Abstract
At 739 amino acids, the nucleoprotein (NP) of Ebola virus is the largest nucleoprotein of the nonsegmented negative-stranded RNA viruses, and like the NPs of other viruses, it plays a central role in virus replication. Huang et al. (Y. Huang, L. Xu, Y. Sun, and G. J. Nabel, Mol. Cell 10:307-316, 2002) previously demonstrated that NP, together with the minor matrix protein VP24 and polymerase cofactor VP35, is necessary and sufficient for the formation of nucleocapsid-like structures that are morphologically indistinguishable from those seen in Ebola virus-infected cells. They further showed that NP is O glycosylated and sialylated and that these modifications are important for interaction between NP and VP35. However, little is known about the structure-function relationship of Ebola virus NP. Here, we examined the glycosylation of Ebola virus NP and further investigated its properties by generating deletion mutants to define the region(s) involved in NP-NP interaction (self-assembly), in the formation of nucleocapsid-like structures, and in the replication of the viral genome. We were unable to identify the types of glycosylation and sialylation, although we did confirm that Ebola virus NP was glycosylated. We also determined that the region from amino acids 1 to 450 is important for NP-NP interaction (self-assembly). We further demonstrated that these amino-terminal 450 residues and the following 150 residues are required for the formation of nucleocapsid-like structures and for viral genome replication. These data advance our understanding of the functional region(s) of Ebola virus NP, which in turn should improve our knowledge of the Ebola virus life cycle and its extreme pathogenicity.
Ebola and Marburg viruses are filamentous, enveloped, nonsegmented, negative-stranded RNA viruses of the family Filoviridae in the order Mononegavirales, which also includes the Paramyxoviridae, Rhabdoviridae, and Bornaviridae (7, 32). Despite the fact that the severe hemorrhagic fever caused by Ebola virus is associated with extremely high mortality rates in human and nonhuman primates, an effective vaccine or antiviral drugs have yet to be developed.
Ebola virus particles consist of at least seven structural proteins encoded by a single-stranded negative-sense RNA genome. Four of these proteins—nucleoprotein (NP), VP35, VP30, and the RNA-dependent RNA polymerase (L)—are components of the ribonucleoprotein complex that is responsible for the transcription and replication of the viral genome (26). Three others—glycoprotein (GP), VP40, and VP24—are membrane-associated proteins (12, 32), and VP24 is also thought to be involved in nucleocapsid formation (15).
The NP of Ebola virus is the largest (739 amino acid residues) nucleoprotein of the nonsegmented negative-stranded RNA viruses and can be divided into a hydrophobic N-terminal half (approximately 350 amino acids) and a hydrophilic C-terminal half (33). Huang et al. (15) showed that Ebola virus NP is O glycosylated and sialylated and that these modifications are required for its interaction with VP35, which suggests that the modifications are important for viral genome replication. However, the functional region(s) of Ebola virus NP has not been characterized. The nucleoproteins (NP/N) of nonsegmented negative-stranded RNA viruses, including Marburg virus NP, are known to self-assemble and form nucleocapsid-like structures without any other viral proteins (2, 4, 9, 11, 22, 24). Huang et al. (15) used transmission electron microscopy to show that Ebola virus NP, VP35, and VP24 are necessary and sufficient for the formation of nucleocapsid-like structures in a mammalian expression system. In addition, we recently found that Ebola virus NP also self-assembles to form helical tubes that are morphologically distinct from nucleocapsids (T. Noda and Y. Kawaoka, unpublished data).
In order to better understand Ebola virus NP, we have examined its protein modifications and self-assembly. Using deletion mutants of Ebola virus NP, we were able to determine the functional region(s) of this protein that is responsible for NP-NP interaction, the formation of nucleocapsid-like structures, and replication of the viral genome.
MATERIALS AND METHODS
Plasmids.
The open reading frame encoding Zaire ebolavirus NP was cloned into the expression vector pCAGGS/MCS (21, 30) as described previously (37). The resulting construct was designated pCEboZNP (37). To generate the NP deletion constructs, the NP open reading frame was cloned into pT7BlueBlunt vector (Novagen), and the mutated NP genes were amplified by inverse PCR (primer sequences are available on request). The PCR products were then cloned into pCAGGS/MCS. The resulting constructs were designated pCEboZNPΔ2-150, pCEboZNPΔ151-300, pCEboZNPΔ301-450, pCEboZNPΔ451-600, pCEboZNPΔ601-739, and pCEboZNPΔ451-739 (e.g., NPΔ2-150 denotes deletion of amino acids 2 to 150 of NP). To produce NP constructs with the FLAG tag or the six-histidine (His) tag at the C terminus, cDNA fragments were amplified by PCR with the appropriate primers and the PCR products were cloned into pT7BlueBlunt vector and then subcloned into pCAGGS/MCS. The resulting constructs were designated pCEboZNPCFLAG and pCEboZNPCHis, respectively. NP deletion constructs with the FLAG tag at the C terminus of the protein were generated by the same procedure. The resulting constructs were designated pCEboZNPΔ2-150CFLAG, pCEboZNPΔ151-300CFLAG, pCEboZNPΔ301-450CFLAG, pCEboZNPΔ451-600CFLAG, pCEboZNPΔ601-739CFLAG, and pCEboZNPΔ451-739CFLAG. To produce a construct to express the transcription factor Sp1, we extracted total cellular RNA from 293T cells. The Sp1 open reading frame with a His tag at its N terminus was amplified by reverse transcription-PCR and was cloned into pCAGGS/MCS. The resulting construct was designated pCSp1NHis. All constructs were sequenced to ensure that unwanted mutations were not present. Plasmids pCEboZGP, pCZGP643HIS, pCEboZ-VP35, pCEboZ-VP30, pCEboZ-L, p3E5EGFP, and pC-T7pol have been described previously (28, 36, 37).
