Abstract
Translation of picornavirus plus-strand RNA genomes occurs via internal ribosomal entry at highly structured 5′ untranslated regions. In addition to canonical translation factors, translation rate is likely influenced by supplementary host and viral trans-acting factors. We previously reported that insertion of a heterologous human rhinovirus type 2 internal ribosomal entry site (IRES) into the poliovirus (PV) genome, generating the chimeric virus PV-RIPO, selectively abrogates viral translation and propagation in neurons, which eliminate poliovirus's signature neuropathogenicity. While severely deficient in cells of neuronal lineage, the rhinovirus IRES promotes efficient propagation of PV-RIPO in cancer cells. Tumor-specific IRES function can be therapeutically exploited to direct viral cytotoxicity to cancer cells. Neuron-glioma heterokaryon analysis implicates neuronal trans-dominant inhibition in this effect, suggesting that host trans-acting factors repress IRES function in a cell-type-specific manner. We identified a set of proteins from neuronal cells with affinity for the rhinovirus IRES, including double-stranded RNA-binding protein 76 (DRBP76). DRBP76 associates with the IRES in neuronal but not in malignant glioma cells. Moreover, DRBP76 depletion in neuronal cells enhances rhinovirus IRES-driven translation and virus propagation. Our observations suggest that cell-type-specific association of DRBP76 with the rhinovirus IRES represses PV-RIPO translation and propagation in neuronal cells.
Posttranscriptional regulation constitutes an important level of control of gene expression. The global rate of protein synthesis is influenced by modification of translation initiation factors. In addition, the translation of many mRNAs is independently regulated by processes targeting their untranslated regions (UTRs). In many instances, specific UTRs bind ribonucleoprotein complexes that alter conditions for translation of individual messages.
The plus-strand RNA genomes of Picornaviridae feature complex UTRs involved in the regulation of viral translation and genome replication. Translation of viral RNA occurs in competition with cellular mRNA in infected host cells (3). Accordingly, picornavirus genomes have adopted unconventional features enabling efficient viral translation while limiting host cell protein synthesis. In contrast to cellular mRNAs, picornaviral genomic RNAs are uncapped (34), and their uncommonly large and highly structured 5′UTRs contain internal ribosomal entry sites (IRESs) that mediate translation initiation in a 5′-end, cap-independent manner (26, 39). IRES-mediated translation is unimpeded by virus-induced cleavage of canonical initiation factors eukaryotic initiation factor 4G and poly(A)-binding protein (13, 27), indicating that their involvement in initiation at the IRES deviates from capped mRNAs. Moreover, divergent means of translation initiation imply the involvement of noncanonical translation factors or IRES trans-acting factors (ITAFs) (4). Such factors, by virtue of their cell and organ-specific distribution, may determine viral translation, propagation, and pathogenesis. For example, neuropathogenicity of the picornavirus Theiler's murine encephalomyelitis virus was reported to be influenced by cell-type-specific distribution of an ITAF in mouse brain (41).
Pathogenesis of poliovirus (PV), the archetypal picornavirus, is characterized by flaccid paralysis resulting from spread of the virus from the primary propagation site in the gastrointestinal tract to spinal cord motor neurons. Apart from host determinants of viral tropism and pathogenicity such as the PV receptor (21), specificity for motor neurons is encrypted within the viral IRES element. This is evident from neuroattenuating mutations mapping to the IRES in the live attenuated (Sabin) vaccine strains of PV (20), as well as from genetically engineered viruses with reduced neurovirulence (2). The most drastic change in neuropathogenicity results from exchange of the entire PV IRES. Insertion of the human rhinovirus type 2 (HRV2) IRES in place of its PV counterpart, generating the chimera PV-RIPO, depresses viral translation and propagation in neuron-like cell lines (e.g., Sk-N-Mc and HEK-293) (9, 17) and attenuates neurovirulence in both mice transgenic for the human PV receptor CD155 (17, 18) and nonhuman primates (18). While the heterologous HRV2 IRES precludes virus propagation in spinal cord motor neurons and prevents poliomyelitis in PV-susceptible organisms, it has no effect on rapid viral growth in nonneuronal malignant cell types, e.g., those derived from malignant glioma cells (19, 33).
Cell-type-specific incompetence of the HRV2 IRES maps to stem-loop domains V and VI (sldV/VI) (Fig. 2A) (9, 18) and is codetermined by viral 3′UTR sequences (11). The observed phenotype suggests involvement of ITAFs regulating viral growth in a cell-type-specific manner. Based on in vitro studies, a number of HRV2 ITAFs have been proposed. These include the polypyrimidine tract-binding protein (PTB) (25), upstream of N-ras (24), and poly(rC)-binding protein 2 (45). The influence of these factors on cell- and organ-type-specific activity of the HRV2 IRES is unknown.
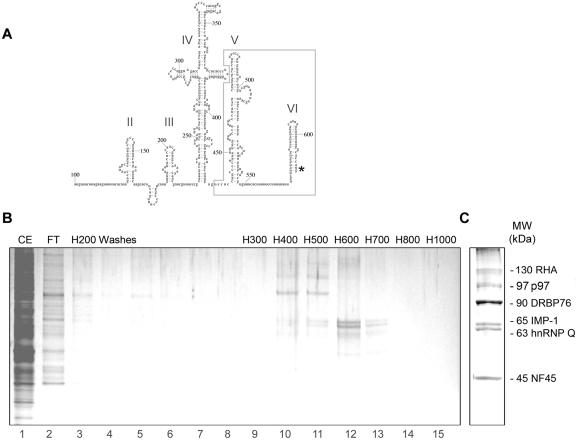
FIG. 2.
sldV/VI RNA affinity chromatography. (A) Predicted secondary structure of the HRV2 IRES element. Numbers refer to the nucleotide positions relative to the 5′ end of the genomic RNA. IRES sld's are marked by roman numerals, and the sldV/VI fragment used for our studies is boxed. The initiation codon is indicated by an asterisk. (B) Neuroblastoma RSW (lane 1, CE) was applied to an sldV/VI RNA affinity column. After collection of flowthrough (lane 2, FT), the column was washed with H200 (lanes 3 to 8) and eluted with a 300 to 1,000 mM KCl gradient (lanes 9 to 15). Column fractions were resolved by electrophoresis through a 4 to 12% SDS-PAGE gel and silver stained. (C) Eluates corresponding to H400 to H600 (panel B, lanes 10 to 12) were concentrated and resolved by electrophoresis through a 10% SDS-PAGE gel, and the protein bands were silver stained and excised for tryptic digestion and peptide sequencing.
Here, we report that the neuronal growth deficit of PV-RIPO is dominant over the permissive glioma phenotype with neuroblast-glioma heterokaryons. This implicates neuronal trans-dominant inhibitors in the cell-type-specific growth phenotype of PV-RIPO. We identified five proteins binding to the HRV2 IRES sldV/VI from a neuron-derived cell line by RNA affinity chromatography. These proteins were identified as double-stranded RNA-binding protein 76 (DRBP76), RNA helicase A (RHA), nuclear factor 45 (NF45), insulin-like growth factor II mRNA-binding protein 1 (IMP-1), and heterogeneous ribonucleoprotein Q1 (hnRNP Q1).
