Abstract
BMS-488043 is a small-molecule human immunodeficiency virus type 1 (HIV-1) CD4 attachment inhibitor with demonstrated clinical efficacy. The compound inhibits soluble CD4 (sCD4) binding to the 11 distinct HIV envelope gp120 proteins surveyed. Binding of BMS-488043 and that of sCD4 to gp120 are mutually exclusive, since increased concentrations of one can completely block the binding of the other without affecting the maximal gp120 binding capacity. Similarly, BMS-488043 inhibited virion envelope trimers from binding to sCD4-immunoglobulin G (IgG), with decreasing inhibition as the sCD4-IgG concentration increased, and BMS-488043 blocked the sCD4-induced exposure of the gp41 groove in virions. In both virion binding assays, BMS-488043 was active only when added prior to sCD4. Collectively, these results indicate that obstruction of gp120-sCD4 interactions is the primary inhibition mechanism of this compound and that compound interaction with envelope must precede CD4 binding. By three independent approaches, BMS-488043 was further shown to induce conformational changes within gp120 in both the CD4 and CCR5 binding regions. These changes likely prevent gp120-CD4 interactions and downstream entry events. However, BMS-488043 could only partially inhibit CD4 binding to an HIV variant containing a specific envelope truncation and altered gp120 conformation, despite effectively inhibiting the pseudotyped virus infection. Taken together, BMS-488043 inhibits viral entry primarily through altering the envelope conformation and preventing CD4 binding, and other downstream entry events could also be inhibited as a result of these induced conformational changes.
It is estimated that >40 million people worldwide are infected with human immunodeficiency virus type 1 (HIV-1). Despite the success of highly active antiretroviral therapies, emergence of resistant viruses, development of adverse side effects, and toxicities from long-term use remain challenging (1, 32, 33, 41). With up to 20% of the new infections involving variants resistant to current medications, antiretroviral agents targeting new steps in the replicative cycle and lacking cross-resistance to existing drug classes are urgently needed for managing HIV infection (24, 44). The recent introduction of a novel fusion inhibitor and other potential entry inhibitors will likely provide an expanded range of treatment options for anti-HIV therapy (7, 20, 29, 31).
HIV envelope, a trimeric protein each part of which consists of an exterior glycoprotein (gp120) and a transmembrane domain (gp41), is required for viral entry into cells (2, 46). The entry process begins with binding of gp120 to the cellular CD4 receptor (22, 34, 40). Resulting conformational changes in gp120 facilitate subsequent binding to an HIV-1 coreceptor and insertion of the fusion peptide of gp41 into the cell membrane. The fusion between the viral and cell membranes occurs via a six-helix bundle formation within gp41 (2, 27). The potential intervention steps in the entry process (and their respective inhibitors) are gp120-CD4 binding (BMS-488043, PRO-542), gp120-coreceptor interactions (SCH D, UK-427857, GW-873140), and gp41-induced membrane fusion (Enfuvirtide). These inhibitors have exhibited clinical efficacy in HIV-infected patients (7, 12, 20, 26).
BMS-488043 is an HIV CD4 attachment inhibitor (AI). Like its predecessor BMS-378806 (13, 23, 43), it targets the initial gp120-CD4 interaction of viral entry and possesses potent antiviral activity against T- and M-tropic laboratory and clinical isolates (13, 23, 43). With improved pharmacokinetic properties, BMS-488043 demonstrated antiviral efficacy and a favorable safety profile in HIV-infected subjects (14, 15). The X-ray crystal structure of an unliganded form of simian immunodeficiency virus (SIV) gp120 (4) was recently reported, and mapping of the BMS-378806 resistance residues identified a potential compound binding cavity in this structure. Numerous CD4 contact residues were also located in the same cavity, supporting previous findings that this series of inhibitors blocks CD4 binding (13). However, Si et al. suggested post-CD4 inhibition as the mechanism for BMS-378806 (38). In this study, we further characterized the inhibition mechanism of HIV-1 AIs against monomeric gp120 proteins and the trimeric virion envelope. We also studied the effects of these inhibitors on gp120 conformation by using conformation-sensitive antibodies and two other independent approaches. To understand the impact of the gp120 conformation on compound action, we also investigated the activity of inhibitors against the HIV-1 variant containing a deletion in gp41 of the envelope (ΔCT) that was utilized by Si et al. (38). This truncation of gp41 affects the gp120 conformation, resulting in greater exposure of receptor-coreceptor binding regions and elevated viral sensitivity to neutralizing antibodies and HIV-positive serum (8, 9). The present study reveals the complex mechanism of inhibition by this class of compounds and provides insight into the different observations made by Si. et al. (38).