Cells and antibodies.
Human embryonic kidney 293T cells were cultured in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal calf serum. To obtain monoclonal antibodies against Zaire ebolavirus NP, we employed gene gun immunization. Six-week-old female BALB/c mice were immunized with 2 μg of pCEboZNP, using particle-mediated DNA immunization twice at 4-week intervals, and then boosted 2 months later with 293T cells expressing Zaire ebolavirus NP. The subsequent steps were done according to standard procedures. Rabbit anti-Ebola virus NP and anti-Ebola virus GP/secretory GP (sGP) polyclonal antibodies were generously provided by A. Sanchez (Centers for Disease Control and Prevention). Rabbit anti-Sp1 (H225) (Santa Cruz Biotechnology), mouse anti-O-linked N-acetylglucosamine (Affinity BioReagent), rabbit and mouse (M2) anti-FLAG (Sigma), mouse anti-His6 (Roche), and rabbit anti-A.v. peptide (against green fluorescent protein [GFP]) (BD Biosciences) antibodies are commercially available.
Purification of His-tagged proteins. (i) Transcription factor Sp1.
Forty-eight hours after the transfection of 293T cells with pCSp1NHis, the cells were washed with Tris-buffered saline (50 mM Tris-Cl [pH 7.5] and 150 mM NaCl) and collected by low-speed centrifugation. They were then resuspended in hypotonic buffer (10 mM Tris-Cl [pH 7.5], 3 mM MgCl2, 10 mM KCl, and 0.5% NP-40) containing protease inhibitors (Complete protease inhibitor cocktails [Roche] and 1 mM phenylmethylsulfonylfluoride) and incubated on ice for 10 min. They were then gently vortexed for 3 s and centrifuged for 15 s at 12,000 rpm to pellet the nuclei. The supernatant was discarded, and the nuclei were resuspended in wash buffer (20 mM Tris-Cl [pH 7.5], 300 mM NaCl, and 20 mM imidazole) containing 1% Triton X-100 and protease inhibitors. The resuspension was intermittently homogenized by pipetting during a 30-min incubation on ice and then centrifuged for 15 min at 12,000 rpm at 4°C. The supernatant was collected and kept on ice until purification.
(ii) Ebola virus NP.
Forty-eight hours after being transfected with pCEboZNPCHis, 293T cells were washed with Tris-buffered saline and collected by low-speed centrifugation. They were then resuspended in hypotonic buffer (20 mM Tris-Cl [pH 7.5]) and incubated for 30 min on ice. After incubation, the cells were disrupted by repeated passage (25 times) through a 25-gauge needle. Unbroken cells and nuclei were removed by centrifugation at 1,000 × g for 5 min at 4°C. The supernatant was collected and kept on ice until purification.
(iii) Soluble form of Ebola virus GP.
Forty-eight hours after being transfected with pCZGP643HIS, 293T cells were similarly processed to obtain supernatant that was centrifuged to remove cell debris and kept on ice until purification. All His-tagged proteins were purified using Ni-nitrilotriacetic acid agarose (QIAGEN) according to the manufacturer's instructions.
Lectin binding.
Purified His-tagged proteins were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to a nitrocellulose membrane. To assess the interaction of these proteins with lectins, Sambucus nigra agglutinin, Maackia amurensis agglutinin, and peanut agglutinin, we used a digoxigenin glycan differentiation kit (Roche) according to the manufacturer's instructions. The membrane was developed with 4-nitroblue tetrazolium chloride-5-bromo-4 chloro-3-indolylphosphate (Roche).
Immunoprecipitation.
293T cells cotransfected with plasmids expressing wild-type His-tagged NP and wild-type FLAG-tagged NP, or FLAG-tagged deletion mutant NPs, were collected 48 h posttransfection, lysed in radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-Cl [pH 7.5], 150 mM NaCl, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS), and maintained for 30 min on ice. After clarification by centrifugation, the supernatants were incubated with a rabbit anti-FLAG antibody or a mouse anti-His6 antibody (1:100 dilution). After overnight incubation at 4°C, protein G- or protein A-Sepharose beads were added to the reaction mixtures, which were then incubated at 4°C for 6 h. The Sepharose beads were washed four times with RIPA buffer and then suspended in sample buffer. After the Sepharose beads were removed by centrifugation, the samples were subjected to SDS-PAGE, followed by Western blot analysis with a rabbit or mouse anti-FLAG antibody or mouse anti-His6 antibody.
Radioisotope labeling of carbohydrates.
293T cells (1 × 106 cells) were transfected with pCEboZNPCHIS, pCEboZGP, or pCAGGS/MCS. Sixteen hours posttransfection, the cells were labeled with 100 μCi of [3H]glucosamine overnight. The cells were then lysed in RIPA buffer and immunoprecipitated with an anti-GP/sGP antibody or mouse anti-NP monoclonal antibodies as described above. The samples were then subjected to SDS-PAGE.
Lectin precipitation assay.
Precipitation of proteins with a lectin was performed as described by Huang et al. (15). Briefly, cell lysates expressing Ebola virus NP were mixed with 2× binding buffer, and the sample volume was adjusted to contain a final binding buffer concentration of 1× (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM MgCl2, 1 mM MnCl2, and 1 mM CaCl2). Digoxigenin-labeled lectin (Datura stramonium agglutinin; Roche) was used for the reaction. After incubation for 1 h with the lectin, antidigoxigenin magnetic particles were added to the reaction. After a further 1-h incubation, the particles were washed four times with 1× binding buffer, suspended with Laemmli's sample buffer, and subjected to SDS-PAGE, followed by Western blotting using an anti-NP polyclonal antibody.