The subcellular distribution of DRBP76 in neuronal and nonneuronal malignant cells differs, and HRV2 IRES binding occurs only with DRBP76 from neuronal cell extracts. DRBP76 specifically associates with the HRV2 IRES in PV-RIPO-infected neuronal cells but not with wild-type PV genomic material. RNA interference-mediated DRBP76 depletion induces both PV-RIPO propagation and HRV2 IRES-driven reporter translation in neuronal cells, suggesting that DRBP76 inhibits PV-RIPO propagation at the level of translation.
MATERIALS AND METHODS
cDNA cloning and in vitro transcription.
Vectors for in vitro synthesis of IRES RNAs were constructed as follows: the HRV2 IRES fragment encompassing the T7 promoter and sldV/VI (nucleotides 434 to 613) was PCR amplified with primers 1 (5′-CCGGATCCTATAATACGACTCACTATAGGGATGATCCTCCGGCCCCTGAATGTGG-3′) and 2 (5′-CCGAATTCTGTGCACCCATGGTGCC-3′). The fragment was digested with BamHI and EcoRI for insertion into pBluescript II SK(+). IRES Renilla luciferase (rLuc), and capped firefly (fLuc) reporter constructs were generated as previously described (8). To synthesize nonspecific multiple-cloning site RNA, pBluescript II SK(+) digested with XhoI was used as a template for in vitro transcription with T3 RNA polymerase (Promega, Madison, WI).
pDRBP76-flag (a gift from G. Sen, Cleveland Clinic) (35) was used to derive pDRBP76(pTyb2), a rDRBP76 expression vector with a C-terminal intein tag. A PCR product corresponding to the DRBP76 open reading frame (ORF) was amplified from pDRBP76-flag using primers 3 (5′-GGATTCCATATGCGTCCAATGCGAATTTTTGT-3′) and 4 (5′-TCCCCCGGGTGAAGACCAAAATCATGAT-3′), digested with NdeI and SmaI, and ligated into pTyb2 (New England Biolabs). pshDRBP76, encoding a retroviral vector expressing short hairpin RNA (shRNA) complementary to DRBP76 mRNA, was generated by annealing primers 5 (5′-phos-GATGTGGATGGACAGGAGTTCCAAGGTGTTCAAGAGTACCTTGGAACTTCTGTCCATCCACTTTTTG-3′) and 6 (5′-phos-AGCTCAAAAAGTGGATGGACAGAAGTTCCAAGGTATCTCTTGAACACCTTGGAACTCCTGTCCATCCACGGG-3′), followed by ligation with pSUPER (7) digested with EcoRI and HindIII. The ligation product was digested with XbaI and ClaI and ligated with pNL-SIN to generate shRNA-expressing lentiviruses (all materials used to generate retroviral vectors were kindly provided by B. Cullen, Duke University) (28). pDRBP76mut, bearing silent mutations in the region targeted by short hairpin DRBP76 (shDRBP76) RNA, was generated by PCR amplification of contiguous fragments with primers 7 (5′-CCAGGATCCATGCGTCCAATGCGAATTTT-3′) and 8 (5′-GGTTTGTTCGAACCAGCACCTCGGAATTTCTGGCC-3′) and 9 (5′-GGTGCTGGTTCGAACAAGAAGGTGGCG-3′) and 10 (5′-GCGAATTCGATGAACAGCAGCAGTAGG-3′). Underlined sequences represent endonuclease cleavage sites. The resulting PCR products were digested with BamHI and SfuI or with SfuI and EcoRI, respectively, and ligated with pcDNA3.1+ (Invitrogen, Carlsbad, CA).
Heterokaryon analysis.
HEK-293 cells transfected with pdsRed-mito (BD Biosciences), HTB-14 cells transfected with pEGFP-N1 (BD Biosciences), or a 50:50 combination of these transfected cell lines was plated overnight in 35-mm dishes. The following day, the cells were washed with phosphate-buffered saline (PBS) and fused for 10 min at room temperature with 50% polyethylene glycol (Sigma), according to established procedures (42). Cells were infected 2 h postfusion with PV-RIPO at a multiplicity of infection of 10. Cell morphology and fusion were monitored with an Olympus IX50 fluorescence microscope with a 40× dry objective. Cells were harvested at specified time points and processed for one-step growth curve analysis by plaque assays of HeLa cell monolayers.
Cell extract preparation and Western blotting.
Nuclear, S10, and ribosomal salt wash (RSW) extracts of Sk-N-Mc, HEK-293, HTB-14, and DU54 cells were prepared essentially as described previously (7). Cell extracts (each, 0.5 mg/ml) were purified over 3-ml heparin-Sepharose columns (CL4B heparin Sepharose; Roche) before use in RNA affinity chromatography. Western blots were performed as described previously (11) with the following antibodies: primary α-DRBP76 (BD Transduction Laboratories), α-RHA (a gift from J. Hurwitz, Memorial Sloan Kettering Cancer Center), α-hnRNP Q (18E4, a gift from G. Dreyfuss, University of Pennsylvania), α-IMP-1 (a gift from F. Nielsen, Copenhagen University Hospital), α-NF45 (a gift from S. Behrens, Fox Chase Cancer Center), α-tubulin (Sigma), and α-2C (a gift from E. Wimmer, Stony Brook University).
RNA affinity chromatography, mass spectrophotometry, and peptide identification.
RNA affinity chromatography with HRV2 IRES sldV/VI on CNBr-activated Sepharose (Pharmacia) was performed essentially as described previously (6). Fractionated column flowthrough and eluates were collected and assayed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) with 4 to 12% Bis-Tris NuPAGE gels (Invitrogen) and silver staining. Proteins from 400 to 600 mM KCl eluates were precipitated with trichloracetic acid and analyzed by SDS-PAGE through a 10% SDS-PAGE gel, followed by silver staining. Individual bands were excised and sent to the Mass Spectrometry Core Facility, University of Massachusetts (Worcester) for identification by tryptic digestion and peptide sequencing; peptides spanning approximately 25% of each protein were sequenced (Table 1).
TABLE 1.
