechanistic insights important for HIV-1 transmission and persistence.
Approximately 1% of Caucasians are homozygous for a nonfunctional CCR5 allele containing a 32-bp deletion (CCR5Δ32) (26). The most compelling argument that supports a central role for CCR5 in the transmission of human immunodeficiency virus type 1 (HIV-1) stems from the observation that the homozygous CCR5Δ32 genotype confers a high degree of protection against HIV-1 infection (6, 15, 22, 25, 27, 39, 54). However, rare cases of HIV-1 infection in CCR5Δ32 homozygotes have been reported (2, 3, 21, 23, 28, 33, 41, 44). In all but one of the nine reported cases, exclusive use of CXCR4 by virus isolates or the presence of env sequences typical of CXCR4-using (X4) viruses was observed. We recently reported dual-tropic (R5X4) HIV-1 in an individual homozygous for the CCR5Δ32 allele (subject C2) (21). Despite the lack of functional CCR5 receptors, HIV-1 isolated 1 year after seroconversion maintained the ability to use CCR5 to enter transfected cell lines and to replicate in primary monocyte-derived macrophages (MDM). In the present study, we undertook a genetic and functional analysis of R5X4 envelope glycoproteins (Env) cloned from HIV-1 isolated from subject C2 and of R5X4 Envs cloned directly from the blood of a newly identified CCR5Δ32 homozygote infected with HIV-1 (subject DR).
Subjects.
The clinical history of subject C2 and the results of laboratory studies, including genotypes at CCR2, SDF1, interleukin-10, CCR5, and HLA alleles, have been described previously (21). Subject DR is a homosexual male with a history of injecting drug use and first tested seropositive for HIV-1 in September 1991. He has congenital deafness caused by intrauterine rubella virus infection and has tested positive for hepatitis C virus RNA and hepatitis B virus core antibody. Genetic analysis demonstrated carriage of CCR2 64I (wild type), SDF1 3′A (wild type), and CCR5 (Δ32/Δ32) alleles. Extended HLA haplotype analysis demonstrated carriage of HLA A*24, B*0702, B*5501, C*0304, C*0703, DRB1*04, DRB1*13, DQB1*03, and DQB1*0604 alleles. After informed consent was obtained in accordance with guidelines endorsed by the Royal Perth Hospital human ethics committee, HIV-1 Envs were cloned directly from blood taken in August 2003, approximately 12 years after subject DR first tested seropositive for HIV-1.
Biological activities of HIV-1 Env clones.
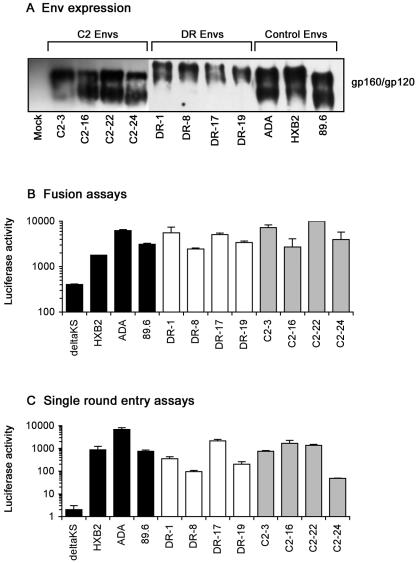
A 2.1-kb fragment spanning the KpnI-to-BamHI restriction sites in HIV-1 env was amplified directly from peripheral blood mononuclear cells (PBMC) purified from the blood of subject DR or from cultured PBMC infected with primary HIV-1 isolated from subject C2 (21) and cloned into the pSVIII-HXB2 Env expression vector (17), as described previously (34). The biological activities of Envs predicted to contain uninterrupted gp160 coding regions were determined by analysis of gp160/gp120 expression in 293T cells transfected with each plasmid. Env clones expressing distinct gp160 and gp120 proteins were detected in four clones from both subjects (Fig. 1A). Envs derived from subject DR had slower mobility by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) than Envs derived from subject C2 and control Envs (Fig. 1A). Treatment of cell lysates with PNGase-F prior to SDS-PAGE and Western blotting resulted in mobilities of DR Env proteins that were similar to the mobilities of Envs derived from C2 and control Envs (data not shown), suggesting that DR Envs are more glycosylated than C2 and control Envs. The correctly processed DR and C2 Envs were functional in cell-cell fusion assays (Fig. 1B) and single-round infection assays (Fig. 1C) using target cells that express CD4, CCR5, and CXCR4. Thus, four correctly processed and functional Env clones were obtained from each subject.
FIG. 1.