Electron microscopy.
Ultrathin-section electron microscopy was done as described previously (31). Briefly, the plasmid-transfected 293T cells were fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer and postfixed with 2% osmium tetroxide in the same buffer. They were dehydrated with a series of ethanol gradients, followed by propylene oxide treatment; embedded in Epon 812 resin mixture; and polymerized at 70°C. Thin sections were stained with uranyl acetate and lead citrate and examined with a HITACHI H-7500 electron microscope at 80 kV.
Minigenome assay.
GFP expression from the artificial Ebola virus minigenome was performed as described previously (37) with minor modifications. 293T cells (4 × 105 cells) were transfected with 0.25 μg of a plasmid encoding either wild-type or mutant NP, along with 0.25 μg of pCEboZ-VP35, 0.15 μg of pCEboZ-VP30, 2 μg of pCEboZ-L, 0.5 μg of p3E5EGFP, and 0.5 μg of pC-T7pol, for the production of the Ebola virus Zaire VP35, VP30, and L; the Ebola virus minigenome; and T7 polymerase, respectively. To examine the dominant-negative effects of NP mutants on GFP expression, we transfected cells with the same amounts of plasmid described above and with different amounts of an NP mutant (0.12, 0.25, 0.5, and 1 μg). Forty-eight hours after transfection, the cells were harvested, put through four cycles of freezing and thawing, and then subjected to SDS-PAGE.
RESULTS
Glycosylation of Ebola virus NP.
Huang et al. (15) have reported that Ebola virus NP is O glycosylated and sialylated. Of the several types of O glycosylation (35), two are well characterized in mammalian cells: the so-called “mucin-type” glycosylation and “nucleocytoplasmic” glycosylation (6, 13). Mucin-type glycosylation occurs in the lumen of the Golgi compartment, where N-acetylgalactosamine (GalNAc) is linked to serine and/or threonine residues of the protein and several oligosaccharides are added to GalNAc. Cell surface glycoproteins, such as Ebola GP, are modified by this type of glycosylation (10). Nucleocytoplasmic glycosylation occurs in the nucleus and cytoplasm of cells, and N-acetylglucosamine (GlcNAc) alone is attached to serine and/or threonine residues of the protein. Nucleocytoplasmic proteins, such as the transcription factor Sp1, are modified by this type of glycosylation (16).
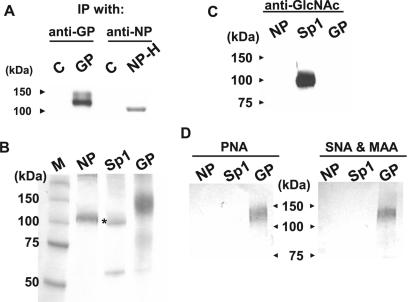
To investigate the nature of the glycosylation of Ebola virus NP, we first repeated the [3H]glucosamine labeling experiment and the lectin precipitation assay reported by Huang et al. (15). Upon expression of Ebola virus GP and His-tagged NP in the presence of [3H]glucosamine and immunoprecipitation with anti-GP antibodies and anti-NP monoclonal antibodies, respectively, we detected 3H-labeled GP and NP (Fig. 1A), suggesting that the NP is glycosylated. However, we detected only nonspecifically precipitated NP in the lectin precipitation assay (data not shown).
FIG. 1.
Characterization of Ebola virus NP glycosylation. (A) Detection of [3H]glucosamine-labeled Ebola virus NP. IP, immunoprecipitation. (B) Purification of Ebola virus NP, transcription factor Sp1, and a soluble form of Ebola virus GP. Purified proteins were subjected to SDS-PAGE and stained with Coomassie brilliant blue. The asterisk indicates Sp1; the lower band is probably a protein nonspecifically bound to and eluted from the column. (C) No nucleocytoplasmic O glycosylation was detected on Ebola virus NP with an anti-GlcNAc antibody. (D) No mucin-type O glycosylation or sialylation was detected on Ebola virus NP by using lectins: peanut agglutinin (PNA), Sambucus nigra agglutinin (SNA), and Maackia amurensis agglutinin (MAA).
Since Huang et al. ruled out the possibility of NP N glycosylation (15), we attempted to determine the type of O glycosylation of NP by partially purifying His-tagged NP, truncated Ebola virus GP (consisting of amino acids 1 to 643 of GP), and the transcription factor Sp1 using a nickel-agarose column (Fig. 1B). GP and Sp1 were used as positive controls of mucin-type and nucleocytoplasmic O glycosylation, respectively. We also confirmed the identities of these purified proteins using the appropriate antibodies (data not shown). Mucin-type and nucleocytoplasmic O glycosylation were assessed by using peanut agglutinin lectin and an anti-GlcNAc antibody, respectively (Fig. 1C and D, left). Peanut agglutinin, which recognizes mucin-type O glycosylation, reacted with GP, but not Sp1 or NP, and the anti-GlcNAc antibody, which recognizes nucleocytoplasmic O glycosylation, reacted with only Sp1. Thus, neither the lectin nor the antibody reacted with Ebola virus NP. We also examined sialylation of NP by using mixtures of lectins, Sambucus nigra agglutinin and Maackia amurensis agglutinin, specific for sialic acid terminally linked to galactose by α(2-3) and α(2-6) linkages, respectively (Fig. 1D, right). These lectins reacted with GP, but not with Sp1 or NP. Taken together, these results suggested that Ebola virus NP is glycosylated, but we were unable to identify the type of O glycosylation or sialylation.
Ebola virus NP interacts with itself.