HRV2 IRES binding proteins identified by mass spectrometry
| Peptide | Measured mass (m/z, Da) | Computed mass (MH+, Da) | Residues | Peptide sequence |
|---|---|---|---|---|
| DRBP76 | ||||
| 1 | 707.4100 | 707.3629 | 215-219 | (K)WFQAR(A) |
| 2 | 878.4800 | 878.4327 | 6-12 | (I)FVNDDR(H) |
| 3 | 984.6500 | 984.5631 | 324-332 | (R)LAAFGQLHK(V) |
| 4 | 1,020.6100 | 1,020.5730 | 578-587 | (K)AYAALAALEK(L) |
| 5 | 1,056.7000 | 1,056.5763 | 333-342 | (K)VLGMDPLPSK(M) |
| 6 | 1,301.8300 | 1,301.7581 | 409-419 | (R)LNQLKPGLQYK(L) |
| 7 | 1,328.8200 | 1,328.6455 | 397-408 | (K)AEPPQAMNALMR(L) |
| 8 | 1,443.9400 | 1,443.7266 | 461-474 | (K)VLQDMGLPTGAEGR(D) |
| 9 | 1,780.9200 | 1,780.9054 | 309-323 | (R)QQREDITQSAQHALR(L) |
| 10 | 1,910.0900 | 1,909.9871 | 183-200 | (K)VLAGETLSVNDPPDVLDR(Q) |
| 11 | 2,671.2100 | 2,671.2422 | 18-40 | (K)HSSVYPTQEELEAVQNMVSHTER(A) |
| NF45 | ||||
| 1 | 886.5100 | 886.4899 | 320-327 | (R)ILSHGGFR(K) |
| 2 | 1,041.6800 | 1,041.6421 | 197-206 | (K)VLQSALAAIR(H) |
| 3 | 1,236.8700 | 1,236.7680 | 175-185 | (K)ILITTVPPNLR(K) |
| 4 | 1,888.1100 | 1,888.0180 | 44-60 | (R)VKPAPDETSFSEALLKR(N) |
| 5 | 2,099.1600 | 2,099.0984 | 61-80 | (R)NQDLAPNSAEQAILALVTK(I) |
| 6 | 2,582.2800 | 2,582.3830 | 81-103 | (K)INNVIDNLIVAPGTFEVQIEEVR(Q) |
| RHA | ||||
| 1 | 758.4400 | 758.4273 | 25-311 | (R)AVGNKNR(Q) |
| 2 | 758.4400 | 758.4313 | 900-905 | (R)LGYIHR(N) |
| 3 | 777.3900 | 777.4115 | 1,131-1,136 | (R)MLNMIR(Q) |
| 4 | 986.5400 | 986.5060 | 1,155-1,163 | (R)YGDGPRPPK(M) |
| 5 | 1,003.5900 | 1,003.5940 | 810-819 | (R)LGGIGQFLAK(A) |
| 6 | 1,025.5600 | 1,025.5420 | 64-71 | (R)DFVNYLVR(I) |
| 7 | 1,075.6200 | 1,075.5536 | 315-323 | (K)LAQFEPSQR(Q) |
| 8 | 1,087.6600 | 1,087.6224 | 447-456 | (R)RISAVSVAER(V) |
| 9 | 1,186.6300 | 1,186.5857 | 142-151 | (R)GANLKDYYSR(K) |
| 10 | 1,186.6300 | 1,186.5605 | 200-209 | (K)YTQVGPDHNR(S) |
| 11 | 1,464.8500 | 1,464.8327 | 681-692 | (R)YQILPLHSQIPR(E) |
| 12 | 1,714.9000 | 1,714.9016 | 1,075-1,089 | (K)VQSDGQIVLVDDWIK(L) |
| 13 | 1,741.8900 | 1,741.8608 | 838-853 | (R)ELDALDANDELTPLGR(I) |
| 14 | 1,872.0000 | 1,872.1033 | 1,058-1,074 | (K)GMTLVPPLQLLLFASKK(V) |
| 15 | 1,872.0000 | 1,872.0125 | 1,137-1,154 | (R)QISRPSAAGINLMIGSTR(Y) |
| 16 | 1,971.0300 | 1,971.0551 | 820-837 | (K)AIEPPPLDAVIEAEHTLR(E) |
| 17 | 2,050.0000 | 1,050.0245 | 418-434 | (K)TTQVPQFILDDFIQNDR(A) |
| hnRNP Q1 | ||||
| 1 | 860.6100 | 860.5246 | 245-252 | (R)LFVGSIPK(S) |
| 2 | 860.6100 | 860.5722 | 337-343 | (K)VKVLFVR(N) |
| 3 | 927.5800 | 927.5052 | 185-192 | (K)AGPIWDLR(L) |
| 4 | 1,058.6200 | 1,058.5383 | 222-229 | (K)LYNNHEIR(S) |
| 5 | 1,182.7300 | 1,182.6271 | 357-366 | (K)AFSQFGKLER(V) |
| 6 | 1,260.7500 | 1,260.6444 | 193-203 | (R)LMMDPLTGLNR(G) |
| 7 | 1,311.7600 | 1,311.6654 | 131-142 | (R)TGYTLDVTTGQR(K) |
| 8 | 1,473.9100 | 1,473.7801 | 344-356 | (R)NLANTVTEEILEK(A) |
| 9 | 1,593.9500 | 1,593.8052 | 172-184 | (R)DLFEDELVPLFEK(A) |
| IMP-1 | ||||
| 1 | 736.4500 | 736.3854 | 168-174 | (R)RGGFGSR(G) |
| 2 | 772.4800 | 772.4681 | 291-297 | (R)LIGKEGR(N) |
| 3 | 837.5100 | 837.4947 | 87-93 | (R)NIPPQLR(W) |
| 4 | 1,096.6600 | 1,096.6115 | 191-199 | (K)QQQVDIPLR(L) |
| 5 | 1,140.7300 | 1,140.6278 | 281-290 | (K)ILAHNNFVGR(L) |
| 6 | 1,309.8000 | 1,309.7116 | 441-452 | (K)IAPPETPDSKVR(M) |
| 7 | 1,430.8500 | 1,430.7717 | 453-465 | (R)MVHTGPPEAQFK(A) |
| 8 | 1,521.8300 | 1,521.7636 | 539-551 | (K)HGHFYASQMAQR(K) |
| 9 | 1,853.0500 | 1,852.9961 | 21-36 | (K)VFAEHKISYSGQFLVK(S) |
| 10 | 1,853.0500 | 1,852.8356 | 331-345 | (K)GAIENCCRAEQEIMK(K) |
| 11 | 1,853.0500 | 1,853.0133 | 509-525 | (K)TVNELQNLTAAEVVVPR(D) |
| 12 | 2,029.0500 | 2,029.0103 | 151-168 | (K)VSYIPDEQIAQGPENGRR(G) |
| 13 | 2,131.2100 | 2,131.1624 | 179-199 | (R)QGSPVAAGAPAKQQQVDIPLR(L) |
Immunoprecipitation-RT-PCR analysis.
HEK-293 cells were either transfected with subgenomic viral RNAs or infected with PV-RIPO at a multiplicity of infection of 100. Subgenomic PV and PV-RIPO RNAs were generated from cDNA clones by digestion with SfuI, thereby linearizing the plasmid in the VP0 coding region, followed by in vitro transcription with T7 RNA polymerase. At 2 h posttransfection or 4 h postinfection (hpi), the cells were rinsed, detached by being scraped in PBS, and collected by centrifugation. Immunoprecipitation was carried out at 4°C; the cells were lysed in 200 μl radioimmunoprecipitation buffer (50 mM Tris-HCl [pH 8.0], 150 mM KCl, 1 mM EGTA, 5 mM MgCl2, 0.25% NP-40, 0.05% sodium deoxycholate, and 100-U/ml RNase OUT) with gentle rocking for 15 min, followed by centrifugation for 10 min at 14,000 rpm. For preclearing, the supernatant was treated with 100 μl of 50% protein A-Sepharose beads (Amersham-Pharmacia) in PBS per ml of lysate for 10 min, followed by pulse centrifugation. The supernatant was diluted to 1 mg/ml in PBS and incubated overnight with α-DRBP76 antibody at 5 μg/ml. A total of 100 μl of 50% protein A-Sepharose beads in PBS was added, followed by incubation for 1 h with gentle rocking. The beads were collected by pulse centrifugation, followed by two washes with ice-cold detergent-free radioimmunoprecipitation buffer and two washes with ice-cold water. Total RNA was extracted from the beads with TRIZOL LS reagent (Invitrogen) according to the manufacturer's protocol. RNA was analyzed by reverse transcription-PCR (RT-PCR) analysis as described previously (19). RT-PCR amplification of both PV- and HRV2 IRES-containing RNAs was performed with primers 11 (5′-CGCCTGTTTTATACTCCCTTCCC-3′) (annealing to the cloverleaf) and 12 (5′-CATGTGCGCCCACTTTCTGTG-3′) (annealing to the 5′ proximal viral ORF). For RT-PCR amplification of c-myc mRNA, we used primers 13 (5′-GCGGATCCTAATACGACTCACTATAGGGGAGGACCCCCGAGCTGTG-3′) and 14 (5′-GTTTTCCACTACCCGAAAAAAATCC-3′).
