Abstract
To develop effective intervention strategies that prevent breast milk transmission of human immunodeficiency virus (HIV), we must understand the specific viral properties and mechanisms responsible for infant infection. We have used lactating rhesus macaques infected with a pathogenic simian immunodeficiency virus (SIV) stock to analyze the viral genotypes expressed in plasma and milk throughout the disease course and to identify those variants ultimately transmitted to infants through breastfeeding. In these studies we observed mother-to-infant transmission of SIV/DeltaB670 by eight females during the chronic phase of disease, and we analyzed by heteroduplex tracking assays and sequence analysis the distribution and fluctuations in viral genotypes expressed. Each female expressed multiple V1 envelope genotypes in milk near the time of transmission, while a single genotype was found in each of the infants. Variants transmitted to infants were not expressed throughout the maternal disease course but were only detected near the time of transmission. The emergence of the transmitted genotype in the dam typically occurred in plasma before milk and was coincident with increased milk viral loads. Transmitted genotypes tended to be longer and more glycosylated and had a less negative charge over the V1 region compared to viral genotypes expressed in milk but not transmitted. These observations demonstrate that specific viral genotypes are selectively transmitted to infants through breastfeeding and support the hypothesis that transmission occurs as genotypes adapt for efficient expression in milk.
Breast milk transmission of human immunodeficiency virus (HIV) is a problem of global importance. In many resource-limited areas with high rates of HIV infection, the benefits of breastfeeding often outweigh the risk of transmission (27). In these areas, the use of replacement feeding is often hindered by access to clean water, cost, availability of formula, and cultural expectations to breastfeed (33). Although short-course antiretroviral therapy (ART) given during pregnancy and parturition has significantly reduced the risk of in utero and peripartal HIV transmission (10, 12, 13), ART is a less feasible option for lactating women (16). This is due in part to the long period of breastfeeding, but the risk of infant toxicity and transmission of drug-resistant virus are important concerns that have not been fully addressed (15, 30). Effective intervention strategies that allow HIV-infected mothers to safely breastfeed are urgently needed. To develop these interventions, we must understand the mechanisms involved in transmission as well as the protective factors that allow a large number of HIV-exposed infants to remain uninfected.
Studies of HIV-infected women worldwide have established that higher plasma viral load and lower CD4+ T-cell counts increase the risk of mother-to-infant transmission in utero, at parturition, and postnatally through breastfeeding (11, 17, 20, 38). The risk of breast milk transmission is additionally correlated with higher cell-free and cell-associated milk viral load (34). The specific viral and immunological factors contributing to these correlates, however, remain unknown. While maternal plasma viral load is often predictive of transmission, mothers with a wide range of plasma viral loads have transmitted virus to their infants, suggesting that additional viral and immunological factors are responsible for transmission (11, 17). Several studies have shown that viral genotypes transmitted to infants tend to be macrophage tropic, utilize CCR5 as a coreceptor, and have rapid in vitro replication kinetics (25, 28). Phylogenetic analysis of maternal and infant sequences indicates that most infants are infected with a homogeneous virus population, despite the heterogeneous virus population in the mother (1, 14, 26, 37, 45, 46). It has also been shown that HIV isolates from infants are resistant to neutralization by maternal plasma (23). These observations suggest that specific viral genotypes are selectively transmitted from mother to infant. However, other studies have identified infants with heterogeneous virus populations (8, 24, 26, 29, 42, 43). In one report, phylogenetic analysis of maternal and infant sequences did not support selective transmission but suggested that the viral population in the infant reflected the inoculum size and timing of exposure (42). Additional studies are needed to more thoroughly characterize genotypes transmitted from mother to infant.
The small number of samples evaluated and the inability to identify the timing of infant infection often limits studies of HIV transmission from mother to infant. Due to the difficulties involved in defining whether transmission occurred during delivery or as a result of breastfeeding, few studies have been able to characterize genotypes transmitted through breastfeeding. Kampinga et al. evaluated HIV genotypes transmitted through breast milk in a small group of Rwandan women that became HIV infected postpartum (22). In this study, five of six breastfed infants seroconverted within the same 3-month period as their mother, and each was infected with a homogeneous virus population that represented the major genotype identified in maternal plasma samples. Multiple viral variants were identified in one infant who seroconverted more than 12 months after the mother. The V3 sequences identified in all of the infants displayed characteristics of macrophage tropism. These limited observations support the hypothesis that selective transmission does occur through breastfeeding.
Infection of lactating rhesus macaques with a pathogenic simian immunodeficiency virus (SIV) inoculum just after delivery provides an excellent model of HIV transmission through breastfeeding (3, 4). This natural transmission model allows evaluation of oral infant infection via breast milk in a controlled environment without potential in utero or peripartal exposure. Although the SIV disease course is accelerated and there is a higher rate of transmission compared to that of HIV, infant infection occurs throughout the entire breastfeeding period and is associated with increased and persistent expression of virus in milk, similar to observations in HIV infected mothers. As previously reported, lactating dams were inoculated with SIV/DeltaB670 after normal vaginal delivery of their infant. This primary SIV stock is composed of a quasispecies and has been used in several studies to evaluate the influence of specific viral genotypes on pathogenesis (2, 5, 6, 39-41). In the current studies, 10 of 14 dams inoculated with SIV/DeltaB670 transmitted virus to their infant via breast milk, while four failed to transmit despite disease progression and continued lactation. Eight of the infants became infected during the chronic phase of maternal infection. Although plasma viral load and CD4+ T-cell counts did not predict transmission status, infant infection was correlated with milk viral loads exceeding 500 copies/ml. Total immunoglobulin and SIV envelope-specific antibody were also not predictive of transmission in these animals (35). Two of the infants became SIV positive during the period of peak viremia in the mother (14 days postinoculation). Transmission at this early time point was associated with rapid disease progression, high plasma viral loads, and lack of specific antibody responses in the dam. Results obtained using this model are similar to what has been reported in breastfeeding cohorts of HIV-infected women, providing an animal model to decipher the correlates and mechanisms of breast milk transmission of HIV. In our initial studies, we observed homogeneous viral populations in five macaque infants infected through breastfeeding, despite the presence of a heterogeneous viral population in the milk of each dam. In this report we extend these studies by examining the expression of envelope genotypes in plasma and milk samples obtained from eight transmitting females over the course of disease.
MATERIALS AND METHODS
Animals.
Fourteen female rhesus macaques (Macaca mulatta) were inoculated intravenously with SIV/DeltaB670 7 to 54 days after the delivery of their infant. The lactating dams were housed at the Tulane National Primate Research Center with their infant and allowed to breastfeed normally as previously described (4). All animal protocols were approved by the Tulane and Louisiana State University Institutional Animal Care and Use Committees and were in accordance with the Guide for the Care and Use of Laboratory Animals (http://www.nap.edu/readingroom/books/labrats/).
Virus.
The SIV/DeltaB670 stock used in these studies was grown from primary rhesus peripheral blood mononuclear cells (PBMC) and is internally referenced as the Rhesus II stock. This stock has been previously characterized and is composed of a quasispecies (2, 40). In vitro titrations performed prior to inoculation confirmed that there was no loss in infectivity after long-term storage in liquid nitrogen. All animals in this study were intravenously inoculated with four 50% tissue culture infectious doses.
Sample collection.