To assess whether Ebola virus NP self-assembles, we constructed two plasmids expressing differentially epitope-tagged NP. Either a FLAG or a His epitope was added to the C terminus of NP (NP-F and NP-H, respectively). Expression of these tagged proteins was confirmed by Western blotting using an anti-Ebola virus NP antibody (data not shown). These proteins were also specifically detected by anti-FLAG and anti-His antibodies without cross-reactivity (Fig. 2, top). Immunoprecipitation of the cell lysates with the anti-His antibody, followed by Western blotting with the anti-FLAG antibody, showed that NP-F was detected only in cells coexpressed with NP-H (Fig. 2, lower left), and vice versa (Fig. 2, lower right).
FIG. 2.
Ebola virus NP interacts with itself. Plasmids encoding His- or FLAG-tagged Ebola virus NP were transfected separately (NP-H or NP-F) or together (NP-H+NP-F) into 293T cells. Proteins were detected with an anti-His or an anti-FLAG antibody either before (Total cell lysate) or after immunoprecipitation with the indicated antibody. IP denotes immunoprecipitation.
To exclude the possibility that the NP detected by immunoprecipitation in the above-mentioned experiment was an aggregate, cell lysates separately expressing NP-F or NP-H were mixed and subjected to immunoprecipitation with the anti-FLAG or anti-His antibody, followed by Western blotting with the anti-His or anti-FLAG antibody, respectively. In this case, no specific band of NP was detected (data not shown). Thus, these results indicate that NP interacts with itself.
The N-terminal half of NP is important for NP-NP interaction.
To identify the region(s) of Ebola virus NP that is important for NP-NP interaction, we generated a series of internal-deletion mutants with the FLAG tag at their C termini and examined their interaction with wild-type His-tagged NP using the procedure described above. Ebola virus NP consists of 739 amino acids; we sequentially deleted 150 amino acids from the N terminus as shown in Fig. 3A. Expression of these mutants was confirmed by Western blotting using an anti-Ebola virus NP (data not shown) or an anti-FLAG antibody (Fig. 3B). Differences in molecular weight among these mutants may reflect their amino acid compositions. NPΔ2-150F exhibited two bands on Western blotting for reasons that are unclear.
FIG. 3.
Interaction of wild-type NP with its mutants. (A) Schematic diagram of NP deletion mutants. A series of NP deletion mutants was constructed by sequential deletion of 150 N-terminal amino acids. A FLAG tag was added to the C-terminal end of the wild type and the mutants. (B) Expression of Ebola virus NP mutants. 293T cells were transfected with a plasmid encoding either wild-type FLAG-tagged NP or a FLAG-tagged mutant NP. Cell lysates were subjected to SDS-PAGE, followed by Western blotting using an anti-FLAG antibody. (C) Interaction of wild-type His-tagged NP with FLAG-tagged mutant NPs. A plasmid encoding wild-type His-tagged NP was cotransfected with a plasmid encoding either wild-type FLAG-tagged NP or FLAG-tagged mutant NP into 293T cells. Cell lysates were subjected to immunoprecipitation with either anti-FLAG or anti-His antibody. The immunoprecipitated proteins were analyzed by Western blotting using anti-FLAG and anti-His antibodies. IP denotes immunoprecipitation.
To identify which region(s) of NP was important for NP-NP interaction, plasmids expressing wild-type FLAG-tagged NP or a FLAG-tagged mutant and wild-type His-tagged NP were cotransfected into 293T cells. Forty-eight hours after transfection, the cells were lysed and the cell lysates were subjected to immunoprecipitation with an anti-FLAG or an anti-His antibody, followed by Western blotting with anti-FLAG or anti-His antibodies, respectively. When we used the anti-FLAG antibody for both immunoprecipitation and Western blotting, we detected FLAG-tagged wild type and all mutants (Fig. 3C, upper left), indicating that this antibody indeed precipitates all FLAG-tagged proteins. In cells expressing NPΔ2-150F, we again detected two bands as described above. The lower band of this mutant was detected in a larger amount by Western blotting after immunoprecipitation than when cell lysates were subjected to Western blotting prior to immunoprecipitation (lanes NPΔ2-150F in Fig. 3B and C), although we were unable to consistently obtain this result using different batches of cell lysates. Using the anti-His antibody for Western blotting after immunoprecipitation of NPs with the anti-FLAG antibody, we found that wild-type His-tagged NP interacted with wild-type FLAG-tagged NP, NPΔ451-600F, and NPΔ601-739F (Fig. 3C, lower left). Similar results were obtained by immunoprecipitating samples with the anti-His antibody, followed by Western blotting with either the anti-His or anti-FLAG antibody (Fig. 3C, right). These results indicated that the region from amino acids 1 to 450 is important for NP-NP interaction.
The FLAG-tagged mutant NPΔ451-739F, which has a deletion from amino acid 451 to the end of the molecule (Fig. 4A and B), was assessed for its interaction with wild-type His-tagged NP. This mutant was precipitated with wild-type His-tagged NP, and wild-type His-tagged NP was precipitated with the mutant (Fig. 4C). We also detected a protein at a higher molecular mass (around 100 kDa) in the NPΔ451-739F lane (Fig. 4C, lower right). The apparent molecular mass of this molecule suggests it is most likely a dimer of NPΔ451-739F. Together, these results indicated that the N-terminal half (from amino acids 1 to 450) of NP is important for NP-NP interaction.
FIG. 4.