Expression and functional activities of Env clones. (A) Western blot analysis of 293T cells cotransfected with 5 μg of pSVIIIenv plasmid and 1 μg of pSVTat using Lipofectamine 2000 (Invitrogen, Carlsbad, CA), following electrophoresis in 8.5% (wt/vol) SDS-PAGE gels. Western blot analysis of cell lysates was performed at 72 h posttransfection using rabbit anti-gp120 polyclonal antisera. pSVIIIenv plasmids containing ADA, HXB2, or 89.6 Env were included as controls. The data are representative of two independent experiments. (B) Fusion assays were conducted by coculturing 1 × 106 TZM-bl cells that express CD4, CCR5, and CXCR4 on the cell surface and stably express the luciferase gene under the control of the HIV-1 long terminal repeat (9, 37, 45) with 1 × 105 293T cells cotransfected with each pSVIIIenv plasmid plus pSVTat for 8 h at 37°C in 500 μl of culture medium, followed by measurement of luciferase activity (Promega, Madison, WI) in cell lysates, as described previously (20, 34). HXB2, ADA, and 89.6 Envs were included as positive controls, and a nonfunctional Env, ΔKS (deltaKS), was included as a negative control to determine the background level of luciferase activity. The data are representative of two independent experiments and are expressed as means from duplicate experiments, and the error bars represent standard deviations. (C) Single-round entry assays were conducted by infection of Cf2th-CD4/CCR5/CXCR4 cells, which were constructed by transduction of the Cf2th-CD4/CCR5 cell line (47) with pBABE-puro vectors expressing CXCR4 (8, 31), followed by selection and expansion in culture medium containing 1 μg of puromycin per ml, with equivalent amounts of Env-pseudotyped luciferase reporter virus that was produced using pCMVΔP1ΔenvpA and pHIV-1Luc plasmids as described previously (46-48). Luciferase activity was measured in cell lysates at 72 h postinfection (Promega). Luciferase reporter viruses pseudotyped with HXB2, ADA, or 89.6 Env were included as positive controls, and luciferase reporter virus pseudotyped with ΔKS Env was included as a negative control to determine the background level of luciferase activity. The data are representative of two independent experiments and are expressed as means from duplicate infections, and the error bars represent standard deviations.
Coreceptor usage.
Luciferase reporter viruses pseudotyped with each Env were produced using pCMVΔP1ΔenvpA and pHIV-1Luc plasmids as described previously (46-48) and characterized for the ability to use CCR5, CXCR4, or alternative coreceptors for virus entry into transfected Cf2th cells (Table 1). Reporter viruses pseudotyped with ADA, HXB2, or 89.6 Envs were included as controls and entered cells expressing CCR5, CXCR4, or both CCR5 and CXCR4, respectively, as well as cells expressing various alternative coreceptors in a pattern consistent with the results of previous studies (4, 5, 7, 8, 11-14, 16, 19, 24, 38). C2 and DR Envs used CCR5 and CXCR4 for virus entry. In addition, C2 Envs used CCR3, Gpr15, and Strl33 as alternative coreceptors for virus entry, consistent with the repertoire of coreceptors used by the primary HIV-1 isolate (21). These results demonstrate that C2 and DR Envs are of R5X4 phenotype, with expanded usage of alternative coreceptors by C2 Envs.
TABLE 1.
Coreceptor usage by primary and reference HIV-1 Envs
| Env | Coreceptor usagea
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CD4 only | CCR2b | CCR3 | CCR5 | CCR8 | CXCR4 | CX3CR1 | Gpr1 | Gpr15 | Strl33 | Apj | |
| Control | |||||||||||
| ΔKS | − | − | − | − | − | − | − | − | − | − | − |
| HXB2 | − | − | − | − | − | +++ | − | − | − | − | + |
| ADA | − | − | ++ | +++ | + | − | +/− | +/− | + | + | − |
| 89.6 | − | + | ++ | +++ | − | +++ | − | − | − | − | + |
| C2 | |||||||||||
| C2-3 | − | − | + | +++ | − | +++ | − | − | + | +/− | − |
| C2-16 | − | − | + | +++ | − | +++ | − | − | + | +/− | − |
| C2-22 | − | − | + | +++ | − | +++ | − | − | + | +/− | − |
| C2-24 | − | − | + | +++ | − | +++ | − | − | + | +/− | − |
| DR | |||||||||||
| DR-1 | − | − | − | +++ | − | +++ | − | − | − | − | − |
| DR-8 | − | − | − | +++ | − | +++ | − | − | − | − | − |
| DR-17 | − | − | − | +++ | − | +++ | − | − | − | − | − |
| DR-19 | − | − | − | +++ | − | +++ | − | − | − | − | − |
Coreceptor usage of HIV-1 Envs was determined by infecting Cf2th cells (5) that were transfected with plasmids expressing CD4 and the indicated coreceptor with Env-pseudotyped luciferase reporter viruses, as described previously (34). Reporter viruses pseudotyped with the X4 HXB2, R5 ADA, and R5X4 89.6 Envs were used as positive controls, and reporter virus pseudotyped with the nonfunctional ΔKS Env was used as a negative control. Entry was determined by measurement of luciferase activities in cell lysates at 72 h postinfection. Entry levels were scored as +++, ++, +/++, +, and +/−, which correspond to luciferase activity >7,500-fold, 100- to 500-fold, 50- to 100-fold, 20- to 50-fold, and 10- to 20-fold above background levels, respectively, as described previously (34). Results less than 10-fold above the background level obtained with ΔKS Env were considered negative.
Our results differ from those of previous studies that demonstrated X4-like V3 Env sequence changes and/or CXCR4-restricted virus isolated from other CCR5Δ32 homozygotes infected with HIV-1 (2, 3, 23, 28, 33, 41, 44). In addition, unlike previous studies, C2 Envs could use the alternative coreceptors CCR3, Gpr15, and Strl33 for virus entry (28, 32, 41). The reasons for the discrepant results between our studies and those of other investigators remain to be determined, but they may reflect technical differences between assays used to assess coreceptor usage that could measure such usage differently (28, 32, 33, 41). For example, differences in cell surface CCR5 expression levels and CCR5 densities that may exist between stably transfected target cells used in other studies (28, 32, 41) and transiently transfected cells used here could impact coreceptor engagement by HIV-1 Env (37). Furthermore, the reliance on V3 Env sequence and MT-2 cell infection assays in previous studies (2, 3, 23, 44), which do not readily discriminate between X4 and R5X4 HIV-1 variants, could have underestimated the prevalence of R5X4 HIV-1 variants and those able to use alternative coreceptors in HIV-1-infected CCR5Δ32 homozygotes.