Samples of blood and milk were collected every 2 weeks for 8 weeks and then monthly throughout the 1-year study period. Blood was collected in acid citrate dextrose anticoagulant tubes and centrifuged at 255 × g for 15 min for separation of the cellular fraction. Plasma was removed and stored at −80°C. Peripheral blood mononuclear cells were purified from blood samples using Lymphocyte Separation Medium (ICN, Aurora, OH). DNA was isolated from this population using a Genomic DNA Isolation kit (Bio-Rad) and quantified by A260 measurement. Approximately 1 ml of milk was collected by manual expression from both breasts, pooled, and stored immediately on ice. Milk samples were separated into cellular and supernatant fractions by centrifugation at 255 × g for 15 min. The fat layer was suctioned off, and the supernatant was stored at −80°C.
Reverse transcription and PCR amplification.
Virus particles contained in plasma and milk samples were purified by high-speed centrifugation of plasma and milk supernatant at 22,000 × g for 1 h. Viral pellets were lysed in Trizol reagent, and RNA was isolated per the manufacturer's instructions (Life Technologies, Rockville, MD). The RNA sample was reverse transcribed into cDNA using Multiscribe Reverse Transcriptase (Applied Biosystems) with 1/10th of the total RNA used in each reaction. Envelope variable region 1 (V1) sequences were amplified by PCR from cDNA samples obtained from maternal plasma and milk at several matched time points during the disease course, including the last available milk sample taken 3 to 8 weeks before the infant tested positive for SIV. Proviral sequences were PCR amplified from infant PBMC DNA obtained at the time point when SIV was first detected. For envelope V1 amplification a nested PCR was performed as described previously (4). In this assay, a 482-bp fragment encompassing V1 and V2 of the SIV envelope gene was amplified in two to three independent reactions from 1 μg of DNA (PBMC) or the cDNA prepared from one-tenth of the total RNA obtained from plasma and milk samples. First-round primers that correspond to nucleotides 6709 to 6728 and 7406 to 7385 and second-round primers that correspond to nucleotides 6845 to 6868 and 7327 to 7305 of SIVmac239 were used.
Heteroduplex tracking assay (HTA).
Single-stranded 32P-labeled probes were made from the V1 sequence identified in each infected infant. The single-stranded probe was generated in two steps. In the first step, V1 PCR products were generated from a plasmid clone containing envelope sequence from each infant or a B670 genotype of interest. The amplification reaction contained 150 mM Tris-HCl (pH 8.0), 500 mM KCl, 25 mM MgCl2, 0.01 mM of each deoxynucleoside triphosphate, 20 nM of 5′ and 3′ envelope V1 primers described above, and 2.5 U of AmpliTaqGold Polymerase (Applied Biosystems, Branchburg, NJ). The resulting PCR products were separated by agarose gel electrophoresis and purified by gel extraction (Qiagen, Valencia, CA). In the second step, 10 ng of purified V1 PCR product from each infant was used to create a single-stranded 32P-labeled product through asymmetrical PCR. The amplification reaction was similar to that described above, except only the 5′ primer was used and 10 μCi 32P-dATP was added to the mixture. The single-stranded probe was purified using the Wizard DNA purification kit (Promega). Samples analyzed by HTA were obtained by pooling products from two to three independent V1 PCR amplifications, and 1 μl of 32P-labeled probe was mixed with approximately 500 μg of pooled V1 PCR products amplified from plasma or milk samples in an annealing buffer of 1 M NaCl, 100 mM Tris-HCl (pH 7.6), 20 mM EDTA in a total volume of 20 μl. This mixture was heated at 90°C for 3 min, chilled on ice for 3 min, and then separated on a mutation detection enhancement gel solution (Cambrex Bioscience Rockland, Inc., Rockland, ME). After electrophoresis for 1,200 V-h, DNA was visualized by autoradiography. The resulting image was captured with a Kodak imaging system and formatted for publication in Microsoft PowerPoint. With this assay, genotypes identical to the probe can be detected if it comprises at least 10% of the total population.
Viral envelope sequences.
Sequences of V1 genotypes expressed in milk and plasma were obtained by cloning of PCR products from two to three independent amplifications, each containing 1 μg of PBMC DNA or cDNA prepared from 1/10th of the total RNA purified from milk or plasma samples. PCR products were pooled and cloned into the TOPO TA vector per the manufacturer's instructions (Invitrogen, Carlsbad, CA). Seven to 22 colonies containing appropriately sized inserts were selected for sequencing from each sample time point as described previously (2). Sequences were aligned and compared using MacVector (Accelrys Inc., Madison, WI) and Clustal W and were formatted for publication using SeqPublish (Los Alamos National Laboratory; http://www.hiv.lanl.gov). Infant sequences were derived similarly from PCR products obtained from PBMC DNA. Deduced amino acid sequences of SIV variants identified in the dams and infants were compared using Mac Vector to SIV/DeltaB670 genotypes found in the stock inoculum. In the event that a sequence from an infant or dam differed by three or more amino acids (>5% divergent) from the most closely aligned V1-B670 genotype, the sequence was considered a variant of that B670 stock genotype.
Viral load quantitation.
SIV RNA copy number in plasma and milk samples was quantitated by real-time reverse transcriptase PCR (RT-PCR) amplification based on a previously described assay that amplifies a region in the SIV ltr (4). Briefly, virions expressed in milk and plasma were purified and reverse transcribed into cDNA in a 10-μl total reaction volume as described above. Quantitation reactions were done with duplicate RNA samples, and SIV copy number was determined by comparison to a standard curve generated from serial dilutions of an RNA standard amplified in triplicate.
Statistical analysis.
Length, charge, and number of potential N-linked glycosylation sites were determined from a region of V1 between a conserved tryptophan residue at position 122 of SIVmac239 up to, but not including, a conserved methionine residue at position 165 of SIVmac239. In order to perform the statistical test, charge was transformed to a positive numeric value. Comparisons of length and charge were performed using the nonparametric Wilcoxon rank-sum test. Since all of the sequences identified in milk either had two or three potential N-linked glycosylation sites within this region, the nonparametric Fisher exact test was used to determine the probability associated with the number of glycosylation sites in transmitting versus nontransmitting sequences.
Nucleotide sequence accession numbers.
The envelope sequences described in this study have been deposited in GenBank under accession numbers AY971377 to AY971509. Sequences of SIV/DeltaB670 genotypes comprising the stock inoculum were obtained from GenBank accession numbers AY118200 to AY118221.
RESULTS
SIV envelope genotypes are selectively transmitted through breastfeeding.