The N-terminal half of Ebola virus NP is important for NP-NP interaction. (A) Schematic diagram of a deletion mutant lacking amino acid residues at positions 451 to 739 (Δ451-739F). (B) Expression of a mutant NPΔ451-739F. 293T cells were transfected with a plasmid encoding either wild-type FLAG-tagged NP or FLAG-tagged NPΔ451-739. Cell lysates were subjected to SDS-PAGE, followed by Western blotting using an anti-FLAG antibody. (C) Interaction of wild-type His-tagged NP with FLAG-tagged NPΔ451-739. A plasmid encoding wild-type His-tagged NP was cotransfected with a plasmid encoding either wild-type FLAG-tagged NP or FLAG-tagged NPΔ451-739 into 293T cells. Cell lysates were subjected to immunoprecipitation with either an anti-FLAG or an anti-His antibody. Immunoprecipitated proteins were analyzed by Western blotting using anti-FLAG and anti-His antibodies. The band marked by an asterisk in panels B and C likely represents a dimeric form of the mutant FLAG-tagged NPΔ451-739. IP denotes immunoprecipitation.
Morphological analyses of self-assembled NP and nucleocapsid-like structure formation.
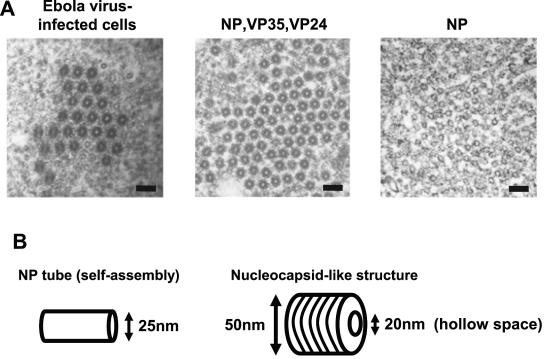
Our laboratory recently demonstrated that expression of Ebola virus NP alone results in the formation of helical structures (Fig. 5A, right) (Noda and Kawaoka, unpublished) that are unlike nucleocapsid-like structures. The diameter of these NP helical tubes is approximately 20 nm, which is almost the same as that of the central portion of the nucleocapsids. We confirmed the findings of Huang et al. (15) that expression of NP with VP35 and VP24 results in the formation of structures similar to authentic nucleocapsids (Fig. 5A, left and middle). Schematic diagrams of the NP tube (expression of NP alone) and nucleocapsid-like (expression of NP with VP35 and VP24) structures are depicted in Fig. 5B. While the diameters of the NP tube structures in this study are 25 nm, the nucleocapsid-like structures have a diameter of 50 nm with an internal hollow space of 20 nm.
FIG.5.
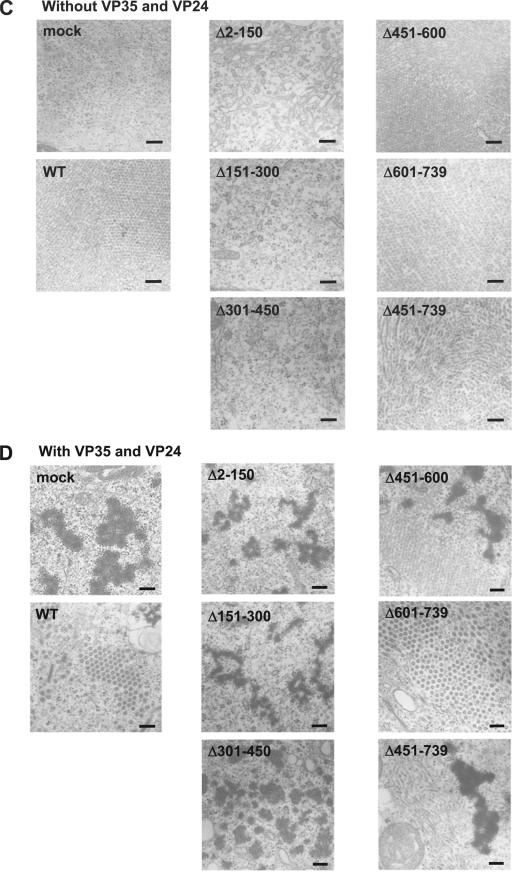
Electron microscopy of NP deletion mutants. (A) Electron micrographs showing Ebola virus-infected cells containing nucleocapsids; cells expressing NP, VP35, and VP24 and thus possessing nucleocapsid-like structures; and cells expressing NP alone, resulting in the formation of helical-ring-like structures. Bars, 100 nm. (B) Schematic diagram of NP tube and nucleocapsid-like structures. (C and D) Formation of NP tube and nucleocapsid-like structures in NP mutant-expressing cells. Wild-type or individual mutants were expressed in the absence (C) or presence (D) of VP35 and VP24 in 293T cells. WT, wild type. Bars, 200 nm.
Which region(s) of NP are important for the formation of these NP tube and nucleocapsid-like structures? Fig. 5C and D shows the morphology in sections of cells transfected with a plasmid expressing an NP mutant in the absence (C) or presence (D) of VP35 and VP24. We were unable to detect either NP tubes or nucleocapsid-like structures in cells transfected with a plasmid expressing NPΔ2-150, NPΔ151-300, or NPΔ301-450 either alone or in the presence of VP35 and VP24, although expression of these mutants in cells was confirmed by immunoelectron microscopy using the anti-NP antibody (data not shown). NP tube structures, however, were observed in cells expressing the other mutants, NPΔ451-600, NPΔ601-739, and NPΔ451-739, indicating self-assembly of these mutants. These results demonstrate that the N-terminal half (from amino acids 1 to 450) of NP is important for the formation of NP tube structures and are consistent with the results of NP-NP interaction experiments described earlier (Fig. 3 and 4).
By contrast, nucleocapsid-like structures were detected only with the NPΔ601-739 mutant in the presence of VP35 and VP24; for mutants NPΔ451-600 and NPΔ451-739, only the ring-like structures were obtained even in the presence of VP35 and VP24. These results indicated that the region of NP from amino acids 451 to 600, as well as the region required for NP-NP interaction (i.e., amino acids 1 to 450), is required for the formation of nucleocapsid-like structures.
The formation of nucleocapsid-like structures is required for replication of the Ebola virus genome.