Cell culture, virus infections, and reporter assays.
Cell culture and propagation, virus infections, and growth assays were performed as described previously (17-19). DNA and RNA transfections were performed with Lipofectamine 2000 and DMRIE-C reagent, respectively (Invitrogen). DNA was incubated with Lipofectamine complexes in OPTI-MEM (Invitrogen) for 20 min and then added to 90% confluent cells in a 10-cm dish containing 15 ml of growth medium with 10% fetal bovine serum. Transfection of reporter RNAs, luc reporter assays, and RNA stability evaluations were performed as described previously (11).
RESULTS
Neuronal cell culture models of PV neurovirulence.
Attenuated neurovirulence of the oral PV (Sabin) vaccines (1, 28) and PV-RIPO (17) can be recapitulated with neuroblastoma cells. We have frequently used Sk-N-Mc neuroblastoma cells in our previous studies (17, 18), including the generation of RSW used in affinity chromatography assays reported here. During the course of our studies, HEK-293 cells, derived from primary human embryonic kidney, were determined to be of neuronal lineage (43). Empirical evidence suggests that mixed primary embryonic kidney cultures contain cells of neuronal lineage, which are selected for by transformation with adenoviral genomic DNA (43). HEK-293 cells have certain advantages over neuroblastoma cells as tissue culture models for neuronal virus growth phenotypes. They faithfully recapitulate neuronal tropism of PV observed in experimental animals and do not spontaneously interconvert from neuron-like to fibroblast-like phenotypes, a notorious property of neuroblastoma cell lines (9). Therefore, during the course of this study, we switched to exclusive use of HEK-293 cells (American Type Culture Collection; passage 53).
Heterokaryon analyses.
Despite unimpeded function in cancer cells, e.g., HeLa, breast cancer, or malignant glioma cells, the HRV2 IRES is severely deficient in neuron-like cells (19, 35). This suggests that either cancer-specific inducers or neuronal inhibitors modulate IRES activity in a cell-type-specific manner. We performed heterokaryon analysis to distinguish cancer- from neuron-specific causes for the selective PV-RIPO growth phenotype. To this end, nonpermissive neuronal HEK-293 cells (9, 43) and permissive glioma HTB-14 cells (19) were fused either to themselves or to each other. Fusion was evident by merging of HEK-293 and HTB-14 compartments containing the fluorescent gene products red fluorescent protein and green fluorescent protein, respectively: incorporation of HEK-293 mitochondria into HTB-14 cytoplasm was detected at 2 h postfusion (Fig. 1A). Approximately 80% of cocultured HEK-293 and HTB-14 cells formed heterokaryons after fusion. Fused HEK-293, HTB-14, or HEK-293/HTB-14 heterokaryons were infected with PV-RIPO at 2 h postfusion; virus propagation was monitored by one-step growth curves (Fig. 1B and C). Fused HTB-14 cells support robust PV-RIPO propagation resembling growth in untreated glioma cells (19); as expected, viral growth was negligible in fused HEK-293 cells, reflecting the nonpermissive neuronal phenotype. Intriguingly, HEK-293/HTB-14 heterokaryons exhibited severely reduced viral propagation, yielding titers of >100 times below those of fused HTB-14 cells and only slightly elevated over those of fused HEK-293 cells. Residual growth in treated cocultures likely occurred in HTB-14 cells that were not fused with HEK-293 cells. These findings indicate trans-dominant inhibition of PV-RIPO growth in neuronal cells.
FIG. 1.
HEK-293/HTB-14 heterokaryon analyses. (A) Fluorescence microscopy of fused HTB-14 cells, fused HEK-293 cells, and HEK-293/HTB-14 heterokaryons at 2 h postfusion. HTB-14 and HEK-293 cells express green fluorescent protein (GFP) throughout the cytoplasm or red fluorescent protein (RFP) targeted to mitochondria, respectively; note overlapping fluorescent compartments in heterokaryons. (B) One-step growth curve analysis of PV-RIPO in fused HTB-14 cells (⧫), fused HEK-293 cells (▪), and HEK-293/HTB-14 heterokaryons (▴). (C) Plaque assay of cell lysate diluents from fused HTB-14, HEK-293, and HEK-293/HTB-14 heterokaryons at 0 hpi and 6 hpi with PV-RIPO on HeLa cell monolayers.
Isolation and identification of HRV2 IRES sldV/VI-binding proteins.
Since heterokaryon analyses implicated neuronal inhibitors in HRV2 IRES incompetence, we searched for HRV2 IRES-binding proteins from neuroblastoma cells. We employed IRES sldV/VI for our binding analysis, because genetic experiments mapped neuronal dysfunction to this portion of the IRES (9, 17, 18). To identify proteins present in neuroblastoma RSW interacting with HRV2 IRES sldV/VI (Fig. 2A), we performed RNA affinity chromatography. RSW was loaded onto a Sepharose column coupled to sldV/VI. After extensive washes, bound proteins were eluted with a stepwise 300 to 1,000 mM KCl-buffer gradient, separated by SDS-PAGE, and silver stained (Fig. 2B). Six proteins with approximate masses of 130, 90, 65, 63, and 45 kDa were eluted from the column in the 400 mM to 600 mM KCl range at levels permitting recovery for identification by mass spectrophotometric analysis of tryptic peptide fragments (Fig. 2C). The deduced peptide sequences (Table 1) and approximate molecular weights enabled unequivocal identification of p130 (RHA), p90 (DRBP76), p65 (IMP-1), p63 (hnRNP Q1), and p45 (NF45). RHA, DRBP76, IMP-1, and hnRNP Q1 contain RNA-binding motifs and have known roles in mRNA stability, localization, and translation.
DRBP76 subcellular distribution is cell type specific.
Involvement of an ITAF in the cell-type-specific deficit of the HRV2 IRES implies differences in expression or availability of this protein in neuronal versus glioma cells. Since all steps of the picornaviral life cycle take place in the cytoplasm, the subcellular localization of IRES-binding proteins may have profound influence on IRES function. Therefore, we performed Western blot analyses of nuclear, cytoplasmic, and RSW extracts from cells of either origin to probe the cell-type-specific abundance of our candidate IRES-binding proteins. To assure even loading, protein abundance was evaluated by Bradford analysis (data not shown).
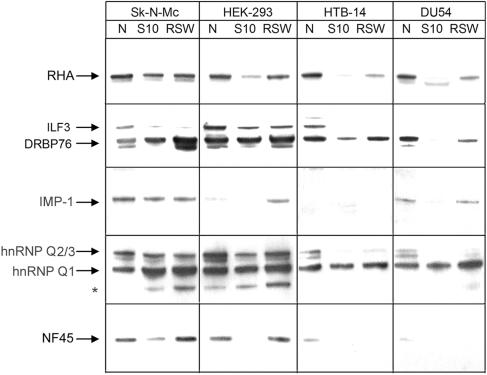
Distribution among cell types and subcellular compartments was fairly even for RHA and hnRNP Q1 (Fig. 3). The hnRNP Q2/3 isoforms are more abundant in neuronal cells (Fig. 3), but they were not identified to bind to the IRES in our search. IMP-1 distribution varied among neuronal and glioma subcellular extracts, respectively, making it an unattractive neuron-specific ITAF candidate (Fig. 3).