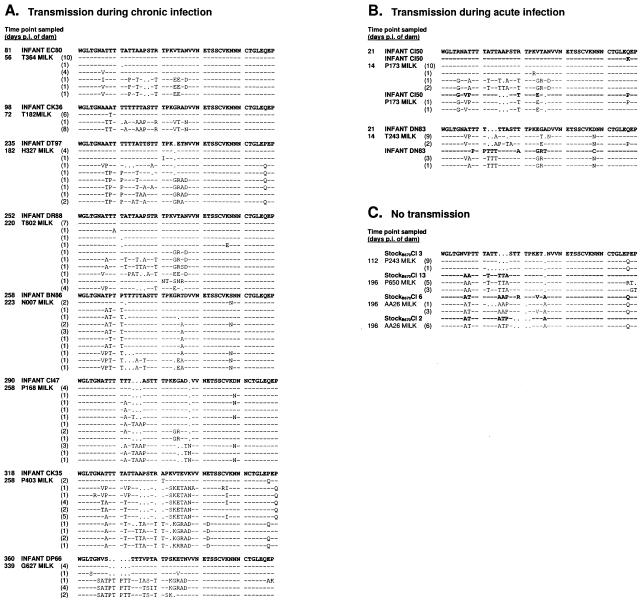
In this study, we analyzed the first hypervariable region of SIV envelope sequences (V1) expressed in the milk of 14 female rhesus macaques inoculated with SIV/DeltaB670 (B670). Ten dams transmitted SIV to their infants at time points throughout the disease course, including both the acute and chronic phase of infection. Viral genotypes expressed in milk near the time of transmission were compared to sequences found in the infant. Due to its extensive diversity, the V1 region is frequently used to identify viral variants within the B670 quasispecies. Twenty-two different V1 genotypes have been identified from acute infection with this stock and designated B670 Cl 1 to Cl 22 (2, 40). Eight infants infected during the chronic phase of maternal SIV disease were found to have a homogeneous virus population. This was in contrast to the multiple genotypes expressed in milk samples from each of the eight dams (Fig. 1A). In every case, the sequence identified in the infant could be found in milk samples from each of the eight dams 25 to 60 days prior to detection in the infants. Five of eight dams expressed a V1 genotype in milk that was identical to the sequence in the infant, while the remaining three females expressed sequences highly similar to those found in their infants, differing only by 1 to 2 amino acids. Some dams transmitted minor variants expressed in the milk, while others transmitted the dominant milk variant. These observations demonstrate that a single genotype is transmitted during the chronic phase of maternal infection despite expression of multiple genotypes in milk.
FIG. 1.
Deduced amino acid sequences from the envelope V1 region of virus expressed in milk of dams inoculated with SIV/DeltaB670. Viral sequences were amplified from milk supernatant by RT-PCR at the indicated time point postinoculation, cloned, and sequenced. The numbers of clones with identical amino acid sequences are indicated in parentheses. Infant sequences were amplified from PBMC DNA at the first time point of SIV detection, shown as days p.i. of the dam. (A) Alignments of milk virus sequences found in eight females that transmitted virus to their infants during the chronic phase of disease. Sequences are aligned with the viral genotype found in the infant. (B) Milk sequences found in two rapid transmitting-rapid progressor dams at 14 days p.i. aligned with genotypes found in the infant. (C) Sequences found in milk of three nontransmitting females at time points later in the disease course aligned with sequences from the SIV/DeltaB670 stock.
Two infants in this study were infected during the acute viremic phase of maternal disease. The dams of these infants displayed a rapid SIV disease course, had high plasma viral levels at set point, and failed to develop SIV-specific humoral immune responses (4, 35). These clinical features represent a well-characterized rapid progressor phenotype observed in a small percentage of SIV-infected macaques (18). As shown in Fig. 1B, infants CI50 and DN83 had two viral genotypes in peripheral blood samples at the time SIV was first detected (21 days postinoculation of dam). The genotypes found in the infant CI50 reflected the distribution of genotypes expressed in the milk of dam P173 7 days prior to detection of SIV in the infant. The two viral genotypes identified in infant DN83 were not found in milk samples from dam T243 7 days prior to detection of SIV in the infant, but these genotypes have been identified in milk samples from other SIV-infected dams (see P168 milk and N007 milk), indicating these genotypes are present in the B670 inoculum. In this model, breast milk transmission of SIV during the period of acute maternal infection is associated with a rapid progressor phenotype and occurs without selection of specific genotypes.
The remaining four female macaques inoculated in our studies failed to transmit SIV despite disease progression and continued lactation. Previously we have associated transmission with milk viral loads exceeding 500 copies/ml (4). Since this level of viral expression was not observed in the milk of the four nontransmitting animals, a complete analysis of viral diversity in these milk samples was difficult. Given these limitations, we analyzed the predominant viral genotypes expressed in milk samples from the chronic stage of infection, when viral RNA was detectable in three of four females. The predominant viral genotypes identified in the nontransmitting dams are shown in Fig. 1C, aligned to the most similar B670 stock genotype. Two nontransmitting dams, P243 and P650, expressed a homogeneous virus population in milk, while monkey AA26 had a more heterogeneous virus population in milk.
Similar viral genotypes are expressed in plasma and milk.
To determine if there was selective expression of certain viral genotypes in milk, the V1 sequences expressed in milk and plasma of lactating females were compared to the predominant sequences that comprise the B670 stock (Table 1). Samples were available for detailed V1 characterization from four transmitting dams and three nontransmitting dams at the time of peak viremia and near the time of transmission or late in the disease course. The predominant genotypes expressed during peak viremia, Cl 2, Cl 3, Cl 6, and Cl 12, are the genotypes that comprise more than 80% of the SIV/Delta stock used for inoculation (2). The distribution of genotypes identified in milk samples generally reflected the populations expressed in plasma. Significant differences were not observed between transmitters and nontransmitters. These results indicate that similar genotypes are expressed in milk and plasma during peak viremia, and unique genotypes were not identified in milk.
TABLE 1.
SIV-B670 genotypes identified in four transmitting and three nontransmitting dams at peak viremia (14 days p.i.) and during chronic diseasea
| B670 genotype | B670 inoculationb (n = 50) | Peak viremia
|
Chronic disease
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Transmitters
|
Nontransmitters
|
Transmitters
|
Nontransmitters
|
||||||||
| Plasma (n = 35) | Milk (n = 23) | PBMC (n = 25) | Plasma (n = 33) | Milk (n = 14) | PBMC (n = 24) | Plasma (n = 43) | Milk (n = 69) | Plasma (n = 32) | Milk (n = 31) | ||
| Cl 2 | 12 | 13 | 8 | 21 | 64 | 42 | 14 | 22 | 3 | ||
| Cl 2varc | 25 | 23 | |||||||||
| Cl 3 | 62 | 46 | 26 | 40 | 6 | 14 | 7 | 22 | 32 | ||
| Cl 4 | 6 | ||||||||||
| Cl 5 | 4 | ||||||||||
| Cl 6 | 4 | 21 | 22 | 25 | 21 | 23 | 3 | 19 | |||
| Cl 7 | 3 | ||||||||||
| Cl 8 | 2 | 8 | |||||||||
| Cl 10 | 3 | ||||||||||
| Cl 12 | 10 | 51 | 61 | 36 | 43 | 33 | 16 | 32 | 19 | ||
| Cl 12var | 16 | 6 | |||||||||
| Cl 13 | 4 | 25 | 26 | ||||||||
| Cl 14 | 6 | 1 | 3 | ||||||||
| Cl 14var | 26 | 13 | |||||||||
| Cl 22 | 4 | 3 | |||||||||
The distribution of clones is shown as a percentage of the total number of clones sequenced from each sample type. The number of clones analyzed (n = x) is indicated in each column. Chronic disease sample time points were obtained from transmitters prior to infant infection (72 to 258 days p.i. of dam) and just prior to end stage disease for nontransmitters (112 or 198 days p.i. of dam).
Distribution of genotypes in SIV inoculum used to infect animals.
var indicates that the genotype differed by three or more amino acids from the stock sequence (>5% divergence).