Ebola virus NP forms the RNP complex, together with Ebola virus proteins VP35, VP30, and L and the viral genome. This RNP complex is the minimal unit required for replication of the viral genome (26). We asked which region(s) of NP is important for replication of the Ebola virus genome by using the Ebola virus minigenome containing the GFP gene (37).
A plasmid for the expression of either the untagged wild type or an NP mutant was transfected into 293T cells with plasmids for the expression of Ebola virus VP35, VP30, L, and the Ebola virus minigenome containing the GFP gene. A plasmid for expression of T7 polymerase was also transfected into these cells to drive the expression of the Ebola virus minigenome under the control of the T7 promoter. Forty-eight hours posttransfection, expression of wild-type NP, mutant NPs, and GFP was confirmed by Western blotting (Fig. 6). With the exception of mutant NPΔ451-739, all of the NP mutants were detected by using an anti-NP polyclonal antibody (Fig. 6A, left). Although expression of NPΔ451-739 was barely detectable with this polyclonal antibody, we were able to detect it by using a mouse anti-NP monoclonal antibody (Fig. 6A, right). The lower bands in the wild-type NP lane, detected by both the polyclonal and monoclonal antibodies, likely represent degraded NP. We then examined whether these NPs supported the replication of the Ebola virus genome. As shown in Fig. 6B, GFP was detected only when the wild type or the NPΔ601-739 mutant was expressed. Since NPΔ601-739 was the only mutant able to form nucleocapsid-like structures (Fig. 5), these results suggest that the formation of the nucleocapid-like structures is needed for the expression of proteins from the Ebola virus genome.
FIG. 6.
The region of NP important for GFP expression from the Ebola virus minigenome. The wild type or individual mutants were expressed, together with VP35, VP30, L, the Ebola virus minigenome possessing the GFP gene, and the T7 polymerase. GFP expression was detected by Western blotting. Ab and mAb denote antibody and monoclonal antibody, respectively. The asterisk indicates expression of NPΔ451-739, which was barely detectable with this polyclonal antibody.
The dominant-negative effect of NP mutants on the function of wild-type NP.
Our finding that the region of NP from amino acids 1 to 450 was required for NP-NP interaction but was unable to support viral-genome replication, as demonstrated by mutants NPΔ451-600 and NPΔ451-739 (Fig. 4, 5, and 6), prompted us to test the dominant-negative potential of this region.
Various amounts of plasmid for the expression of either mutant NPΔ451-600 or NPΔ451-739 were transfected, together with plasmids for the expression of wild-type NP, VP35, VP30, L, T7 polymerase, and the Ebola virus minigenome, as described above. Forty-eight hours posttransfection, we examined the expression of wild-type and mutant NPs and GFP in cell lysates by Western blotting (Fig. 7). Although the expression of wild-type NP was not affected by the expression of either NPΔ451-600 or NPΔ451-739, GFP expression was inhibited in a dose-dependent manner by NPΔ451-600 and to a lesser extent by NPΔ451-739, indicating that these mutants indeed have a dominant-negative effect on the function of wild-type NP.
FIG. 7.
Dominant-negative effect of NP mutants on GFP expression from the Ebola virus minigenome. Various amounts of NP mutants (NPΔ451-600 or NPΔ451-739) were expressed with a constant amount of wild-type NP (WT), along with VP35, VP30, L, the Ebola virus minigenome possessing the GFP gene, and the T7 polymerase. GFP expression was detected by Western blotting.
DISCUSSION
By using biochemical and electron microscopic analyses, we showed that the N-terminal 450 amino acids of Ebola virus NP are important for NP-NP interaction and for the formation of NP tube structures (Fig. 3 to 5). Several studies have demonstrated that the N-terminal regions of the NP/N molecules of paramyxoviruses are generally important for NP-NP (N-N) interaction (1, 4, 19, 20, 27, 29), therefore our data support the functional conservation of this region of NP for NP-NP interactions among the members of Mononegavirales. The fact that a small region (residues 230 to 370) of the N terminus of Ebola virus NP shares appreciable amino acid homology with the NPs/Ns of paramyxoviruses and, to a lesser extent, with those of rhabdoviruses (34) further supports this notion.
The expression of Ebola virus NP alone results in the generation of inclusions in Ebola virus-infected cells (26), which have been shown by electron microscopy to be Ebola virus NP tube structures (Noda and Kawaoka, unpublished) (Fig. 5). These structures differ from the nucleocapsid-like structures that are generated by NP in the presence of VP35 and VP24 or nucleocapsids in Ebola virus-infected cells (Fig. 5) (15). NP mutants lacking the N-terminal region (e.g., NPΔ2-150) demonstrated that unless NP tube structures were formed, nucleocapsid-like structures were not formed (Fig. 3C and 5). Thus, our results suggest that the Ebola virus NP tube forms a scaffold for the nucleocapsid-like structures. Marburg virus NP alone is sufficient to form nucleocapsid-like structures (22, 24), which suggests that the nucleocapsid structures of these two viruses are different.
The region from amino acids 601 to 739 of NP was not required for nucleocapsid formation or replication of the minigenome. Recently, it was reported that the last 50 amino acids of Ebola virus NP are required for incorporation of NP into virus-like particles generated by Ebola virus VP40 (23). Therefore, this region may be important for the interaction of NP with VP40 for nucleocapsid incorporation into virions. Further analyses of this region will likely shed light on its functional role in the Ebola virus replication cycle.