FIG. 3.
Expression profile of IRES-binding proteins in cultured cell lines. Sk-N-Mc neuroblastoma and HEK-293 neuronal cell extracts and HTB-14 and DU54 glioma cell extracts (5 μg/lane) were resolved by SDS-PAGE and analyzed by Western blotting with α-RHA, α-DRBP76, α-IMP-1, α-hnRNP Q, and α-NF45 antibodies as indicated. N, nuclear fraction; S10, crude cytoplasmic fraction.
DRBP76 and NF45, while similarly abundant in nuclear fractions of all cell lines, are significantly enriched in cytoplasmic and RSW fractions of neuronal cells (Fig. 3). DRBP76 is capable of binding both single-stranded RNA and double-stranded RNA (dsRNA) (31) and as such is a satisfactory ITAF candidate. Interleukin enhancer-binding factor 3 (ILF3) is a predominantly nuclear, larger isoform of DRBP76 (12), which did not bind the HRV2 IRES in our assay. Since NF45 does not contain RNA-binding motifs and has been shown to bind DRBP76 (10, 31), its interaction with the HRV2 IRES is likely to occur by virtue of association with DRBP76. Based on these considerations, we investigated the possibility that DRBP76 may act as an inhibitory ITAF governing the non-neuronal-growth phenotype of PV-RIPO.
Binding of DRBP76 to the HRV2 IRES is neuron specific.
DRBP76 is expressed in neuronal and glioma cells at different levels and with distinct intracellular distribution. Therefore, we investigated whether IRES binding of DRBP76 is cell type specific by performing comparative affinity chromatography analysis using cytoplasmic extracts from HEK-293 and HTB-14 cells (Fig. 4). S10 cytoplasmic lysates were loaded onto sldV/VI RNA columns, and protein was eluted with a stepwise KCl gradient. The protein elution profile was monitored by silver staining of SDS-PAGE gels (Fig. 4). In accordance with previous observations of a neuroblastoma RSW (Fig. 2), DRBP76 from HEK-293 cells efficiently bound to sldV/VI (Fig. 4A). Interestingly, however, we did not observe binding of DRBP76 from HTB-14 cells, although it was clearly present in the flowthrough (Fig. 4B). This suggests that the IRES-binding ability or availability of DRBP76 varies in a cell-type-specific manner.
FIG. 4.
Comparative RNA affinity chromatography. HEK-293 (A) and HTB-14 (B) S10 cytoplasmic lysates were applied to sldV/VI RNA affinity columns. After collection of flowthrough (lane 1, FT), the column was washed with H200 (lanes 2 to 3) and eluted with a 400 to 1,000 mM KCl gradient (lanes 4 to 11). Column fractions were analyzed by SDS-PAGE and silver stain or Western blot analysis with α-DRBP76.
DRBP76 associates with the HRV2 IRES in vivo.
DRBP76 specifically interacts with the HRV2 IRES in neuronal, but not malignant, glioma cell extracts (Fig. 4). We investigated whether this association occurs in vivo and whether it is specific for the HRV2 IRES. This was accomplished through RT-PCR analyses of viral RNA coimmunoprecipitated with DRBP76 in HEK-293 cells (Fig. 5). To demonstrate specific binding of DRBP76, we compared in vivo affinity for the HRV2 and PV IRESs. Side-by-side analysis of nonpropagating PV-RIPO with PV, which generates vast amounts of viral RNA in infected HEK-293 cells (9), would introduce bias. Therefore, we transfected nonreplicating subgenomic RNAs differing only with regard to IRES origin (Fig. 5A and B). RT-PCR amplified a viral cDNA fragment from RNA coimmunoprecipitating with DRBP76 in cells transfected with PV-RIPO subgenomic RNA (Fig. 5A); no RT-PCR product was obtained from the DRBP76 immunoprecipitate from cells transfected with PV subgenomic RNA (Fig. 5B). Since only the IRES region differed in both constructs, we can deduce that DRBP76 does not interact with any part of the viral subgenome other than the IRES and that the association occurs with the HRV2 but not the PV IRES. Moreover, amplification of IRES cDNA did not occur in the absence of RT enzyme or after immunoprecipitation with nonspecific immunoglobulin G (IgG) (Fig. 5A and B).
FIG. 5.
Immunoprecipitation-RT-PCR of DRBP76 from HEK-293 cells transfected with PV-RIPO (A) and PV (B) subgenomic RNAs, as well as cells infected with PV-RIPO (C and D). Total RNA or immunoprecipitates generated with α-DRBP76, nonspecific IgG, or α-2C antibodies as indicated were subjected to RT-PCR amplification of the HRV2 (A and C), PV (B), or a c-myc 5′UTR (D). −RT and +RT, reactions carried out in the absence and presence of reverse transcriptase, respectively. (E) Western blot detection of DRBP76 in the immunoprecipitates generated with α-DRBP76 and nonspecific IgG antibodies as indicated.
To examine the HRV2 IRES-DRBP76 interaction in the context of the intact viral genome, we performed DRBP76 immunoprecipitation from PV-RIPO-infected HEK-293 cells. As with subgenomic RNA, PV-RIPO cDNA was amplified from DRBP76 immunoprecipitate (Fig. 5C). In a positive control, IRES cDNA was amplified from RNA coimmunoprecipitated with the nonstructural PV protein 2C (Fig. 5C), which associates with viral RNA in infected cells (5). In contrast, the endogenous cellular IRES-containing c-myc mRNA did not coimmunoprecipitate with DRBP76, while c-myc cDNA was readily amplified from total cellular RNA (Fig. 5D). Amplification of c-myc cDNA from total RNA required RT enzyme (Fig. 5D), indicating that cDNA amplification was due to the presence of c-myc mRNA and not to contaminating genomic DNA. We conclude that DRBP76 associates specifically with PV-RIPO genomic RNA in infected HEK-293 cells.
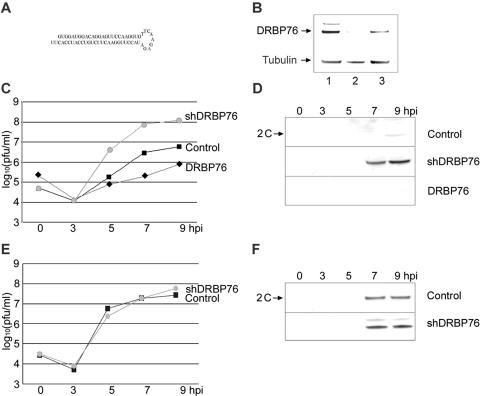
DRBP76 knockdown induces PV-RIPO propagation.