MATERIALS AND METHODS
Reagents.
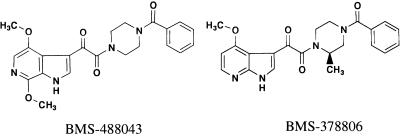
AI BMS-488043 (Fig. 1), its photoactive analog, and the tritium-labeled inhibitors were prepared at Bristol-Myers Squibb Company. BMS-488043 is compound 5ao in a patent by T. Wang et al. (42); experimental procedures for complete synthesis from commercially available materials is described on line 1 of column 87 through line 49 of column 90. HIV-1 proviral DNA and isolates JRFL, JRCSF, YU2, SF162, BAL, LAI, NL4-3, and 92US715.6 were obtained from the National Institutes of Health AIDS Research and Reference Reagent Program. Additional envelope-expressing plasmids (NA420B33, NA420LN40, and AD8) were from the University of Massachusetts or were prepared at Bristol-Myers Squibb Company. Soluble CD4 (sCD4) was from ImmunoDiagnostics, Inc. (Woburn, MA), and CD4-IgG was from Retro-Tech GmbH (Unterschleissheim, Germany). Conformation-sensitive human anti-gp120 monoclonal antibodies (MAbs) were from the Tulane University School of Medicine (New Orleans, LA), and sheep polyclonal anti-gp120 D7324 was from Cliniqa (Fallbrook, CA). Anti-N36-C34 complex mouse MAb NC-1 (18) and the C34 peptide came from the New York Blood Center (New York, NY) and Macromolecular Resources (Fort Collis, CO). Anti-CD4 MAb OKT4 was purified from an OKT4-producing hybridoma cell line (ATCC CRL-8002).
FIG. 1.
Structures of CD4 attachment inhibitors.
[125I]sCD4 was prepared by labeling sCD4 with 125I-labeled Bolton-Hunter reagent (specific activity, 4,400 Ci/mmol; Perkin-Elmer Life Sciences) as described in the product literature. In brief, 1 mCi of 125I-labeled Bolton-Hunter reagent was taken to dryness under a stream of nitrogen gas and then mixed with 25 μg of sCD4 (adjusted to pH 8.0 with potassium phosphate). After 40 min at room temperature, the reaction was quenched by addition of Tris-glycine buffer, pH 8.3, and then purified on a Micro Bio-spin 6 column (Bio-Rad Laboratories). The purified [125I]sCD4 was eluted in a volume of ∼300 μl at 150 μCi/ml with a specific activity estimated to be 250 Ci/mmol.
sCD4-gp120 binding.
Expression and purification of recombinant gp120 and enzyme-linked immunosorbent assay (ELISA) to determine gp120-CD4 binding were done as previously described (13).
[125I]sCD4-gp120 binding study.
[125I]sCD4 binding to gp120 was assayed in 25-μl reaction mixtures containing 25 mM Tris-Cl (pH 7.6), 125 mM NaCl, 0.02% Tween 20, and 8 μg/ml gp120. Binding was initiated by addition of [125I]sCD4 (14,000 cpm/reaction mixture), followed, 5 min later, by the addition of 0 to 300 μM BMS-488043. In an alternative assay, the order of addition of [125I]sCD4 and BMS-488043 was reversed. Reactions were quenched by adding 250 μl of a 20% slurry of lentil lectin Sepharose 4B (Amersham Biosciences), followed immediately by centrifugation through an empty Micro Bio-Spin column (Bio-Rad Laboratories). The dried columns, containing lentil lectin-bound gp120, were counted directly in a gamma counter.
Inhibitor-gp120 binding.
Gel filtration assays using Micro Bio-Spin 6 columns (Bio-Rad Laboratories) to measure [3H]AI binding to gp120 were done as previously described (13). To evaluate the ability of sCD4 to compete out inhibitor binding with gp120, mixtures containing [3H]BMS-488043 (200 nM) and various concentrations of sCD4 were incubated with recombinant gp120 (30 nM) at ambient temperature for 15 min and then subjected to Micro Bio-Spin 6 column separation. 3H activities associated with gp120 were eluted and determined with a Beckman scintillation counter and normalized to that obtained from control reaction mixtures containing only gp120 and [3H]BMS-488043 without sCD4 added.
MAb-gp120 binding ELISA.
The MAb-gp120 binding ELISA used was adapted from the previously described gp120-CD4 binding ELISA (13) by replacing sCD4 with a designated anti-gp120 MAb and coupling to a corresponding secondary antibody-peroxidase conjugate for signal detection.