Posttranslational modifications, such as glycosylation and phosphorylation, are often essential structural alterations for the functions of proteins. For example, phosphorylation is critical for proteins involved in signal transduction in interferon signaling (14, 38). Huang et al. (15) have previously reported that O glycosylation and sialylation of Ebola virus NP are required for interaction of NP with VP35. Immunofluorescence (25) and immunoelectron microscopy (not shown) have shown that Ebola virus NP is localized in the cytoplasm but not in the Golgi apparatus, indicating that mucin-type O glycosylation is unlikely to occur. On the other hand, nucleocytoplasmic O glycosylation involves the attachment of GlcNAc to serine and/or threonine residues of proteins (6), and interestingly, all known O-linked GlcNAc proteins are also phosphoproteins, because phosphate is also linked to serine and/or threonine residues (17). Indeed, the dynamic interplay between O-linked GlcNAc modification and phosphorylation of nucleocytoplasmic proteins has been well documented (3, 5, 18). Since Ebola virus NP is phosphorylated (8), it was possible that nucleocytoplasmic O glycosylation might occur. Like Huang et al. (15), we detected [3H]glucosamine-labeled NP, suggesting that Ebola virus NP is glycosylated; however, none of our analyses identified the types of O-linked glycosylation or sialylation (Fig. 1). Thus, further studies are essential to understand the nature of NP glycosylation.
In conclusion, the region from amino acids 1 to 450 of Ebola virus NP is important for NP-NP interaction, while this region and the next 150 amino acids are required for the formation of nucleocapsid-like structures and for replication of the viral genome. Another functional region(s) of Ebola virus NP required for its association with other viral proteins has yet to be characterized. Given that NP is likely involved in all steps of viral replication after uncoating of the viral genome, including the formation of nucleocapsid-like structures, replication of the viral genome, and incorporation of nucleocapsid into virions, a better understanding of the functional region(s) of this protein will enhance our knowledge of the life cycle of Ebola virus and provide insight into its extreme pathogenicity.
Acknowledgments
We thank Elke Mühlberger and Hans-Dieter Klenk for providing us with Ebola virus genes and Anthony Sanchez for antiserum to GP and NP. We also thank Krisna Wells and Martha McGregor for excellent technical assistance and Susan Watson for editing the manuscript.
This work was supported by Public Health Service research grants from the National Institute of Allergy and Infectious Diseases; by Grants-in-Aid for Scientific Research on Priority Areas from the Ministries of Education, Culture, Sports, Science, and Technology, Japan; and by CREST (Japan Science and Technology Agency). The work was also sponsored by the NIH/NIAID Regional Center for Excellence for Biodefense and Emerging Infectious Diseases Research (RCE) Program. We acknowledge membership of and support from the Region V “Great Lakes” RCE (NIH award 1-U54-AI-057153).
REFERENCES
- 1.Bankamp, B., S. M. Horikami, P. D. Thompson, M. Huber, M. Billeter, and S. A. Moyer. 1996. Domains of the measles virus N protein required for binding to P protein and self-assembly. Virology 216:272-277. [DOI] [PubMed] [Google Scholar]
- 2.Bhella, D., A. Ralph, L. B. Murphy, and R. P. Yeo. 2002. Significant differences in nucleocapsid morphology within the Paramyxoviridae. J. Gen. Virol. 83:1831-1839. [DOI] [PubMed] [Google Scholar]
- 3.Brownlee, M. 2001. Biochemistry and molecular cell biology of diabetic complications. Nature 414:813-820. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz, C. J., D. Spehner, R. Drillien, W. J. Neubert, and H. E. Homann. 1993. The conserved N-terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J. Virol. 67:5803-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chou, T. Y., C. V. Dang, and G. W. Hart. 1995. Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. USA 92:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Comer, F. I., and G. W. Hart. 2000. O-glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J. Biol. Chem. 275:29179-29182. [DOI] [PubMed] [Google Scholar]
- 7.Curran, J., and D. Kolakofsky. 1999. Replication of paramyxoviruses. Adv. Virus Res. 54:403-422. [DOI] [PubMed] [Google Scholar]
- 8.Elliott, L. H., M. P. Kiley, and J. B. McCormick. 1985. Descriptive analysis of Ebola virus proteins. Virology 147:169-176. [DOI] [PubMed] [Google Scholar]
- 9.Errington, W., and P. T. Emmerson. 1997. Assembly of recombinant Newcastle disease virus nucleocapsid protein into nucleocapsid-like structures is inhibited by the phosphoprotein. J. Gen. Virol. 78:2335-2339. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann, H., S. T. Nichol, H. D. Klenk, C. J. Peters, and A. Sanchez. 1994. Characterization of filoviruses based on differences in structure and antigenicity of the virion glycoprotein. Virology 199:469-473. [DOI] [PubMed] [Google Scholar]
- 11.Fooks, A. R., J. R. Stephenson, A. Warnes, A. B. Dowsett, B. K. Rima, and G. W. Wilkinson. 1993. Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J. Gen. Virol. 74:1439-1444. [DOI] [PubMed] [Google Scholar]
- 12.Han, Z., H. Boshra, J. O. Sunyer, S. H. Zwiers, J. Paragas, and R. N. Harty. 2003. Biochemical and functional characterization of the Ebola virus VP24 protein: implications for a role in virus assembly and budding. J. Virol. 77:1793-1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanisch, F. G. 2001. O-glycosylation of the mucin type. Biol. Chem. 382:143-149. [DOI] [PubMed] [Google Scholar]