Tropism of Envs for entry into primary cells.
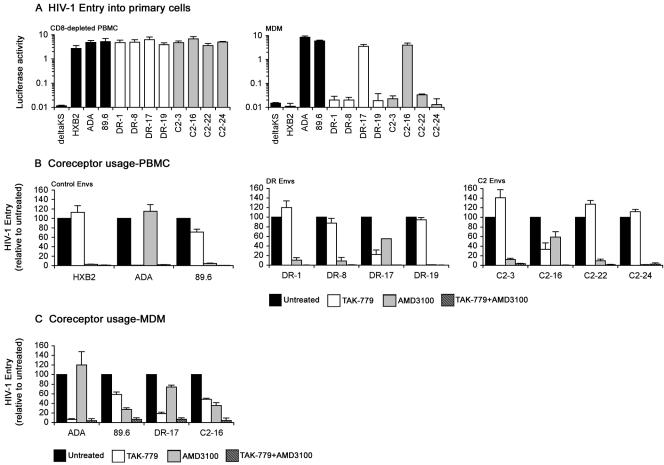
An R5X4 phenotype does not necessarily reflect dual tropism for primary cell types (36). Therefore, luciferase reporter viruses pseudotyped with each Env were characterized for the ability to enter CD8-depleted PBMC and MDM (Fig. 2A). Consistent with previous studies, ADA and 89.6 Envs entered PBMC and MDM, whereas HXB2 Env entered PBMC but not MDM (18, 19, 34, 42, 43, 49-51). All C2 and DR Envs entered PBMC, but only C2-16 and DR-17 Envs entered MDM. These studies demonstrate that the majority of the R5X4 Envs are monotropic for T lymphocytes, and a subset are dual-tropic for both T lymphocytes and macrophages. Based on the most up-to-date HIV-1 phenotype classification system, which takes into account coreceptor usage for entry into transfected cells, as well as tropism for primary cell types (18), DR-1, DR-8, DR-19, C2-3, C2-22, and C2-24 Envs can be classified as T-R5X4 Envs and C2-16 and DR-17 Envs as D-R5X4 Envs.
FIG. 2.
Tropism of Envs and coreceptor preference for HIV-1 entry into primary cells. (A) Single-round entry assays were conducted by infection of 0.5 × 106 phytohemagglutinin-activated, interleukin 2-stimulated PBMC that were depleted of CD8+ cells with anti-CD8-conjugated magnetic beads (Invitrogen) or infection of confluent monolayers of MDM cultured for 5 days in 48-well tissue culture plates in medium containing 10% (vol/vol) human serum and 12.5 ng/ml macrophage colony-stimulating factor that were purified from PBMC by plastic adherence with equivalent amounts of Env-pseudotyped luciferase reporter virus for 3 h at 37°C. After washing the cells to remove the virus inoculum and replacement of culture medium, luciferase activity was measured in cell lysates at 72 h postinfection (Promega). Luciferase reporter viruses pseudotyped with HXB2, ADA, 89.6, or ΔKS Env were included as controls. The data are representative of two independent experiments using cells obtained from different donors and are expressed as means from duplicate infections, and the error bars represent standard deviations. CD8-depleted PBMC (B) or MDM (C) that were prepared as described above were left untreated or preincubated with 50 nM TAK-779 (1) and/or 1.2 μM AMD3100 (10, 40) for 1 h at 37°C prior to infection with equivalent amounts of Env-pseudotyped luciferase reporter viruses containing the same concentration(s) of inhibitor. Infections proceeded for 3 h at 37°C. After washing the cells to remove the virus inoculum and replacement of culture medium, luciferase activity was measured in cell lysates at 72 h postinfection (Promega). Luciferase reporter viruses pseudotyped with ADA, HXB2, or 89.6 Env were used as controls in CD8-depleted PBMC infections, and luciferase reporter viruses pseudotyped with ADA or 89.6 Env were used as controls in MDM infections. Luciferase measurements were normalized to the values obtained in untreated infections, which were set at 100. The data are representative of two independent experiments using cells obtained from different donors and are expressed as means from duplicate infections, and the error bars represent standard deviations.
Coreceptor preferences of Envs for entry into primary cells.
The coreceptor preferences of Envs for entry into CD8-depleted PBMC and MDM were next determined by measuring the sensitivities of Env-pseudotyped luciferase reporter viruses to an inhibitor of CCR5 (TAK-779) or CXCR4 (AMD3100).
In CD8-depleted PBMC (Fig. 2B), TAK-779 reduced entry by ADA, 89.6, DR-17, and C2-16 Envs by 99%, 30%, 78%, and 66%, respectively, but had no effect on entry by HXB2, DR-1, DR-8, DR-19, C2-3, C2-22, and C2-24 Envs. AMD3100 reduced entry by 89.6, HXB2, DR-1, DR-8, DR-19, C2-3, C2-22, and C2-24 Envs by 95 to 99% and reduced entry by DR-17 and C2-16 Envs by 45% and 40%, respectively, but had no effect on entry by ADA Env. Combinations of TAK-779 and AMD3100 completely inhibited entry by control, DR, and C2 Envs.