Specific association of DRBP76 from neuronal cells with the HRV2 IRES suggests a potential role in trans-dominant repression of PV-RIPO propagation (Fig. 1). To investigate such a role for DRBP76, we analyzed the effect of its depletion on viral growth in neuronal cells. shRNA was employed to knock down DRBP76 expression by a previously established lentivirus delivery strategy (Fig. 6A) (8, 30). Lentivirus infection and ensuing blasticidin selection did not produce morphological abnormalities or affect the viability of shDRBP76 cells (data not shown), but DRBP76-ILF3 protein levels were dramatically reduced in shDRBP76 cells relative to those of control HEK-293 cells infected with empty retroviral vector (Fig. 6B). shDRBP76 and control cells were subjected to synchronized infection with PV-RIPO to establish the kinetics of viral propagation and HRV2-IRES-driven translation (Fig. 6C and D). DRBP76 depletion substantially elevated viral growth; progeny at 7 hpi in shDRBP76 cells exceeded that in the controls by ∼20 fold (Fig. 6C), accompanied by markedly increased viral translation (Fig. 6D). In contrast, DRBP76 depletion had no effect on virus replication or translation of PV (Fig. 6E and F). To exclude nonspecific or off-target effects of shRNA-mediated knockdown, we reconstituted the protein in shDRBP76 cells by transfecting a DRBP76 cDNA modified by silent mutagenesis in the region targeted by the shRNA. DRBP76 complementation yielded wild-type levels of the protein (Fig. 6B) and reversed the enhancing effects of DRBP76 depletion on PV-RIPO propagation (Fig. 6C) and translation (Fig. 6D).
FIG. 6.
DRBP76 depletion enhances PV-RIPO propagation in HEK-293 cells. (A) Schematic depiction of shRNA targeting the DRBP76 mRNA. (B) Western blot analysis of control cells (lane 1), shDRBP76 cells (lane 2), and shDRBP76 cells transfected with DRBP76mut DNA (lane 3) using α-DRBP76 and α-tubulin antibodies as indicated. One-step growth curve analysis of PV-RIPO (C) and PV (E) propagation in control cells (▪), shDRBP76 cells (•), or shDRBP76 cells transfected with pDRBP76mut DNA (⧫) is shown. Western blot analysis of PV-RIPO (D) and PV (F) proteins in infected cell lysates at specified hours postinfection using α-2C antibody is shown.
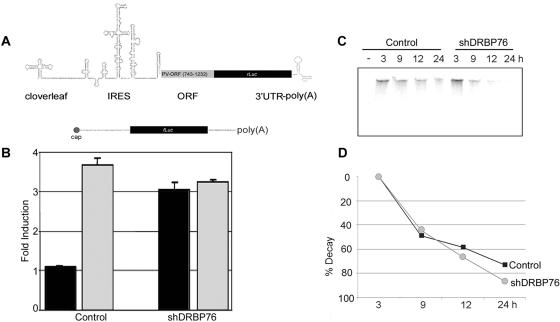
DRBP76 knockdown induces HRV2 IRES activity.
Translation and replication of picornavirus genomic RNAs are intertwined processes involving the same template. Hence, ITAFs conceivably can influence virus propagation at multiple levels. To assess a role for DRBP76 in translation control at the HRV2 IRES, we analyzed the effect of DRBP76 knockdown on translation of HRV2 IRES-driven rLuc reporters. To separately evaluate an influence of DRBP76 on PV translation, we constructed analogous PV-IRES-containing reporters. The expression constructs contain the known determinants of picornavirus type 1 IRES-driven translation, including the 3′UTR, poly(A) tail, cloverleaf, and a portion of the 5′ proximal viral ORF (Fig. 7A) (E. Y. Dobrikova et al., unpublished data). shDRBP76 and control cells were cotransfected with IRES reporter RNA and m7GTP-capped fLuc RNA (Fig. 7A). IRES driven translation was analyzed relative to cap-dependent translation by a dual rLuc/fLuc reporter assay at 6 h posttransfection. While DRBP76 knockdown did not dramatically affect PV IRES-mediated reporter expression, HRV2 IRES-driven translation was induced 3.5 fold (Fig. 7B). DRBP76 depletion did not alter reporter RNA stability in HEK-293 cells (Fig. 7C and D), suggesting that induction of HRV2 IRES-driven translation after DRBP76 knockdown is not due to mRNA stabilization.
FIG. 7.
DRBP76 represses HRV2 IRES-driven reporter translation. (A) Schematic depiction of the HRV2 IRES rLuc reporter expression construct (top) and the capped fLuc reporter construct containing the β-globin 5′UTR (bottom). (B) Effect of DRBP76 knockdown on HRV2 (black columns) or PV (gray columns) IRES-driven rLuc translation relative to capped fLuc translation in shDRBP76 and control HEK-293 cells. The data are the average of three independent assays plus standard error and are expressed as the fold induction of rLuc activity relative to fLuc activity. (C) DRBP76 depletion does not affect the stability of the HRV2 IRES reporter construct in vivo. 32P-labeled HRV2 IRES reporter RNAs recovered from transfected or untransfected (−) shDRBP76 and control cells were analyzed on a denaturing polyacrylamide gel. (D) Kinetics of labeled HRV2 IRES reporter RNA decay by phosphorimager quantification in control (▪) and shDRBP76 (•) cells.
DISCUSSION
Viruses have adapted to the intracellular milieu of specific host cells to successfully complete their replication cycle. The specific host cell environment determines the ability of viral UTRs to control viral translation and genome replication. To dissect virus-host interactions mediating these functions, host factors have been identified which interact with viral 5′ or 3′UTRs. Such searches have been conducted biochemically for many plus- and minus-strand RNA viruses and yielded a rich repertoire of potential candidates. Generally, there is scarce evidence for a functional role of most such host factors in viral cell type specificity or pathogenesis.
The first host factors determined to interact with type 1 picornavirus IRESs (PV and HRV) were the La autoantigen (32) and PTB (23, 25). The IRES stimulatory activity of PTB was demonstrated in vivo by transient overexpression (16) and targeted depletion (14). In addition, variable IRES trans activity of PTB and its neural isoform nPTB have been implicated in the neuroattenuation phenotype of the serotype 3 Sabin strain (22). Cell-type-specific growth of the picornavirus hepatitis A virus is determined by the relative abundance of PTB, a stimulatory ITAF, and glyceraldehyde 3-phosphate dehydrogenase, an inhibitory ITAF (46), demonstrating that cell-type-specific viral propagation can be regulated by IRES-binding proteins with opposing effects on translation.
Efficient PV propagation in the human spinal cord and the propensity to cause paralytic poliomyelitis are partly the result of optimized IRES performance in motor neurons. Insertion of a heterologous HRV2 IRES into the PV genome abrogates viral translation and propagation in motor neurons (17) but does not affect growth in malignant cell types, implicating cell-type-specific host factors in the control of IRES activity. trans-Dominant inhibition of PV-RIPO propagation in neuroblast-glioma heterokaryons suggests that a factor(s) in neuronal cells may repress the HRV2 IRES. We report that DRBP76 bound to HRV2 IRES sldV/VI specifically in neuronal and not glioma cell extracts. This association also occurred in vivo with the intact HRV2 IRES but not with its PV counterpart or a cellular IRES element.
DRBP76 contains two dsRNA-binding motifs and is almost identical to M-phase phosphoprotein 4, NF90, translation control protein 80 (TCP80), and NF associated with dsRNA-1 (designated NFAR-1). It has been assigned several disparate functions (36), but its effects on translation stand out. DRBP76 binds to acid β-glucosidase mRNA and inhibits translation (46, 47). Moreover, it is a regulator and substrate of the dsRNA-dependent protein kinase PKR, whose capacity to inhibit translation initiation is a key component of the innate antiviral response (29, 36, 38).