To evaluate the V1 genotypes expressed during the chronic stage of infection, sequences were obtained from paired plasma and milk samples from transmitting females 3 to 8 weeks prior to the identification of SIV infection in the infant (as indicated in Fig. 1). Similar genotypes were expressed in milk and plasma of these females during chronic disease; however, several differences were observed compared to data from 14 days postinfection (p.i.). Interestingly, B670 stock Cl 3, the predominant variant observed at 14 days p.i., was rarely expressed in both milk and plasma near the time of transmission. B670 stock Cl 7, Cl 13, and Cl 14 sequences were not identified in milk at 14 days p.i., but these genotypes were found in the milk of transmitters during chronic disease near the time of transmission. During the chronic stage of disease, transmitting dams continued to express the major B670 genotypes Cl 2, 6, and 12 in both milk and plasma. Nontransmitting females also displayed changes over the disease course. Samples were collected from nontransmitting females at 112 to 198 days p.i., near the end stage of disease for these analyses. These animals also expressed similar genotypes in milk and plasma during the chronic phase of disease; however, some genotypes expressed in milk of nontransmitters (Cl 3 and Cl 13) were not found in milk of transmitting females. These results suggest there may be differences in the expression of viral genotypes in the milk of nontransmitting dams compared to late transmitting dams; however, the low level of virus expression in nontransmitters makes a more detailed analysis difficult.
Sequence-specific characteristics of transmitted genotypes.
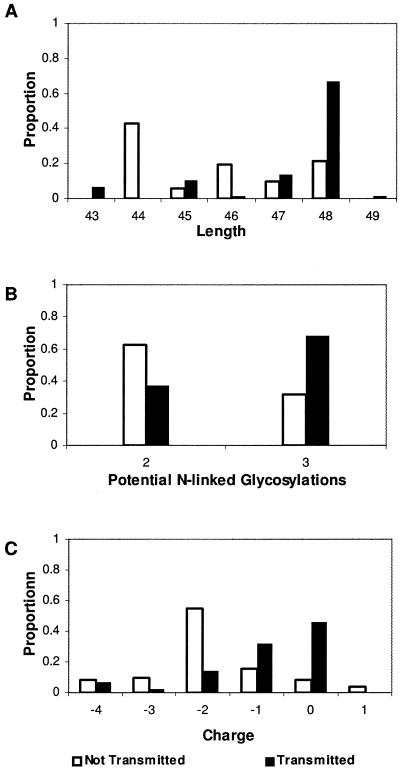
Because the viral populations expressed in milk of transmitting dams consisted of transmitted and nontransmitted genotypes and the viral load in these samples was >500 copies ml, we compared the characteristics of these two groups of sequences. Using 130 sequences identified in milk samples (represented in Fig. 1A), transmitted and nontransmitted genotypes were identified and compared. The V1 genotypes were designated transmitted genotypes if they were identical to sequences found in infants or differed by no more than two amino acids from infant sequences. Nontransmitted genotypes were the remaining sequences expressed in milk that differed from infant sequences by three or more amino acid changes, since this represents greater than 5% divergence. The V1 length of transmitted genotypes ranged from 41 to 49 amino acids, which was broader than that observed for nontransmitted sequences (Fig. 2A). Sequences identified in six of eight infected infants were longer than the median length of nontransmitted genotypes. When considered as a group, transmitted genotypes expressed in milk were significantly longer than nontransmitted genotypes (P < 0.001; Wilcoxon rank sum test). Transmitted genotypes expressed in the milk were more likely to have three potential N-linked glycosylation sites over the V1 region than nontransmitted genotypes (P < 0.0006; Fisher exact test), with six of eight infants having genotypes with three potential N-linked glycosylation sites in V1 (Fig. 2B). Although the charge of the V1 region had a similar range in each group, transmitted genotypes had a less negative charge than nontransmitted genotypes. This observation was significant when the two groups were compared (P < 0.001; Wilcoxon rank sum test). Seven of the eight infant sequences had a charge at or above the median charge of nontransmitted genotypes (Fig. 2C). Therefore, longer V1 sequences with three N-linked glycosylation sites and a less negative charge were more likely to be transmitted.
FIG. 2.
Comparison of V1 sequence characteristics of transmitted genotypes (black bars) and nontransmitted genotypes (white bars) expressed in milk of eight transmitting dams near the time of infant infection (sequences shown in Fig. 1A). V1 sequences transmitted to the infant were compared to genotypes expressed in milk that were not transmitted to evaluate (A) the number of amino acids in the V1 loop, (B) the number of potential N-linked glycosylation sites, and (C) the net charge of the V1 region.
Transmitted genotypes are not expressed throughout the disease course.
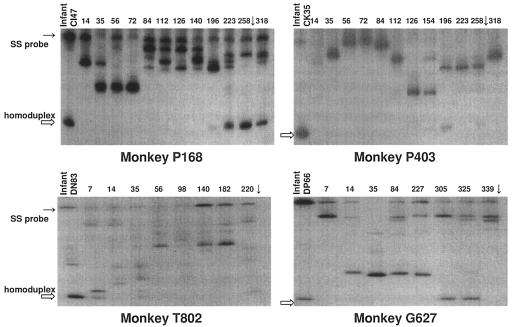
To determine time points when transmitted genotypes were expressed in the dams, heteroduplex tracking assays (HTA) were used to monitor envelope variants expressed in milk samples from 14 days p.i. through the time of infant infection. In this assay, single-stranded 32P-labeled probes were made from V1 genotypes of each infant and mixed with PCR products from the corresponding maternal milk samples, followed by electrophoresis on nondenaturing acrylamide gels. This assay estimates the number of V1 genotypes that comprise at least 10% of the total population and determines their relative divergence from the probe sequence by the degree of retarded migration from the homoduplex (probe bound to a PCR product with identical sequence). This assay provides a means of easily monitoring changes in the viral populations of several animals over multiple time points. Shown in Fig. 3 are representative tracking gels from four of the eight dams that transmitted during the chronic phase of infection. Several heteroduplex bands were observed at multiple time points in milk samples, indicating a heterogeneous virus population throughout the disease course. In milk from each dam, a homoduplex band, or a band indicating close sequence identity to the infant probe became detectable at time points just prior to transmission. In each case, the variant transmitted to the infant was expressed in the milk of the dam just prior to transmission but not at detectable levels throughout the disease course. The homoduplex band is less pronounced in milk samples of dams P403 and T802, confirming that the infant genotype was a minor variant in the population at the time point analyzed. Due to the length of time between sampling, it is possible that the variant transmitted to the infant was more dominant at the exact time of transmission. The remaining four animals (data not shown) displayed similar patterns, with expression of the transmitted genotype detectable only near the time of transmission.
FIG. 3.
Heteroduplex tracking assay gels of envelope V1 genotypes expressed in milk samples throughout the disease course of four transmitting females. Single-stranded 32P-labeled probes (SS probe) were made from genotypes identified in infants. Homoduplex bands, indicating sequence identity to the probe, are shown in lane 1 of each gel. The time when transmission to the infant occurred is indicated (↓).
Increases in viral load are associated with changes in the viral population.