CD analysis.
Circular-dichroism (CD) data were generated with a Jasco J-720 Spectropolarimeter at 20°C with a quartz cell with a 1-mm path length (19). The CD spectra were smoothed with a Savitzky-Golay algorithm, and peak minima were identified by 4rt derivative resolution enhancement. The gp120 solution was at 4.0 μM in 20 mM Tris-HCl (pH 8.0), aliquots of BMS-488043 stock solution were added incrementally, and the protein spectra were scanned at wavelengths of 200 to 245 nm; CD intensity data were corrected for dilution.
Thrombin digestion.
Protease susceptibility testing of gp120 (2.8 nM) was carried out by pretreating gp120 with 5 μM inhibitor or 625 nM sCD4 and then exposing it to 600 nM thrombin at 37°C. The reaction was stopped with a thrombin inhibitor (DUP-714), and the extent of V3 loop cleavage was determined by ELISA with an anti-V3 antibody-peroxidase conjugate (1121-P; ImmunoDiagnostics, Inc.).
Viruses.
Antiviral assays with HIV-1 envelope-pseudotyped viruses were done as previously described (13, 23). HIV-1 LAI virus was prepared by transfecting 293T cells with pLAI proviral DNA and Lipofectamine plus reagent (Invitrogen). The pseudotyped YU2 and JRFL viruses were prepared by cotransfecting 293T cells with pLAI-Luc-ΔEnv and plasmid DNA encoding the designated envelopes. Viruses in supernatant were harvested 48 to 72 h after transfection and kept at −140°C until use.
Pseudotype virus infection assay.
HeLa CD4 CCR5 cells were seeded (104/well; Corning) 1 day prior to assay in selection-free medium (Dulbecco modified Eagle medium [DMEM], 10% fetal bovine serum [FBS]). HIV-1 JRFL or YU-2 pseudotype virus was added to cells in the presence of an inhibitor and incubated for 3 days at 37°C. Luciferase activity was determined with the Roche Luciferase Reporter Gene Assay.
Replication fitness assay.
Peripheral blood mononuclear cells stimulated with 2-μg/ml phytohemagglutinin for 3 days were infected at a multiplicity of infection of 0.005 for 3 h at 37°C. Unbound viruses were removed by washing cells with phosphate-buffered saline. Cells were resuspended in complete medium (RPMI 1640 medium, 20% FBS, 4 mM l-glutamine, 1-ng/ml interleukin-1) at 106 cells/ml and incubated at 37°C for 10 days. Supernatant (200 μl) was collected daily starting at day 0, and p24 levels were measured to determine the viral yield.
Virion binding assays.
All virion binding assays were carried out at ambient temperature. Assay mixtures without sCD4 or sCD4-IgG were included to determine the level of nonspecific virion binding.
Virion-sCD4-IgG binding assay.
HIV-1 LAI (molecular clone) stock obtained from transfected 293T cells were pelleted through a 20% sucrose cushion, resuspended in DMEM in one-eighth of the initial volume, and stored at −140°C until use. For sCD4-IgG binding, purified LAI virion stock was diluted eightfold with DMEM-10% FBS and 100 μl was transferred to each well (Immulon 4HBX 96-well plate; Thermo Electron Corporation) with sCD4-IgG preanchored to the Protein A/G-coated wells (Pierce Biotechnology, Inc., Rockford, IL). Virions were mixed with various concentrations of inhibitor, added to an sCD4-IgG-coated plate, and incubated for 1 h. Unbound virions were removed at the end of incubation period, and bound virions were measured with the Alliance HIV p24 assay kit (Perkin-Elmer, Shelton, CT). Nonspecific virion-IgG binding (determined as that not competed out by excess sCD4) was subtracted during calculation of the compound effect. For time-of-addition studies, BMS-488043 was added to viruses at 30 min prior to and 0, 10, 20, and 40 min after initiation of sCD4-IgG binding.
Virion-sCD4 binding assay.
Virion stock solutions were treated with defined concentrations of inhibitors as specified in each experiment for 0.5 h. Soluble CD4 (5 μg/ml) was then added, and the reaction mixtures (100 μl) were transferred to ELISA plates containing anti-CD4 MAb OKT4 anchored to the Protein A/G coating as already described and incubated for an additional 0.5 or 1 h. Unbound virions were then removed by multiple washes, and the OKT4-captured, sCD4-bound virions were detected by the p24 ELISA.
Virion-sCD4-C34 binding assay.