Association of DRBP76 with the IRES specifically occurs in cytoplasmic extracts of neuron- but not glioma-derived cells, although the protein is present in both. The mechanisms regulating the RNA-binding capacity of DRBP76 have not been characterized, but posttranslational modifications and the extent of engagement in RNPs may control the potential for interaction with viral RNA in a cell-type-specific manner (37, 44, 48). Thus, the intracellular microenvironment may balance the availability of RNA-binding proteins for interaction with target messages and their association with the translation machinery. This balance may vary in a cell-type-specific manner, affecting IRES performance according to cell type, which may be difficult to mimic in crude in vitro translation extracts.
Previous investigations of translation initiation at IRESs have focused on the identification of cellular RNA-binding proteins that stimulate IRES function (40). However, like the overwhelming majority of translation modulators acting through binding to capped mRNA UTRs (15), ITAFs may repress IRES activity. DRBP76 depletion elevates translation at the HRV2 IRES and significantly enhances PV-RIPO growth, suggesting that DRBP76 represses HRV2 IRES-mediated translation in neuronal cells. The relative effect of DRBP76 depletion in HEK-293 cells on HRV2 IRES-driven translation is less than on PV-RIPO propagation. HRV2 IRES repression is likely to affect rate-limiting steps early in the viral life cycle by inhibiting biosynthesis of viral proteins after genome uncoating. Thus, even slightly reduced translation may dramatically affect virus growth. Alternatively, the interaction of DRBP76 with viral RNA may exert effects beyond translation. Picornavirus genome replication and translation are interlaced processes that are difficult to dissect experimentally. Translation repression lowers the yield of nonstructural proteins required for genome replication, and reduced RNA replication hinders accumulation of translation templates. Thus, concomitant effects of DRBP76 on translation and genome replication cannot be categorically excluded. Wild-type PV readily propagates in neuronal cells, reflecting its neuropathogenic properties, and DRBP76 depletion has no effect on viral growth or translation via its cognate IRES.
The ability of DRBP76 to thwart propagation of a genetically engineered PV demonstrates that the intracellular distribution and RNA-binding capacity of translation factors can determine translation, particle propagation, and hence the pathogenic features of viruses. Manipulation of viral noncoding sequences can be exploited to selectively abolish viral translation in certain cells or tissues for therapeutic purposes (19).
Acknowledgments
We thank Haifan Lin, Shelton Bradrick, and Paola Florez (Duke University) for critical reading of the manuscript. We are grateful to Ganes Sen (Cleveland Clinics) for providing the pDRBP76-flag expression plasmid; Bryan Cullen (Duke University) for the pSUPER, pNL-SIN, pTat, pRev and pHIT-G plasmids; Jerard Hurwitz (Memorial Sloan-Kettering Cancer Center) for the α-RHA antibody; Gideon Dreyfuss (University of Pennsylvania) for the α-hnRNP Q 18E4 antibody; Finn Nielsen (Copenhagen University Hospital) for the α-IMP1 antibody; Sven-Erik Behrens (Fox Chase Cancer Center) for the α-NF45 antibody; and Eckard Wimmer (Stony Brook University) for the α-2C antibody.
This work is supported by PHS grant CA87537. M.G. is a recipient of a Burroughs Wellcome Career Award in the Biomedical Sciences. We gratefully acknowledge a Seth Harris Feldman Research Award from the Brain Tumor Society.
REFERENCES
- 1.Agol, V. I., S. G. Drozdov, T. A. Ivannikova, M. S. Kolesnikova, M. B. Korolev, and E. A. Tolskaya. 1989. Restricted growth of attenuated poliovirus strains in cultured cells of a human neuroblastoma. J. Virol. 63:4034-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agol, V. I., E. V. Pilipenko, and O. R. Slobodskaya. 1996. Modification of translational control elements as a new approach to design of attenuated picornavirus strains. J. Biotechnol. 44:119-128. [DOI] [PubMed] [Google Scholar]
- 3.Belsham, G., and R. Jackson. 2000. Translation initiation on picornavirus RNA, p. 869-900. In N. Sonenberg, J. W. B. Hershey, and M. B. Mathews (ed.), Translational control of gene expression. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 4.Belsham, G. J., and N. Sonenberg. 1996. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 60:499-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienz, K., D. Egger, M. Troxler, and L. Pasamontes. 1990. Structural organization of poliovirus RNA replication is mediated by viral proteins of the P2 genomic region. J. Virol. 64:1156-1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borman, A., M. T. Howell, J. G. Patton, and R. J. Jackson. 1993. The involvement of a spliceosome component in internal initiation of human rhinovirus RNA translation. J. Gen. Virol. 74:1775-1788. [DOI] [PubMed] [Google Scholar]
- 7.Brown, B. A., and E. Ehrenfeld. 1979. Translation of poliovirus RNA in vitro: changes in cleavage pattern and initiation sites by ribosomal salt wash. Virology 97:396-405. [DOI] [PubMed] [Google Scholar]
- 8.Brummelkamp, T. R., R. Bernards, and R. Agami. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science 296:550-553. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, S., J. Lin, E. Dobrikova, and M. Gromeier. 2005. Genetic determinants of cell-type specific poliovirus propagation in HEK 293 cells. J. Virol. 79:6281-6290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corthesy, B., and P. N. Kao. 1994. Purification by DNA affinity chromatography of two polypeptides that contact the NF-AT DNA binding site in the interleukin 2 promoter. J. Biol. Chem. 269:20682-20690. [PubMed] [Google Scholar]
- 11.Dobrikova, E., P. Florez, S. Bradrick, and M. Gromeier. 2003. Activity of a type 1 picornavirus internal ribosomal entry site is determined by sequences within the 3′ nontranslated region. Proc. Natl. Acad. Sci. USA 100:15125-15130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Duchange, N., J. Pidoux, E. Camus, and D. Sauvaget. 2000. Alternative splicing in the human interleukin enhancer binding factor 3 (ILF3) gene. Gene 261:345-353. [DOI] [PubMed] [Google Scholar]
- 13.Etchison, D., S. C. Milburn, I. Edery, N. Sonenberg, and J. W. Hershey. 1982. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J. Biol. Chem. 257:14806-14810. [PubMed] [Google Scholar]
- 14.Florez, P. M., O. M. Sessions, E. J. Wagner, M. Gromeier, and M. A. Garcia-Blanco. 2005. The polypyrimidine tract binding protein is required for efficient picornavirus gene expression and propagation. J. Virol. 79:6172-6179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebauer, F., and M. W. Hentze. 2004. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 5:827-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gosert, R., K. H. Chang, R. Rijnbrand, M. Yi, D. V. Sangar, and S. M. Lemon. 2000. Transient expression of cellular polypyrimidine-tract binding protein stimulates cap-independent translation directed by both picornaviral and flaviviral internal ribosome entry sites in vivo. Mol. Cell. Biol. 20:1583-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gromeier, M., L. Alexander, and E. Wimmer. 1996. Internal ribosomal entry site substitution eliminates neurovirulence in intergeneric poliovirus recombinants. Proc. Natl. Acad. Sci. USA 93:2370-23705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gromeier, M., B. Bossert, M. Arita, A. Nomoto, and E. Wimmer. 1999. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J. Virol. 73:958-964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gromeier, M., S. Lachmann, M. R. Rosenfeld, P. H. Gutin, and E. Wimmer. 2000. Intergeneric poliovirus recombinants for the treatment of malignant glioma. Proc. Natl. Acad. Sci. USA 97:6803-6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gromeier, M., and A. Nomoto. 2002. Pathogenesis of Enteroviruses, p. 426-443. In B. L. Semler and E. Wimmer (ed.), Picornaviruses. ASM Press, Washington, DC.