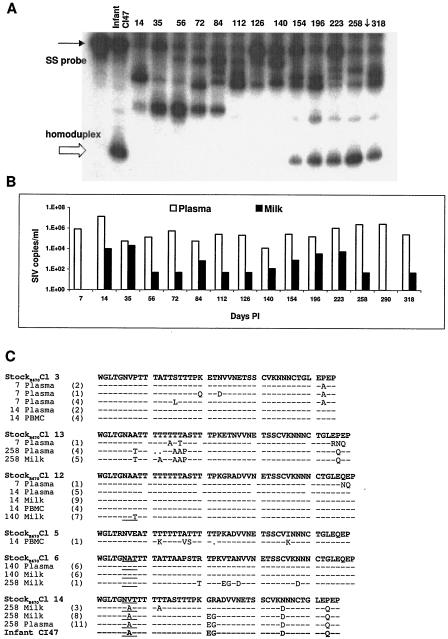
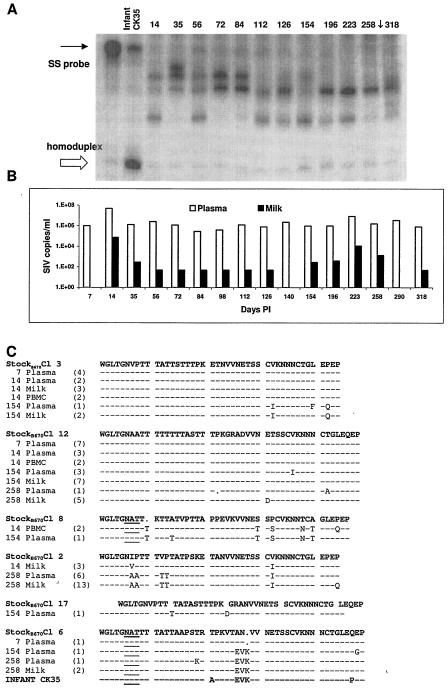
We selected two transmitting dams, monkeys P168 and P403, to more thoroughly evaluate virus expression throughout disease and to monitor the evolution of the genotype transmitted to the infant. Heteroduplex tracking was performed on plasma samples from these dams for comparison with HTA of milk samples and viral V1 sequences in milk and plasma. HTA of plasma from monkey P168 (Fig. 4A) showed that there were distinct shifts in V1 genotypes expressed in plasma over the course of disease, similar to the pattern observed in milk. The genotype that predominates in plasma at days 35 and 56 gradually decreases and is no longer expressed by day 112. Concurrent with the disappearances of this genotype, new variants appear at 72 and 84 days p.i. Several new genotypes can be observed in plasma samples during the middle phase of the disease course (day 112 through day 154). At 154 days p.i, the genotype transmitted to the infant is first detected in plasma and gradually becomes the predominant variant expressed at day 258. This genotype was not detected in milk until day 196.
FIG. 4.
Analysis of viral expression in transmitting female P168. (A) Heteroduplex tracking assays evaluating V1 genotypes expressed in plasma at indicated times p.i. A single-stranded 32P-labeled probe (SS probe) was made from the infant genotype. The time point when transmission to the infant occurred as indicated (↓). (B) SIV RNA copies per milliliter of milk or plasma as determined by real-time RT-PCR amplification at the indicated time points p.i. (C) Deduced amino acid sequences of envelope VI genotypes expressed in milk or plasma and proviral sequences from PBMC. Sequences were obtained by PCR amplification, followed by cloning and sequencing. The numbers of clones containing identical sequences are indicated in parentheses. Sequences identified in infants are also shown. Material and infant sequences are aligned with the most similar B670 stock genotype as determined by Clustal W alignments.
The shifts in plasma and milk genotypes of dam P168 were considered in the context of viral load (Fig. 4B). The first major shift in the plasma genotypes of monkey P168 occurred between 84 and 112 days p.i., when plasma viral loads had reached set point, milk viral loads were low, and the titer of SIV-specific antibody in milk and plasma reached a plateau (98 days p.i.) (4, 35). Milk viral loads begin to increase at 154 days p.i., and by 196 days p.i. they had increased above 500 copies/ml. This increase in milk viral load is coincident with the expression of the infant genotype in milk. A 1-log increase in plasma viral loads is observed at 223 days, coincident with the appearance of new viral genotypes in plasma.
Sequence analysis of the V1 region at representative time points from each phase of the disease course confirmed the appearance and disappearance of genotypes (Fig. 4C). Genotypes expressed during the initial phase of the disease course were similar, if not identical, to the predominant genotypes comprising the stock, B670 Cl 3 and Cl 12. Although B670 Cl 3 sequences predominate early in the disease course, they were not identified at day 140 or later. B670 Cl 12 genotypes persisted through day 140 but were also not found at the end stage of disease. New genotypes appearing during the middle and late stage of disease were also closely related to genotypes known to be in the SIV-B670 stock. At 140 days p.i., a variant identical to SIV-B670 clone 6 was one of the main genotypes expressed in plasma and milk. This genotype was previously not detected in monkey P168. Similarly, a genotype similar to SIV B670 clone 13 was detected at low frequency at 7 days p.i., and then at 258 days a related sequence is detected. The genotype transmitted to the infant first appears at 154 days p.i. and is most similar to SIV-B6mac23970 clone 14, as determined by Clustal W alignment with B670 stock genotypes. These observations suggest that genotypes expressed later in the disease course were sequestered, or their expression was so low it could not be detected by the methods used.
Viral expression in plasma of monkey P403 also displayed changes over the disease course (Fig. 5A), with some genotypes appearing intermittently, some expressed for several weeks, and one genotype persisting throughout. The genotype transmitted to the infant first appears in plasma at day 154 p.i. and is detected only at very low levels through 258 days p.i. Viral loads increase slightly in plasma of monkey P403 between 196 and 223 days p.i.; however, they increase substantially in the milk during this same time (Fig. 5B).
FIG. 5.
Analysis of viral expression in transmitting female P403. See the legend to Fig. 4.
Sequence analysis showed that the predominant SIV-B670 stock genotype Cl 3 was expressed through 154 days p.i., but like that observed in monkey P168, this genotype is not found at end stage. The Cl 12 genotype was expressed throughout the disease course in monkey P403. Similar to that observed in monkey P168, new genotypes appear in plasma and milk at later time points in the disease course. Some genotypes expressed later in disease may have been sequestered, since they were similar in sequence to known inoculum genotypes (clones 2, 6, 8, and 17) but were only detected later in the disease course. At 258 days p.i., several genotypes are expressed in milk and plasma, with the genotype transmitted to the infant being a minor variant of those in both milk and plasma. These analyses show that new genotypes continue to appear in plasma and milk of transmitting dams, with one genotype selectively infecting the infant.
DISCUSSION
We have used SIV-infected lactating macaques to evaluate the expression of viral genotypes in milk and plasma throughout disease and to determine their relationship to variants transmitted to the infant through breastfeeding. The correlates of breast milk transmission in SIV-infected dams are similar to those observed in HIV-infected cohorts, providing an excellent model to evaluate the viral properties associated with transmission. In this report we demonstrate selective transmission of genotypes through breastfeeding and identify common viral characteristics associated with transmission.