Virions were first treated with inhibitor for 30 min, and then C34 (50 μg/ml) and sCD4 (5 μg/ml) were added concomitantly (final volume, 200 μl). Immediately after mixing, two 100-μl aliquots were transferred to an ELISA plate (Immulon 4HBX) containing NC-1 MAb anchored to the Protein A/G that was immobilized on the plate. Following a 1-h incubation, unbound virions were removed and the p24 level was determined as described above to measure the bound virions. For time-of-addition studies, BMS-488043 was added to viruses at 30 min prior to and 0, 10, 20, 40, and 60 min after sCD4-C34 peptide addition.
Levels of p24 detected in the presence of inhibitor were normalized to that detected from the respective no-inhibitor control. The inhibitor IC50 for virion binding was calculated with the Microsoft XLfit software.
RESULTS
Inhibition of gp120 binding to sCD4.
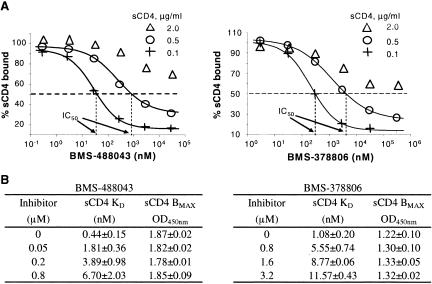
BMS-488043, a close analog of BMS-378806, effectively blocked HIV-1 LAI and JRFL replication with 50% effective concentrations of around 3 nM. Previously, we reported that BMS-378806 inhibited viral replication by interfering with the binding interactions of gp120 with the cellular CD4 receptor (13, 23). Similarly, BMS-488043 inhibited the binding of gp120JRFL to sCD4 (0.1 μg/ml) in a gp120-CD4 binding ELISA with an IC50 of 50 to 100 nM. Inhibition of gp120-sCD4 binding was consistently observed with several structural analogs of BMS-488043 (data not shown). Importantly, inhibition potency was dependent on sCD4 concentrations, since the ability of BMS-4888043 to interfere with binding decreased significantly (IC50, 0.05 → >30 μM) with increasing concentrations of sCD4 (0.1 → 2 μg/ml) (Fig. 2A, left half). This concomitant increase in IC50s with sCD4 concentrations also held true for BMS-378806 (Fig. 2A, right half). The higher concentration of sCD4 used by Si et al. in their ELISA (38) likely masked the BMS-378806 activity. Furthermore, inhibition by either compound was no longer detectable (IC50, >300 μM) when assayed against a preformed complex of gp120JRFL and sCD4 (data not shown). Conversely, like BMS-378806 (Fig. 2B and reference 13), the presence of elevated concentrations of BMS-488043 increased the amount of sCD4 needed to occupy 50% of the gp120JRFL molecules (apparent KD) without affecting the maximal CD4 binding capacity (Bmax) of gp120JRFL (Fig. 2B). Together, these results suggest a competitive inhibition of AIs against sCD4 binding to gp120.
FIG. 2.
Inhibition of gp120JRFL binding to sCD4 by BMS-488043 and BMS-378806. Inhibitor activity was measured with the gp120-sCD4 binding ELISA. (A) Effect of sCD4 concentration on the inhibitory activity of AI against gp120-sCD4 binding (IC50, concentration needed to inhibit gp120-sCD4 binding by 50%). (B) Effect of AI concentration on the apparent affinity (KD) of sCD4 for gp120. BMAX, maximal sCD4 bound to gp120. OD450nm, optical density at 450 nm.
To further show that inhibition of sCD4 binding is the primary consequence of AI binding to viral gp120, we surveyed BMS-488043 activity against 10 additional gp120s of various strain origins (M- and T-tropic viruses from laboratory or clinical isolates) with the gp120-CD4 binding ELISA. BMS-488043 was effective against all of the gp120s tested. The IC50s determined for the gp120s from HIV LAI, BAL, NA420LN40, SF162, NL4-3, NA420B33, YU2, AD8, JRCSF, and 92US15.6 were 0.1, 0.1, 0.3, 0.5, 0.6, 0.7, 0.9, 1.0, 1.1, and 1.6 μM, respectively. A similar observation was also made for BMS-378806 (IC50s ranged from 0.2 to 9.6 μM). Therefore, the primary action of AIs appears to be interference with gp120 binding to cellular sCD4 receptors.
Binding of sCD4 and that of AI to gp120JRFL are mutually exclusive.