- 21.Gromeier, M., D. Solecki, D. D. Patel, and E. Wimmer. 2000. Expression of the human poliovirus receptor/CD155 gene during development of the central nervous system: implications for the pathogenesis of poliomyelitis. Virology 273:248-257. [DOI] [PubMed] [Google Scholar]
- 22.Guest, S., E. Pilipenko, K. Sharma, K. Chumakov, and R. P. Roos. 2004. Molecular mechanisms of attenuation of the Sabin strain of poliovirus type 3. J. Virol. 78:11097-11107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hellen, C. U., G. W. Witherell, M. Schmid, S. H. Shin, T. V. Pestova, A. Gil, and E. Wimmer. 1993. A cytoplasmic 57-kDa protein that is required for translation of picornavirus RNA by internal ribosomal entry is identical to the nuclear pyrimidine tract-binding protein. Proc. Natl. Acad. Sci. USA 90:7642-7646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunt, S. L., J. J. Hsuan, N. Totty, and R. J. Jackson. 1999. unr, a cellular cytoplasmic RNA-binding protein with five cold-shock domains, is required for internal initiation of translation of human rhinovirus RNA. Genes Dev. 13:437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hunt, S. L., and R. J. Jackson. 1999. Polypyrimidine-tract binding protein (PTB) is necessary, but not sufficient, for efficient internal initiation of translation of human rhinovirus-2 RNA. RNA 5:344-359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jang, S. K., H. G. Krausslich, M. J. Nicklin, G. M. Duke, A. C. Palmenberg, and E. Wimmer. 1988. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62:2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joachims, M., P. C. Van Breugel, and R. E. Lloyd. 1999. Cleavage of poly(A)-binding protein by enterovirus proteases concurrent with inhibition of translation in vitro. J. Virol. 73:718-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.La Monica, N., and V. R. Racaniello. 1989. Differences in replication of attenuated and neurovirulent polioviruses in human neuroblastoma cell line SH-SY5Y. J. Virol. 63:2357-2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Langland, J. O., P. N. Kao, and B. L. Jacobs. 1999. Nuclear factor-90 of activated T-cells: a double-stranded RNA-binding protein and substrate for the double-stranded RNA-dependent protein kinase, PKR. Biochemistry 38:6361-6368. [DOI] [PubMed] [Google Scholar]
- 30.Lee, M. T., G. A. Coburn, M. O. McClure, and B. R. Cullen. 2003. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J. Virol. 77:11964-11972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liao, H. J., R. Kobayashi, and M. B. Mathews. 1998. Activities of adenovirus virus-associated RNAs: purification and characterization of RNA binding proteins. Proc. Natl. Acad. Sci. USA 95:8514-8519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meerovitch, K., J. Pelletier, and N. Sonenberg. 1989. A cellular protein that binds to the 5′-noncoding region of poliovirus RNA: implications for internal translation initiation. Genes Dev. 3:1026-1034. [DOI] [PubMed] [Google Scholar]
- 33.Merrill, M. K., G. Bernhardt, J. H. Sampson, C. J. Wikstrand, D. D. Bigner, and M. Gromeier. 2004. Poliovirus receptor CD155-targeted oncolysis of glioma. Neuro-oncology 6:208-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nomoto, A., Y. F. Lee, and E. Wimmer. 1976. The 5′ end of poliovirus mRNA is not capped with m7G(5′)ppp(5′)Np. Proc. Natl. Acad. Sci. USA 73:375-380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ochiai, H., S. A. Moore, G. E. Archer, T. Okamura, T. A. Chewning, J. R. Marks, J. H. Sampson, and M. Gromeier. 2004. Treatment of intracerebral neoplasia and neoplastic meningitis with regional delivery of oncolytic recombinant poliovirus. Clin. Cancer Res. 10:4831-4838. [DOI] [PubMed] [Google Scholar]
- 36.Parker, L. M., I. Fierro-Monti, T. W. Reichman, S. Gunnery, and M. B. Mathews. 2001. Double-stranded RNA-binding proteins and the control of protein synthesis and cell growth. Cold Spring Harb. Symp. Quant. Biol. 66:485-497. [DOI] [PubMed] [Google Scholar]
- 37.Parrott, A. M., M. R. Walsh, T. W. Reichman, and M. B. Mathews. 2005. RNA binding and phosphorylation determine the intracellular distribution of nuclear factors 90 and 110. J. Mol. Biol. 348:281-293. [DOI] [PubMed] [Google Scholar]
- 38.Patel, R. C., D. J. Vestal, Z. Xu, S. Bandyopadhyay, W. Guo, S. M. Erme, B. R. Williams, and G. C. Sen. 1999. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 274:20432-20437. [DOI] [PubMed] [Google Scholar]
- 39.Pelletier, J., and N. Sonenberg. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334:320-325. [DOI] [PubMed] [Google Scholar]
- 40.Pestova, T. V., V. G. Kolupaeva, I. B. Lomakin, E. V. Pilipenko, I. N. Shatsky, V. I. Agol, and C. U. Hellen. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. USA 98:7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pilipenko, E. V., T. V. Pestova, V. G. Kolupaeva, E. V. Khitrina, A. N. Poperechnaya, V. I. Agol, and C. U. Hellen. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 14:2028-2045. [PMC free article] [PubMed] [Google Scholar]
- 42.Pontecorvo, G. 1975. Production of mammalian somatic cell hybrids by means of polyethylene glycol treatment. Somatic Cell Genet. 1:397-400. [DOI] [PubMed] [Google Scholar]
- 43.Shaw, G., S. Morse, M. Ararat, and F. L. Graham. 2002. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 16:869-871. [DOI] [PubMed] [Google Scholar]
- 44.Smith, W. A., B. T. Schurter, F. Wong-Staal, and M. David. 2004. Arginine methylation of RNA helicase A determines its subcellular localization. J. Biol. Chem. 279:22795-22798. [DOI] [PubMed] [Google Scholar]
- 45.Walter, B. L., J. H. Nguyen, E. Ehrenfeld, and B. L. Semler. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5:1570-1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xu, Y. H., C. Busald, and G. A. Grabowski. 2000. Reconstitution of TCP80/NF90 translation inhibition activity in insect cells. Mol. Genet. Metab. 70:106-115. [DOI] [PubMed] [Google Scholar]
- 47.Xu, Y. H., and G. A. Grabowski. 1999. Molecular cloning and characterization of a translational inhibitory protein that binds to coding sequences of human acid beta-glucosidase and other mRNAs. Mol. Genet. Metab. 68:441-454. [DOI] [PubMed] [Google Scholar]
- 48.Xu, Y. H., and G. A. Grabowski. 2005. Translation modulation of acid beta-glucosidase in HepG2 cells: participation of the PKC pathway. Mol. Genet. Metab. 84:252-264. [DOI] [PubMed] [Google Scholar]