Other investigators using the same SIV/DeltaB670 inoculum have characterized expression of V1 genotypes in the plasma, brain, genital tract, spleen, and lymph nodes (5, 6, 32). In most cases, SIV-B670 clones 2, 3, 6, and 12 were identified, indicating the predominance of these variants. However, the dynamics of viral expression are unique in each animal and each tissue compartment. In this study, we were able to specifically distinguish transmissible from nontransmissible variants based on their molecular characteristics and monitor their expression in milk and plasma throughout the disease course. This provides considerable insight into the mechanisms of breast milk transmission that would be difficult to show in HIV-infected cohorts.
The predominant genotype in the SIV-B670 inoculum, Cl 3, was identified in plasma and milk of most dams during the period of peak viremia. This variant was not identified in milk samples from transmitting dams later in the disease course, suggesting it does not adapt well for expression in milk and is less fit for replication during chronic disease. Variants similar to another major SIV-B670 genotype, Cl 12, were identified in dams early in infection and were expressed in milk and plasma of several near the time of transmission, with two dams ultimately transmitting genotypes similar to clone 12. Interestingly, variants similar to clone 12 were identified in the milk of other dams that did not transmit this genotype, suggesting that there are multiple factors involved in transmission. Because only a small region of the envelope gene was used to differentiate these variants, it is possible that specific changes in other regions are also important for infant infection.
Although the dams transmitted different B670 genotypes, common molecular features were identified in the V1 sequences. Transmitted sequences expressed in milk were longer, had a less negative charge, and were more likely to have three potential N-linked glycosylation sites than nontransmitted genotypes expressed in milk. Structural and mutational studies of the SIV envelope indicate that the V1/V2 loop plays a role in CD4 dependence and influences coreceptor binding (9, 21, 31). Additionally, changes in the number and arrangement of N-linked glycosylation sites on the HIV envelope have previously been described as a mechanism of escape from neutralizing antibody responses (44). The molecular features unique to transmitted variants may therefore represent differences in receptor or coreceptor binding, broadening of tropism to include cell types found in the breast, and evasion of local maternal immune responses.
Homogeneous virus populations were identified in all of the infants infected during the chronic phase of the maternal disease course (late transmission) despite the expression of multiple viral genotypes in maternal milk samples. It is possible that infant macaques were infected with multiple variants, but these genotypes were not detectable at the time points we identified infection in the infant. If multiple genotypes were transmitted during chronic maternal infection, only a single genotype was selectively replicated in the infant.
Milk provides a unique milieu for virus replication, since the cell types and immune modulators differ from what is found in plasma or other tissue compartments. Sequence analysis of virus from three HIV-infected lactating women found one of three mothers expressing different virus populations in milk compared to plasma, while in two mothers minor variants expressed in plasma were major variants expressed in the milk (7). A cross-sectional comparison of SIV-B670 genotypes expressed in plasma and milk of lactating dams showed that the distribution of genotypes expressed in milk and plasma was similar, with the diversity in milk increasing later in the disease course. Humoral- and cell-mediated immune responses in the breast have been shown to differ from those in the blood (36), which may promote viral evolution in this compartment. In this study, the V1 genotype transmitted to the infant was found in the plasma and milk of every transmitting dam, and no evidence of selective viral expression in milk was associated with transmission.
Viral genotypes selectively transmitted to infant macaques were similar to variants known to be in the SIV-B670 inoculum; however, their expression was not detected in plasma or milk of the dam until just prior to transmission. The detection of these genotypes was coincident with increases in milk viral load. It is unclear whether expression of the transmitted variant caused this increase in milk viral load or if viral load increases allowed for the expression or detection of these variants. These results indicate that transmitted variants were maintained at undetectable levels in the dams. Their emergence may have been due to immune evasion, adaptation to the breast, or deterioration of the maternal immune response.
Our ability to determine the diversity of envelope genotypes in nontransmitting dams was limited due to the low level of virus expression in milk observed in these animals. Given these limitations, we identified less diverse virus populations in milk of nontransmitters and two of the nontransmitters expressed genotypes not found in transmitting dams near the time of transmission. It is possible that immune responses in nontransmitting dams were able to control replication of certain viral genotypes and prevented the molecular changes required for expression in milk and transmission.
Several studies have characterized HIV isolates transmitted from mother to infant as macrophage tropic, CCR5 utilizing, and non-syncytia inducing (25, 28). Although phenotypes were not directly assessed in this study, other investigators have identified several SIV-B670 genotypes as having a macrophage-tropic phenotype in vivo. SIV-B670 clones 2 and 6 have been found in macrophage-rich areas of the spleen (32), and clones 2 and 12 have been found in microglial cells from brain tissue of macaques with SIV-encephalitis (5). Additionally, clone 12 has been shown to selectively cross the placenta of SIV-infected macaques, suggesting tropism for macrophage-like cell populations (2). Tissue macrophages have been shown to harbor high levels of virus in macaques infected with SIV-HIV chimeras, implicating these cell types as important viral reservoirs (19). Because genotypes similar to B670 clones 2, 6, and 12 were transmitted through breastfeeding, we hypothesize that transmitted variants may have persisted at low levels in tissue macrophages in the dam prior to their appearance in the peripheral circulation.
Two dams characterized as rapid progressors transmitted virus through breastfeeding coincident with the period of peak viremia. The infants of these dams were infected with a heterogeneous virus population that reflected the predominant genotypes expressed in milk. At 14 days p.i., the early transmitters could not be distinguished from other animals based on viral load or the distribution of genotypes expressed in milk. However, they progressed very rapidly to the end stage of disease and subsequently failed to develop a detectable SIV-specific humoral immune response, suggesting poor control over virus expression. Future studies that define the aberrant immune responses in these rapid progressor animals may also identify factors that control breast milk transmission in some dams.
In summary, the common sequence characteristics identified in the infected infants are consistent with changes resulting from immune evasion and adaptation of macrophage-tropic genotypes for efficient replication in cells of breast tissue and/or milk. Although common genotypes are expressed in the milk of all transmitting females, host-specific factors within each animal direct the evolution and expression of genotypes that are ultimately transmitted. By defining the viral and immunological characteristics common to transmitters and understanding the mechanisms that allow some dams to reach the end stage of disease while controlling viral expression and evolution in milk, we may be able to develop strategies and therapeutic approaches to prevent breast milk transmission of HIV.