- 14.Hiscott, J., N. Grandvaux, S. Sharma, B. R. Tenoever, M. J. Servant, and R. Lin. 2003. Convergence of the NF-κB and interferon signaling pathways in the regulation of antiviral defense and apoptosis. Ann. N. Y. Acad. Sci. 1010:237-248. [DOI] [PubMed] [Google Scholar]
- 15.Huang, Y., L. Xu, Y. Sun, and G. J. Nabel. 2002. The assembly of Ebola virus nucleocapsid requires virion-associated proteins 35 and 24 and posttranslational modification of nucleoprotein. Mol. Cell 10:307-316. [DOI] [PubMed] [Google Scholar]
- 16.Jackson, S. P., and R. Tjian. 1988. O-glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55:125-133. [DOI] [PubMed] [Google Scholar]
- 17.Kamemura, K., and G. W. Hart. 2003. Dynamic interplay between O-glycosylation and O-phosphorylation of nucleocytoplasmic proteins: a new paradigm for metabolic control of signal transduction and transcription. Prog. Nucleic Acid Res. Mol. Biol. 73:107-136. [DOI] [PubMed] [Google Scholar]
- 18.Kelly, W. G., M. E. Dahmus, and G. W. Hart. 1993. RNA polymerase II is a glycoprotein. Modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 268:10416-10424. [PubMed] [Google Scholar]
- 19.Kho, C. L., W. S. Tan, B. T. Tey, and K. Yusoff. 2003. Newcastle disease virus nucleocapsid protein: self-assembly and length-determination domains. J. Gen. Virol. 84:2163-2168. [DOI] [PubMed] [Google Scholar]
- 20.Kingston, R. L., W. A. Baase, and L. S. Gay. 2004. Characterization of nucleocapsid binding by the measles virus and mumps virus phosphoproteins. J. Virol. 78:8630-8640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobasa, D., M. E. Rodgers, K. Wells, and Y. Kawaoka. 1997. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J. Virol. 71:6706-6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kolesnikova, L., E. Mühlberger, E. Ryabchikova, and S. Becker. 2000. Ultrastructural organization of recombinant Marburg virus nucleoprotein: comparison with Marburg virus inclusions. J. Virol. 74:3899-3904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Licata, J. M., R. F. Johnson, Z. Han, and R. N. Harty. 2004. Contribution of Ebola virus glycoprotein, nucleoprotein, and VP24 to budding of VP40 virus-like particles. J. Virol. 78:7344-7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mavrakis, M., L. Kolesnikova, G. Schoehn, S. Becker, and R. W. Ruigrok. 2002. Morphology of Marburg virus NP-RNA. Virology 296:300-307. [DOI] [PubMed] [Google Scholar]
- 25.Modrof, J., E. Mühlberger, H. D. Klenk, and S. Becker. 2002. Phosphorylation of VP30 impairs Ebola virus transcription. J. Biol. Chem. 277:33099-33104. [DOI] [PubMed] [Google Scholar]
- 26.Mühlberger, E., M. Weik, V. E. Volchkov, H. D. Klenk, and S. Becker. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J. Virol. 73:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murphy, L. B., C. Loney, J. Murray, D. Bhella, P. Ashton, and R. P. Yeo. 2003. Investigations into the amino-terminal domain of the respiratory syncytial virus nucleocapsid protein reveal elements important for nucleocapsid formation and interaction with the phosphoprotein. Virology 307:143-153. [DOI] [PubMed] [Google Scholar]
- 28.Neumann, G., H. Feldmann, S. Watanabe, I. Lukashevich, and Y. Kawaoka. 2002. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J. Virol. 76:406-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishio, M., M. Tsurudome, M. Ito, M. Kawano, S. Kusagawa, H. Komada, and Y. Ito. 1999. Mapping of domains on the human parainfluenza virus type 2 nucleocapsid protein (NP) required for NP-phosphoprotein or NP-NP interaction. J. Gen. Virol. 80:2017-2022. [DOI] [PubMed] [Google Scholar]
- 30.Niwa, H., K. Yamamura, and J. Miyazaki. 1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108:193-199. [DOI] [PubMed] [Google Scholar]
- 31.Noda, T., H. Sagara, E. Suzuki, A. Takada, H. Kida, and Y. Kawaoka. 2002. Ebola virus VP40 drives the formation of virus-like filamentous particles along with GP. J. Virol. 76:4855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanchez, A., A. S. Khan, S. R. Zaki, G. J. Nabel, T. G. Ksiazek, and C. J. Peters. 2001. Filoviridae: Marburg and Ebola viruses, p. 1279-1304. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott-Raven, Philadelphia, Pa.
- 33.Sanchez, A., M. P. Kiley, B. P. Holloway, J. B. McCormick, and D. D. Auperin. 1989. The nucleoprotein gene of Ebola virus: cloning, sequencing, and in vitro expression. Virology 170:81-91. [DOI] [PubMed] [Google Scholar]
- 34.Sanchez, A., M. P. Kiley, H. D. Klenk, and H. Feldmann. 1992. Sequence analysis of the Marburg virus nucleoprotein gene: comparison to Ebola virus and other non-segmented negative-strand RNA viruses. J. Gen. Virol. 73:347-357. [DOI] [PubMed] [Google Scholar]
- 35.Spiro, R. G. 2002. Protein glycosylation: nature, distribution, enzymatic formation, and disease implications of glycopeptide bonds. Glycobiology 12:43R-56R. [DOI] [PubMed] [Google Scholar]
- 36.Takada, A., S. Watanabe, K. Okazaki, H. Kida, and Y. Kawaoka. 2001. Infectivity-enhancing antibodies to Ebola virus glycoprotein. J. Virol. 75:2324-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Watanabe, S., T. Watanabe, T. Noda, A. Takada, H. Feldmann, L. D. Jasenosky, and Y. Kawaoka. 2004. Production of novel Ebola virus-like particles from cDNAs: an alternative to Ebola virus generation by reverse genetics. J. Virol. 78:999-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoneyama, M., W. Suhara, and T. Fujita. 2002. Control of IRF-3 activation by phosphorylation. J. Interferon Cytokine Res. 22:73-76. [DOI] [PubMed] [Google Scholar]