In MDM (Fig. 2C), TAK-779 inhibited entry by ADA, 89.6, DR-17, and C2-16 Envs by 95%, 42%, 82%, and 50%, respectively. AMD3100 reduced entry by 89.6, DR-17, and C2-16 Envs by 72%, 26%, and 65%, respectively, but had no effect on entry by ADA Env. Combinations of TAK-779 and AMD3100 inhibited entry by control, DR-17, and C2-16 Envs by at least 95%.
These data indicate that DR-1, DR-8, DR-19, C2-3, C2-22, and C2-24 Envs, which are monotropic for T lymphocytes (Fig. 2A), use CXCR4 exclusively for entry into PBMC. These results are consistent with the results of recent studies that demonstrated preferential use of CXCR4 by R5X4 HIV-1 isolates for infection of primary T lymphocytes (51). The data also indicate that DR-17 and C2-16 Envs, which are dual-tropic for T lymphocytes and macrophages (Fig. 2A), can use CCR5 or CXCR4 for entry into PBMC and MDM. Although C2 Envs utilized CCR3, Gpr15, and Strl33 for entry into transfected cells (Table 1), entry of virus pseudotyped with these Envs was abolished by combinations of TAK-779 and AMD3100, indicating that C2 Envs do not utilize coreceptors other than CCR5 and CXCR4 for entry into PBMC or MDM. This supports the results of previous studies showing that infection of primary cells by HIV-1 with expanded coreceptor usage occurs, with few exceptions, exclusively via CCR5 or CXCR4 (30, 52, 53). Taken together, our findings demonstrate the persistence of R5X4 HIV-1 variants in two subjects homozygous for the CCR5Δ32 allele and that a subset of these variants retained the ability to use CCR5 for entry into primary CD4+ cells.
Sequence analysis.
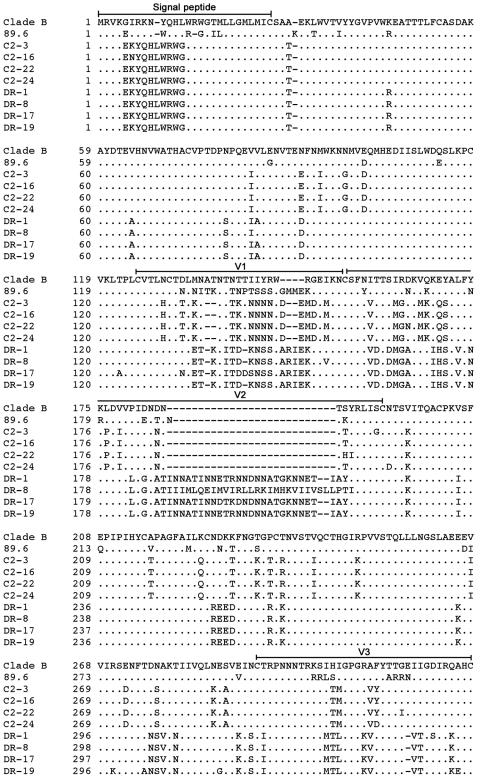
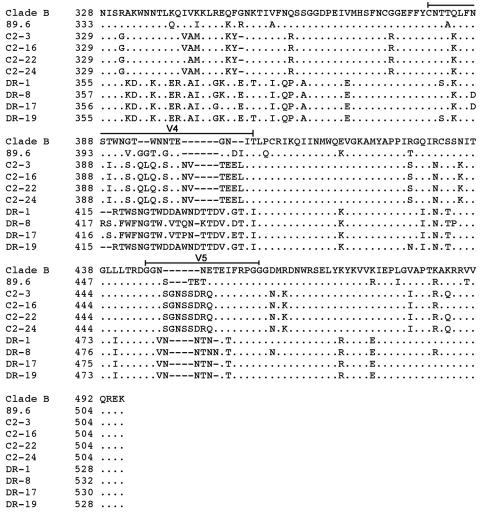
The preceding studies demonstrate persistent CCR5 usage by HIV-1 Envs cloned from two individuals who lack CCR5 expression due to the presence of the homozygous CCR5Δ32 deletion. To better understand the viral determinants that may contribute to persistent CCR5 usage of C2 and DR Envs and to identify genetic changes that may underlie a T-R5X4 or D-R5X4 phenotype, we sequenced the gp120 regions of the eight functional Env clones (Fig. 3).
FIG.3.
Env amino acid sequences. The gp120 amino acid sequences of Env clones were deduced from nucleotide sequences obtained by Big Dye terminator sequencing (Applied Biosystems, Foster City, CA). The amino acid alignments were compared to Env from HIV-1 89.6 and the clade B consensus sequence. The dots indicate residues identical to the clade B consensus sequence, and the dashes indicate gaps.
The net charge of the V3 variable-loop region of C2 Envs was +3, and the net charges of the V3 regions of DR-1, DR-8, DR-17, and DR-19 Envs were +6, +5, +5, and +4, respectively. Although the presence of a basic residue at position 11 or 25 in V3 is strongly associated with CXCR4 usage of HIV-1 (29), all Envs lacked basic residues at either position. Therefore, although C2 and DR Envs are R5X4, they differ from prototypic R5X4 Envs, such as 89.6 and most other blood-derived R5X4 viruses, in that they lack typical V3 features that normally govern coreceptor usage and in fact more closely resemble typical R5 Envs. Thus, it is possible that the ability of DR and C2 Envs to continue to use CCR5 for virus entry despite the absence of CCR5 expression in vivo reflects the persistence of R5-like V3 sequences, despite the Envs being functionally R5X4. The results further suggest that structural features of C2 and DR Envs linked to CCR5 usage are highly stable and might confer a selective advantage in vivo.