REFERENCES
- 1.Ahmad, N., B. M. Baroudy, R. C. Baker, and C. Chappey. 1995. Genetic analysis of human immunodeficiency virus type 1 envelope V3 region isolates from mothers and infants after perinatal transmission. J. Virol. 69:1001-1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amedee, A. M., N. Lacour, J. L. Gierman, L. N. Martin, J. E. Clements, R. Bohm, Jr., R. M. Harrison, and M. Murphey-Corb. 1995. Genotypic selection of simian immunodeficiency virus in macaque infants infected transplacentally. J. Virol. 69:7982-7990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amedee, A. M., N. Lacour, and M. Ratterree. 2003. Mother-to-infant transmission of SIV via breast-feeding in rhesus macaques. J. Med. Primatol. 32:187-193. [DOI] [PubMed] [Google Scholar]
- 4.Amedee, A. M., J. Rychert, N. Lacour, L. Fresh, and M. Ratterree. 2004. Viral and immunological factors associated with breast milk transmission of SIV in rhesus macaques. Retrovirology 1:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Babas, T., D. Munoz, J. L. Mankowski, P. M. Tarwater, J. E. Clements, and M. C. Zink. 2003. Role of microglial cells in selective replication of simian immunodeficiency virus genotypes in the brain. J. Virol. 77:208-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Babas, T., E. Vieler, D. A. Hauer, R. J. Adams, P. M. Tarwater, K. Fox, J. E. Clements, and M. C. Zink. 2001. Pathogenesis of SIV pneumonia: selective replication of viral genotypes in the lung. Virology 287:371-381. [DOI] [PubMed] [Google Scholar]
- 7.Becquart, P., N. Chomont, P. Roques, A. Ayouba, M. D. Kazatchkine, L. Belec, and H. Hocini. 2002. Compartmentalization of HIV-1 between breast milk and blood of HIV-infected mothers. Virology 300:109-117. [DOI] [PubMed] [Google Scholar]
- 8.Briant, L., C. M. Wade, J. Puel, A. J. Brown, and M. Guyader. 1995. Analysis of envelope sequence variants suggests multiple mechanisms of mother-to-child transmission of human immunodeficiency virus type 1. J. Virol. 69:3778-3788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 10.Connor, E. M., R. S. Sperling, R. Gelber, P. Kiselev, G. Scott, M. J. O'Sullivan, R. VanDyke, M. Bey, W. Shearer, R. L. Jacobson, E. Jimenexz, E. O'Neill, B. Bazin, J.-F. Delfraissy, M. Culnane, R. Coombs, M. Elkins, J. Moye, P. Stratton, J. Balsley, et al. 1994. Reduction of maternal-infant transmission of human immunodeficiency virus type 1 with zidovudine treatment. N. Engl. J. Med. 331:1173-1180. [DOI] [PubMed] [Google Scholar]
- 11.Contopoulos-Ioannidis, D. G., and J. P. Ioannidis. 1998. Maternal cell-free viremia in the natural history of perinatal HIV-1 transmission: a meta-analysis. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 18:126-135. [DOI] [PubMed] [Google Scholar]
- 12.Cooper, E. R., M. Charurat, L. Mofenson, I. C. Hanson, J. Pitt, C. Diaz, K. Hayani, E. Handelsman, V. Smeriglio, R. Hoff, and W. Blattner. 2002. Combination antiretroviral strategies for the treatment of pregnant HIV-1-infected women and prevention of perinatal HIV-1 transmission. J. Acquir. Immune Defic. Syndr. 29:484-494. [DOI] [PubMed] [Google Scholar]
- 13.Cooper, E. R., R. P. Nugent, C. Diaz, J. Pitt, C. Hanson, L. A. Kalish, H. Mendez, C. Zorrilla, R. Hershow, J. Moye, V. Smeriglio, M. G. Fowler, et al. 1996. After AIDS clinical trial 076: the changing pattern of zidovudine use during pregnancy, and the subsequent reduction in the vertical transmission of human immunodeficiency virus in a cohort of infected women and their infants. J. Infect. Dis. 174:1207-1211. [DOI] [PubMed] [Google Scholar]
- 14.Dickover, R. E., E. M. Garratty, S. Plaeger, and Y. J. Bryson. 2001. Perinatal transmission of major, minor, and multiple maternal human immunodeficiency virus type 1 variants in utero and intrapartum. J. Virol. 75:2194-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eshleman, S. H., L. A. Guay, A. Mwatha, E. R. Brown, S. P. Cunningham, P. Musoke, F. Mmiro, and J. B. Jackson. 2004. Characterization of nevirapine resistance mutations in women with subtype A vs. D HIV-1 6-8 weeks after single-dose nevirapine (HIVNET 012). J. Acquir. Immune Defic. Syndr. 35:126-130. [DOI] [PubMed] [Google Scholar]
- 16.Gaillard, P., M. G. Fowler, F. Dabis, H. Coovadia, C. Van Der Horst, K. Van Rompay, A. Ruff, T. Taha, T. Thomas, I. De Vincenzi, and M. L. Newell. 2004. Use of antiretroviral drugs to prevent HIV-1 transmission through breast-feeding: from animal studies to randomized clinical trials. J. Acquir. Immune Defic. Syndr. 35:178-187. [DOI] [PubMed] [Google Scholar]
- 17.Garcia, P. M., L. A. Kalish, J. Pitt, H. Minkoff, T. C. Quinn, S. K. Burchett, J. Kornegay, B. Jackson, J. Moye, C. Hanson, C. Zorrilla, J. F. Lew, et al. 1999. Maternal levels of plasma human immunodeficiency virus type 1 RNA and the risk of perinatal transmission. N. Engl. J. Med. 341:394-402. [DOI] [PubMed] [Google Scholar]
- 18.Hirsch, V. M., and P. R. Johnson. 1994. Pathogenic diversity of simian immunodeficiency viruses. Virus Res. 32:183-203. [DOI] [PubMed] [Google Scholar]
- 19.Igarashi, T., C. R. Brown, Y. Endo, A. Buckler-White, R. Plishka, N. Bischofberger, V. Hirsch, and M. A. Martin. 2001. Macrophage are the principal reservoir and sustain high virus loads in rhesus macaques after the depletion of CD4+ T cells by a highly pathogenic simian immunodeficiency virus/HIV type 1 chimera (SHIV): implications for HIV-1 infections of humans. Proc. Natl. Acad. Sci. USA 98:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.John, G. C., R. W. Nduati, D. A. Mbori-Ngacha, B. A. Richardson, D. Panteleeff, A. Mwatha, J. Overbaugh, J. Bwayo, J. O. Ndinya-Achola, and J. K. Kreiss. 2001. Correlates of mother-to-child human immunodeficiency virus type 1 (HIV-1) transmission: association with maternal plasma HIV-1 RNA load, genital HIV-1 DNA shedding, and breast infections. J. Infect. Dis. 183:206-212. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, W. E., J. Morgan, J. Reitter, B. A. Puffer, S. Czajak, R. W. Doms, and R. C. Desrosiers. 2002. A replication-competent, neutralization-sensitive variant of simian immunodeficiency virus lacking 100 amino acids of envelope. J. Virol. 76:2075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kampinga, G. A., A. Simonon, P. Van de Perre, E. Karita, P. Msellati, and J. Goudsmit. 1997. Primary infections with HIV-1 of women and their offspring in Rwanda: findings of heterogeneity at seroconversion, coinfection, and recombinants of HIV-1 subtypes A and C. Virology 227:63-76. [DOI] [PubMed] [Google Scholar]
- 23.Kliks, S., C. H. Contag, H. Corliss, G. Learn, A. Rodrigo, D. Wara, J. I. Mullins, and J. A. Levy. 2000. Genetic analysis of viral variants selected in transmission of human immunodeficiency viruses to newborns. AIDS Res. Hum. Retrovir. 16:1223-1233. [DOI] [PubMed] [Google Scholar]