Our findings are similar to those of previous studies of brain-derived R5X4 Envs that also found no association between charged amino acids in V3 and coreceptor usage (19, 34, 35). The mechanisms underlying the discrepant dependencies on charged amino acids for coreceptor usage between C2, DR, and brain-derived R5X4 Envs and prototypic R5X4 Envs remain to be determined, but it is possible that the absent or low CCR5 expression levels in CCR5Δ32 homozygotes and the brain, respectively, may contribute to an in vivo environment that promotes similar R5X4 Env structures that may have altered requirements for coreceptor usage.
In addition, the tropism of D-R5X4 DR-17 and C2-16 Envs for MDM was not governed by the presence of isoleucine at amino acid position 326 of V3 or by proline or cysteine residues in V1, which are amino acid changes recently shown to be important for the macrophage tropism of 89.6 and other blood-derived R5X4 viruses (18). Further mutagenesis studies are required to identify genetic changes that distinguish the D-R5X4 DR-17 Env from the T-R5X4 DR-1, DR-8, and DR-19 Envs and that distinguish the D-R5X4 C2-16 Env from the T-R5X4 C2-3, C2-22, and C2-24 Envs, but they most likely map to discrete amino acid changes that exist in the V1 and V2 regions of DR and C2 Envs, respectively.
Conclusions.
In this study, we characterized R5X4 Envs cloned from HIV-1 harbored by two infected CCR5Δ32 homozygotes, which comprise 20% of the known cases of HIV-1 infection in subjects with this genotype. Despite the fact that the subjects were infected with HIV-1 for 1 or 12 years and the absence of CCR5 expression in vivo, Envs that retained the ability to use CCR5 efficiently for virus entry into transfected cells, PBMC, and MDM were cloned from both subjects. The results suggest that structural features of C2 and DR Envs linked to persistent CCR5 usage are highly stable and might confer a selective advantage in vivo. A better understanding of these structural determinants may provide mechanistic insights into HIV-1 transmission and persistence and facilitate the development of CCR5 inhibitors and the elicitation of neutralizing antibodies.
Nucleotide sequence accession numbers.
The gp120 nucleotide sequences reported here have been assigned GenBank accession numbers DQ356577 to DQ356584.
Acknowledgments
We thank J. Sodroski for providing Cf2th and Cf2th-CD4/CCR5 cells and the pSVIIIenv, pCMVΔP1ΔenvpA, and pHIV-1Luc plasmids; H. Göttlinger for providing the pSVTat plasmid; and J. Sodroski, R. Doms, and S. Peiper for CD4 and coreceptor plasmids. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: Bicyclam JM-2987 (hydrobromide salt of AMD-3100) and TAK-779.
This study was supported by grants to P.R.G. from the Australian National Health and Medical Research Council (NHMRC) (251520) and NIH/NIAID (AI054207-01-A1) and a grant to D.G. from NIH/NS (NS37277). L.G. is the recipient of a University of Melbourne Postgraduate Research Scholarship. J.S. is the recipient of an NHMRC Dora Lush Biomedical Research Scholarship. P.R.G. is the recipient of an NHMRC R. Douglas Wright Biomedical Career Development Award.
REFERENCES
- 1.Baba, M., O. Nishimura, N. Kanzaki, M. Okamoto, H. Sawada, Y. Iizawa, M. Shiraishi, Y. Aramaki, K. Okonogi, Y. Ogawa, K. Meguro, and M. Fujino. 1999. A small-molecule, nonpeptide CCR5 antagonist with highly potent and selective anti-HIV-1 activity. Proc. Natl. Acad. Sci. USA 96:5698-5703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balotta, C., P. Bagnarelli, M. Violin, A. L. Ridolfo, D. Zhou, A. Berlusconi, S. Corvasce, M. Corbellino, M. Clementi, M. Clerici, M. Moroni, and M. Galli. 1997. Homozygous delta 32 deletion of the CCR-5 chemokine receptor gene in an HIV-1-infected patient. AIDS 11:F67-F71. [DOI] [PubMed] [Google Scholar]
- 3.Biti, R., R. Ffrench, J. Young, B. Bennetts, G. Stewart, and T. Liang. 1997. HIV-1 infection in an individual homozygous for the CCR5 deletion allele. Nat. Med. 3:252-253. [DOI] [PubMed] [Google Scholar]
- 4.Choe, H., M. Farzan, M. Konkel, K. Martin, Y. Sun, L. Marcon, M. Cayabyab, M. Berman, M. E. Dorf, N. Gerard, C. Gerard, and J. Sodroski. 1998. The orphan seven-transmembrane receptor apj supports the entry of primary T-cell-line-tropic and dual-tropic human immunodeficiency virus type 1. J. Virol. 72:6113-6118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choe, H., M. Farzan, Y. Sun, N. Sullivan, B. Rollins, P. D. Ponath, L. Wu, C. R. Mackay, G. LaRosa, W. Newman, N. Gerard, C. Gerard, and J. Sodroski. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135-1148. [DOI] [PubMed] [Google Scholar]
- 6.Dean, M., M. Carrington, C. Winkler, G. A. Huttley, M. W. Smith, R. Allikmets, J. J. Goedert, S. P. Buchbinder, E. Vittinghoff, E. Gomperts, S. Donfield, D. Vlahov, R. Kaslow, A. Saah, C. Rinaldo, R. Detels, S. J. O'Brien, et al. 1996. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Science 273:1856-1862. [DOI] [PubMed] [Google Scholar]
- 7.Deng, H., R. Liu, W. Ellmeier, S. Choe, D. Unutmaz, M. Burkhart, P. Di Marzio, S. Marmon, R. E. Sutton, C. M. Hill, C. B. Davis, S. C. Peiper, T. J. Schall, D. R. Littman, and N. R. Landau. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661-666. [DOI] [PubMed] [Google Scholar]
- 8.Deng, H. K., D. Unutmaz, V. N. KewalRamani, and D. R. Littman. 1997. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature 388:296-300. [DOI] [PubMed] [Google Scholar]
- 9.Derdeyn, C. A., J. M. Decker, J. N. Sfakianos, X. Wu, W. A. O'Brien, L. Ratner, J. C. Kappes, G. M. Shaw, and E. Hunter. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358-8367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Donzella, G. A., D. Schols, S. W. Lin, J. A. Este, K. A. Nagashima, P. J. Maddon, G. P. Allaway, T. P. Sakmar, G. Henson, E. De Clercq, and J. P. Moore. 1998. AMD3100, a small molecule inhibitor of HIV-1 entry via the CXCR4 co-receptor. Nat. Med. 4:72-77. [DOI] [PubMed] [Google Scholar]
- 11.Doranz, B. J., J. Rucker, Y. Yi, R. J. Smyth, M. Samson, S. C. Peiper, M. Parmentier, R. G. Collman, and R. W. Doms. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149-1158. [DOI] [PubMed] [Google Scholar]
- 12.Dragic, T., V. Litwin, G. P. Allaway, S. R. Martin, Y. Huang, K. A. Nagashima, C. Cayanan, P. J. Maddon, R. A. Koup, J. P. Moore, and W. A. Paxton. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667-673. [DOI] [PubMed] [Google Scholar]
- 13.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, B. O'Dowd, and R. W. Doms. 1998. Use of GPR1, GPR15, and STRL33 as coreceptors by diverse human immunodeficiency virus type 1 and simian immunodeficiency virus envelope proteins. Virology 249:367-378. [DOI] [PubMed] [Google Scholar]
- 14.Edinger, A. L., T. L. Hoffman, M. Sharron, B. Lee, Y. Yi, W. Choe, D. L. Kolson, B. Mitrovic, Y. Zhou, D. Faulds, R. G. Collman, J. Hesselgesser, R. Horuk, and R. W. Doms. 1998. An orphan seven-transmembrane domain receptor expressed widely in the brain functions as a coreceptor for human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 72:7934-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eugen-Olsen, J., A. K. Iversen, P. Garred, U. Koppelhus, C. Pedersen, T. L. Benfield, A. M. Sorensen, T. Katzenstein, E. Dickmeiss, J. Gerstoft, P. Skinhoj, A. Svejgaard, J. O. Nielsen, and B. Hofmann. 1997. Heterozygosity for a deletion in the CKR-5 gene leads to prolonged AIDS-free survival and slower CD4 T-cell decline in a cohort of HIV-seropositive individuals. AIDS 11:305-310. [DOI] [PubMed] [Google Scholar]
- 16.Farzan, M., H. Choe, K. Martin, L. Marcon, W. Hofmann, G. Karlsson, Y. Sun, P. Barrett, N. Marchand, N. Sullivan, N. Gerard, C. Gerard, and J. Sodroski. 1997. Two orphan seven-transmembrane segment receptors which are expressed in CD4-positive cells support simian immunodeficiency virus infection. J. Exp. Med. 186:405-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gao, F., S. G. Morrison, D. L. Robertson, C. L. Thornton, S. Craig, G. Karlsson, J. Sodroski, M. Morgado, B. Galvao-Castro, H. von Briesen, S. Beddows, J. Weber, P. M. Sharp, G. M. Shaw, B. H. Hahn, and the World Health Organization and National Institute for Allergy and Infectious Diseases Networks for HIV Isolation and Characterization. 1996. Molecular cloning and analysis of functional envelope genes from human immunodeficiency virus type 1 sequence subtypes A through G. J. Virol. 70:1651-1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghaffari, G., D. L. Tuttle, D. Briggs, B. R. Burkhardt, D. Bhatt, W. A. Andiman, J. W. Sleasman, and M. M. Goodenow. 2005. Complex determinants in human immunodeficiency virus type 1 envelope gp120 mediate CXCR4-dependent infection of macrophages. J. Virol. 79:13250-13261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gorry, P. R., G. Bristol, J. A. Zack, K. Ritola, R. Swanstrom, C. J. Birch, J. E. Bell, N. Bannert, K. Crawford, H. Wang, D. Schols, E. De Clercq, K. Kunstman, S. M. Wolinsky, and D. Gabuzda. 2001. Macrophage tropism of human immunodeficiency virus type 1 isolates from brain and lymphoid tissues predicts neurotropism independent of coreceptor specificity. J. Virol. 75:10073-10089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gorry, P. R., J. Taylor, G. H. Holm, A. Mehle, T. Morgan, M. Cayabyab, M. Farzan, H. Wang, J. E. Bell, K. Kunstman, J. P. Moore, S. M. Wolinsky, and D. Gabuzda. 2002. Increased CCR5 affinity and reduced CCR5/CD4 dependence of a neurovirulent primary human immunodeficiency virus type 1 isolate. J. Virol. 76:6277-6292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gorry, P. R., C. Zhang, S. Wu, K. Kunstman, E. Trachtenberg, J. Phair, S. Wolinsky, and D. Gabuzda. 2002. Persistence of dual-tropic HIV-1 in an individual homozygous for the CCR5 delta 32 allele. Lancet 359:1832-1834. [DOI] [PubMed] [Google Scholar]