- 24.Lamers, S. L., J. W. Sleasman, J. X. She, K. A. Barrie, S. M. Pomeroy, D. J. Barrett, and M. M. Goodenow. 1994. Persistence of multiple maternal genotypes of human immunodeficiency virus type I in infants infected by vertical transmission. J. Clin. Investig. 93:380-390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matala, E., T. Hahn, V. R. Yedavalli, and N. Ahmad. 2001. Biological characterization of HIV type 1 envelope V3 regions from mothers and infants associated with perinatal transmission. AIDS Res. Hum. Retrovir. 17:1725-1735. [DOI] [PubMed] [Google Scholar]
- 26.Mulder-Kampinga, G. A., A. Simonon, C. L. Kuiken, J. Dekker, H. J. Scherpbier, P. van de Perre, K. Boer, and J. Goudsmit. 1995. Similarity in env and gag genes between genomic RNAs of human immunodeficiency virus type 1 (HIV-1) from mother and infant is unrelated to time of HIV-1 RNA positivity in the child. J. Virol. 69:2285-2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nagelkerke, N. J., S. Moses, J. E. Embree, F. Jenniskens, and F. A. Plummer. 1995. The duration of breastfeeding by HIV-1-infected mothers in developing countries: balancing benefits and risks. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 8:176-181. [PubMed] [Google Scholar]
- 28.Ometto, L., C. Zanotto, A. Maccabruni, D. Caselli, D. Truscia, C. Giaquinto, E. Ruga, L. Chieco-Bianchi, and A. De Rossi. 1995. Viral phenotype and host-cell susceptibility to HIV-1 infection as risk factors for mother-to-child HIV-1 transmission. AIDS 9:427-434. [PubMed] [Google Scholar]
- 29.Pasquier, C., C. Cayrou, A. Blancher, C. Tourne-Petheil, A. Berrebi, J. Tricoire, J. Puel, and J. Izopet. 1998. Molecular evidence for mother-to-child transmission of multiple variants by analysis of RNA and DNA sequences of human immunodeficiency virus type 1. J. Virol. 72:8493-8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patel, S. M., S. Johnson, S. M. Belknap, J. Chan, B. E. Sha, and C. Bennett. 2004. Serious adverse cutaneous and hepatic toxicities associated with nevirapine use by non-HIV-infected individuals. J. Acquir. Immune Defic. Syndr. 35:120-125. [DOI] [PubMed] [Google Scholar]
- 31.Puffer, B. A., L. A. Altamura, T. C. Pierson, and R. W. Doms. 2004. Determinants within gp120 and gp41 contribute to CD4 independence of SIV Envs. Virology 327:16-25. [DOI] [PubMed] [Google Scholar]
- 32.Reinhart, T. A., M. J. Rogan, A. M. Amedee, M. Murphey-Corb, D. M. Rausch, L. E. Eiden, and A. T. Haase. 1998. Tracking members of the simian immunodeficiency virus deltaB670 quasispecies population in vivo at single-cell resolution. J. Virol. 72:113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rollins, N., N. Meda, R. Becquet, A. Coutsoudis, J. Humphrey, B. Jeffrey, S. Kanshana, L. Kuhn, V. Leroy, D. Mbori-Ngacha, J. McIntyre, and M. L. Newell. 2004. Preventing postnatal transmission of HIV-1 through breast-feeding: modifying infant feeding practices. J. Acquir. Immune Defic. Syndr. 35:188-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rousseau, C. M., R. W. Nduati, B. A. Richardson, G. C. John-Stewart, D. A. Mbori-Ngacha, J. K. Kreiss, and J. Overbaugh. 2004. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J. Infect. Dis. 190:1880-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rychert, J., and A. M. Amedee. 2005. The antibody response to SIV in lactating rhesus macaques. J. Acquir. Immune Defic. Syndr. 38:135-141. [DOI] [PubMed] [Google Scholar]
- 36.Sabbaj, S. E. B., M. K. Ghosh, K. Semrau, S. Cheelo, D. M. Thea, L. Kuhn, G. D. Ritter, M. J. Mulligan, P. A. Goepfert, and G. M. Aldrovandi. 2002. Human immunodeficiency virus-specific CD8(+) T cells in human breast milk. J. Virol. 76:7365-7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scarlatti, G., T. Leitner, E. Halapi, J. Wahlberg, P. Marchisio, M. A. Clerici-Schoeller, H. Wigzell, E. M. Fenyo, J. Albert, M. Uhlen, and P. Rossi. 1993. Comparison of variable region 3 sequences of human immunodeficiency virus type 1 from infected children with the RNA and DNA sequences of the virus populations of their mothers. Proc. Natl. Acad. Sci. USA 90:1721-1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.St. Louis, M. E., M. Kamenga, C. Brown, A. M. Nelson, T. Manzila, V. Batter, F. Behets, U. Kabagabo, R. W. Ryder, M. Oxtoby, et al. 1993. Risk for perinatal HIV-1 transmission according to maternal immunologic, virologic, and placental factors. JAMA 269:2853-2859. [PubMed] [Google Scholar]
- 39.Trichel, A. M., P. A. Rajakumar, and M. Murphey-Corb. 2002. Species-specific variation in SIV disease progression between Chinese and Indian subspecies of rhesus macaque. J. Med. Primatol. 31:171-178. [DOI] [PubMed] [Google Scholar]
- 40.Trichel, A. M., E. D. Roberts, L. A. Wilson, L. N. Martin, R. M. Ruprecht, and M. Murphey-Corb. 1997. SIV/DeltaB670 transmission across oral, colonic, and vaginal mucosae in the macaque. J. Med. Primatol. 26:3-10. [DOI] [PubMed] [Google Scholar]
- 41.Valli, P. J., V. V. Lukashov, J. L. Heeney, and J. Goudsmit. 1998. Shortening of the symptom-free period in rhesus macaques is associated with decreasing nonsynonymous variation in the env variable regions of simian immunodeficiency virus SIVsm during passage. J. Virol. 72:7494-7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Verhofstede, C., E. Demecheleer, N. De Cabooter, P. Gaillard, F. Mwanyumba, P. Claeys, V. Chohan, K. Mandaliya, M. Temmerman, and J. Plum. 2003. Diversity of the human immunodeficiency virus type 1 (HIV-1) env sequence after vertical transmission in mother-child pairs infected with HIV-1 subtype A. J. Virol. 77:3050-3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wade, C. M., D. Lobidel, and A. J. Brown. 1998. Analysis of human immunodeficiency virus type 1 env and gag sequence variants derived from a mother and two vertically infected children provides evidence for the transmission of multiple sequence variants. J. Gen. Virol. 79:1055-1068. [DOI] [PubMed] [Google Scholar]
- 44.Wei, X., J. M. Decker, S. Wang, H. Hui, J. C. Kappes, X. Wu, J. F. Salazar-Gonzalez, M. G. Salazar, J. M. Kilby, M. S. Saag, N. L. Komarova, M. A. Nowak, B. H. Hahn, P. D. Kwong, and G. M. Shaw. 2003. Antibody neutralization and escape by HIV-1. Nature 422:307-312. [DOI] [PubMed] [Google Scholar]
- 45.Wolinsky, S. M., C. M. Wike, B. T. Korber, C. Hutto, W. P. Parks, L. L. Rosenblum, K. J. Kunstman, M. R. Furtado, and J. L. Munoz. 1992. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science 255:1134-1137. [DOI] [PubMed] [Google Scholar]
- 46.Zhang, H., G. Orti, Q. Du, J. He, C. Kankasa, G. Bhat, and C. Wood. 2002. Phylogenetic and phenotypic analysis of HIV type 1 env gp120 in cases of subtype C mother-to-child transmission. AIDS Res. Hum. Retrovir. 18:1415-1423. [DOI] [PubMed] [Google Scholar]