- 22.Huang, Y., W. A. Paxton, S. M. Wolinsky, A. U. Neumann, L. Zhang, T. He, S. Kang, D. Ceradini, Z. Jin, K. Yazdanbakhsh, K. Kunstman, D. Erickson, E. Dragon, N. R. Landau, J. Phair, D. D. Ho, and R. A. Koup. 1996. The role of a mutant CCR5 allele in HIV-1 transmission and disease progression. Nat. Med. 2:1240-1243. [DOI] [PubMed] [Google Scholar]
- 23.Kuipers, H., C. Workman, W. Dyer, A. Geczy, J. Sullivan, and R. Oelrichs. 1999. An HIV-1-infected individual homozygous for the CCR-5 Δ32 allele and the SDF-1 3′A allele. AIDS 13:433-434. [DOI] [PubMed] [Google Scholar]
- 24.Liao, F., G. Alkhatib, K. W. Peden, G. Sharma, E. A. Berger, and J. M. Farber. 1997. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J. Exp. Med. 185:2015-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu, R., W. A. Paxton, S. Choe, D. Ceradini, S. R. Martin, R. Horuk, M. E. MacDonald, H. Stuhlmann, R. A. Koup, and N. R. Landau. 1996. Homozygous defect in HIV-1 coreceptor accounts for resistance of some multiply-exposed individuals to HIV-1 infection. Cell 86:367-377. [DOI] [PubMed] [Google Scholar]
- 26.Martinson, J. J., N. H. Chapman, D. C. Rees, Y. T. Liu, and J. B. Clegg. 1997. Global distribution of the CCR5 gene 32-basepair deletion. Nat. Genet. 16:100-103. [DOI] [PubMed] [Google Scholar]
- 27.Michael, N. L., G. Chang, L. G. Louie, J. R. Mascola, D. Dondero, D. L. Birx, and H. W. Sheppard. 1997. The role of viral phenotype and CCR-5 gene defects in HIV-1 transmission and disease progression. Nat. Med. 3:338-340. [DOI] [PubMed] [Google Scholar]
- 28.Michael, N. L., J. A. Nelson, V. N. KewalRamani, G. Chang, S. J. O'Brien, J. R. Mascola, B. Volsky, M. Louder, G. C. White II, D. R. Littman, R. Swanstrom, and T. R. O'Brien. 1998. Exclusive and persistent use of the entry coreceptor CXCR4 by human immunodeficiency virus type 1 from a subject homozygous for CCR5 Δ32. J. Virol. 72:6040-6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milich, L., B. H. Margolin, and R. Swanstrom. 1997. Patterns of amino acid variability in NSI-like and SI-like V3 sequences and a linked change in the CD4-binding domain of the HIV-1 Env protein. Virology 239:108-118. [DOI] [PubMed] [Google Scholar]
- 30.Moore, J. P., S. G. Kitchen, P. Pugach, and J. A. Zack. 2004. The CCR5 and CXCR4 coreceptors—central to understanding the transmission and pathogenesis of human immunodeficiency virus type 1 infection. AIDS Res. Hum. Retrovir. 20:111-126. [DOI] [PubMed] [Google Scholar]
- 31.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 18:3587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naif, H. M., A. L. Cunningham, M. Alali, S. Li, N. Nasr, M. M. Buhler, D. Schols, E. de Clercq, and G. Stewart. 2002. A human immunodeficiency virus type 1 isolate from an infected person homozygous for CCR5Δ32 exhibits dual tropism by infecting macrophages and MT2 cells via CXCR4. J. Virol. 76:3114-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Brien, T. R., C. Winkler, M. Dean, J. A. Nelson, M. Carrington, N. L. Michael, and G. C. White II. 1997. HIV-1 infection in a man homozygous for CCR5 delta 32. Lancet 349:1219. [DOI] [PubMed] [Google Scholar]
- 34.Ohagen, A., A. Devitt, K. J. Kunstman, P. R. Gorry, P. P. Rose, B. Korber, J. Taylor, R. Levy, R. L. Murphy, S. M. Wolinsky, and D. Gabuzda. 2003. Genetic and functional analysis of full-length human immunodeficiency virus type 1 env genes derived from brain and blood of patients with AIDS. J. Virol. 77:12336-12345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peters, P. J., J. Bhattacharya, S. Hibbitts, M. T. Dittmar, G. Simmons, J. Bell, P. Simmonds, and P. R. Clapham. 2004. Biological analysis of human immunodeficiency virus type 1 R5 envelopes amplified from brain and lymph node tissues of AIDS patients with neuropathology reveals two distinct tropism phenotypes and identifies envelopes in the brain that confer an enhanced tropism and fusigenicity for macrophages. J. Virol. 78:6915-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Philpott, S. M. 2003. HIV-1 coreceptor usage, transmission, and disease progression. Curr. HIV Res. 1:217-227. [DOI] [PubMed] [Google Scholar]
- 37.Platt, E. J., K. Wehrly, S. E. Kuhmann, B. Chesebro, and D. Kabat. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J. Virol. 72:2855-2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rucker, J., A. L. Edinger, M. Sharron, M. Samson, B. Lee, J. F. Berson, Y. Yi, B. Margulies, R. G. Collman, B. J. Doranz, M. Parmentier, and R. W. Doms. 1997. Utilization of chemokine receptors, orphan receptors, and herpesvirus-encoded receptors by diverse human and simian immunodeficiency viruses. J. Virol. 71:8999-9007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Samson, M., F. Libert, B. J. Doranz, J. Rucker, C. Liesnard, C. M. Farber, S. Saragosti, C. Lapoumeroulie, J. Cognaux, C. Forceille, G. Muyldermans, C. Verhofstede, G. Burtonboy, M. Georges, T. Imai, S. Rana, Y. Yi, R. J. Smyth, R. G. Collman, R. W. Doms, G. Vassart, and M. Parmentier. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382:722-725. [DOI] [PubMed] [Google Scholar]
- 40.Schols, D., S. Struyf, J. Van Damme, J. A. Este, G. Henson, and E. De Clercq. 1997. Inhibition of T-tropic HIV strains by selective antagonization of the chemokine receptor CXCR4. J. Exp. Med. 186:1383-1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sheppard, H. W., C. Celum, N. L. Michael, S. O'Brien, M. Dean, M. Carrington, D. Dondero, and S. P. Buchbinder. 2002. HIV-1 infection in individuals with the CCR5-Δ32/Δ32 genotype: acquisition of syncytium-inducing virus at seroconversion. J. Acquir. Immune. Defic. Syndr. 29:307-313. [DOI] [PubMed] [Google Scholar]
- 42.Simmons, G., J. D. Reeves, A. McKnight, N. Dejucq, S. Hibbitts, C. A. Power, E. Aarons, D. Schols, E. De Clercq, A. E. Proudfoot, and P. R. Clapham. 1998. CXCR4 as a functional coreceptor for human immunodeficiency virus type 1 infection of primary macrophages. J. Virol. 72:8453-8457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh, A., Y. Yi, S. N. Isaacs, D. L. Kolson, and R. G. Collman. 2001. Concordant utilization of macrophage entry coreceptors by related variants within an HIV type 1 primary isolate viral swarm. AIDS Res. Hum. Retrovir. 17:957-963. [DOI] [PubMed] [Google Scholar]
- 44.Theodorou, I., L. Meyer, M. Magierowska, C. Katlama, C. Rouzioux et al. 1997. HIV-1 infection in an individual homozygous for CCR5 delta 32. Lancet 349:1219-1220. [PubMed] [Google Scholar]
- 45.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yang, X., S. Kurteva, S. Lee, and J. Sodroski. 2005. Stoichiometry of antibody neutralization of human immunodeficiency virus type 1. J. Virol. 79:3500-3508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang, X., V. Tomov, S. Kurteva, L. Wang, X. Ren, M. K. Gorny, S. Zolla-Pazner, and J. Sodroski. 2004. Characterization of the outer domain of the gp120 glycoprotein from human immunodeficiency virus type 1. J. Virol. 78:12975-12986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yang, X., R. Wyatt, and J. Sodroski. 2001. Improved elicitation of neutralizing antibodies against primary human immunodeficiency viruses by soluble stabilized envelope glycoprotein trimers. J. Virol. 75:1165-1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yi, Y., S. N. Isaacs, D. A. Williams, I. Frank, D. Schols, E. De Clercq, D. L. Kolson, and R. G. Collman. 1999. Role of CXCR4 in cell-cell fusion and infection of monocyte-derived macrophages by primary human immunodeficiency virus type 1 (HIV-1) strains: two distinct mechanisms of HIV-1 dual tropism. J. Virol. 73:7117-7125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yi, Y., S. Rana, J. D. Turner, N. Gaddis, and R. G. Collman. 1998. CXCR-4 is expressed by primary macrophages and supports CCR5-independent infection by dual-tropic but not T-tropic isolates of human immunodeficiency virus type 1. J. Virol. 72:772-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yi, Y., F. Shaheen, and R. G. Collman. 2005. Preferential use of CXCR4 by R5X4 human immunodeficiency virus type 1 isolates for infection of primary lymphocytes. J. Virol. 79:1480-1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang, Y., B. Lou, R. B. Lal, A. Gettie, P. A. Marx, and J. P. Moore. 2000. Use of inhibitors to evaluate coreceptor usage by simian and simian/human immunodeficiency viruses and human immunodeficiency virus type 2 in primary cells. J. Virol. 74:6893-6910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang, Y. J., T. Dragic, Y. Cao, L. Kostrikis, D. S. Kwon, D. R. Littman, V. N. KewalRamani, and J. P. Moore. 1998. Use of coreceptors other than CCR5 by non-syncytium-inducing adult and pediatric isolates of human immunodeficiency virus type 1 is rare in vitro. J. Virol. 72:9337-9344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zimmerman, P. A., A. Buckler-White, G. Alkhatib, T. Spalding, J. Kubofcik, C. Combadiere, D. Weissman, O. Cohen, A. Rubbert, G. Lam, M. Vaccarezza, P. E. Kennedy, V. Kumaraswami, J. V. Giorgi, R. Detels, J. Hunter, M. Chopek, E. A. Berger, A. S. Fauci, T. B. Nutman, and P. M. Murphy. 1997. Inherited resistance to HIV-1 conferred by an inactivating mutation in CC chemokine receptor 5: studies in populations with contrasting clinical phenotypes, defined racial background, and quantified risk. Mol. Med. 3:23-36. [PMC free article] [PubMed] [Google Scholar]