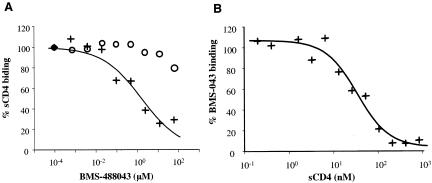
Previous studies showed that HIV AIs selectively bind to gp120 (13). To further understand the interactions of CD4 receptor and AIs with viral envelope gp120, binding assays were performed with [125I]sCD4 or [3H]BMS-488043 in the presence or absence of an unlabeled competing ligand. As expected, inhibition of [125I]sCD4 binding to gp120, in a compound concentration-dependent manner, was observed when gp120 was exposed to unlabeled BMS-488043 simultaneously with or prior to [125I]sCD4 addition (Fig. 3A). If [125I]sCD4 was added prior to the AI, little reduction in [125I]sCD4 binding was detected, even at an AI concentration of 100 μM (Fig. 3A). In a separate experiment, [3H]BMS-488043 was found to bind rapidly to gp120, reaching an equilibrium with a half-time (t1/2) of ≤5 min. This binding could subsequently be reversed via the addition of unlabeled BMS-488043 with a t1/2 of ∼40 min (data not shown). When gp120 was coincubated with [3H]BMS-488043 and sCD4, the binding of [3H]BMS-488043 to gp120 was inhibited in an sCD4 concentration-dependent manner (Fig. 3B), while prebinding gp120 with sCD4 eliminated [3H]BMS-488043 binding (data not shown), agreeing with the aforementioned results from the ELISA studies. Furthermore, when gp120 monomer or envelope trimer covalently linked to a tritiated photoaffinity analog of BMS-488043 was treated with excess sCD4, the formation of a [3H]AI-gp120-sCD4 ternary complex could not be detected with a blue-native polyacrylamide gel electrophoresis gel shift assay (35, 36) with a detection sensitivity of <0.5 fmol complex (data not shown). Thus, AI and sCD4 do not appear to be able to bind gp120 simultaneously and prior binding of the AI is required for sCD4 inhibition.
FIG. 3.
Mutually exclusive binding of BMS-488043 and sCD4 to gp120JRFL. (A) Effect of the order of BMS-488043 and [125I]sCD4 addition. Symbols: +, BMS-488043 added to gp120 along with [125I]sCD4; ○, [125I]sCD4 added to gp120 before BMS-488043. gp120-[125I]sCD4 was bound to lectin-resin and counted in a gamma counter. (B) Blocking of [3H]BMS-488043 binding to gp120 by sCD4. gp120 was treated with a mixture of [3H]BMS-488043 and sCD4 and then subjected to gel filtration, and gp120-bound [3H] activity was determined in a scintillation counter.
Compound binding-induced conformational changes in gp120JRFL.
We propose that AI binding alters the conformation of gp120, explaining how a small molecule can effectively interfere with the extensive protein-protein interactions between viral gp120 and cellular CD4 receptors. To explore this possibility, three independent approaches were utilized.
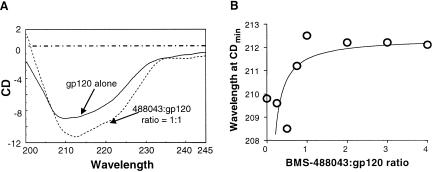
The first approach, using CD analysis (19), showed that BMS-488043 itself has no CD response in the far-UV (200- to 240-nm) range (data not shown) while properly folded gp120 shows a CD spectrum which is typical of a peptide with considerable α-helix character (3, 4, 19). By incrementally increasing the concentrations of BMS-488043, an AI-mediated shift in the gp120 CD spectrum was observed (Fig. 4A). The wavelength of the primary CD minimum of gp120 shifted from 209 nm to 212 nm as increasing amounts of BMS-488043 were added (Fig. 4B). The CD wavelength minimum shift was complete at a [gp120]/[BMS-488043] ratio of 1.0 (Fig. 4B). This observation is consistent with direct binding of BMS-488043 to gp120, and the resulting gp120 conformational changes occur at a 1:1 inhibitor-gp120 stoichiometry. The shift in the CD minimum wavelength from 209 to 212 nm and the accompanying small intensity increase from the unbound to the bound state likely reflect a relatively minor qualitative change in the secondary structures of the gp120 protein. Similar results were also noted for BMS-378806, and no sCD4 spectrum shift was induced by the compound alone (data not shown).
FIG. 4.
CD analysis of inhibitor-bound gp120JRFL. The CD spectra of purified gp120JRFL and that treated with BMS-488043 were scanned with a Jasco J-720 Spectropolarimeter. (A) CD spectra with (dashed line) and without (solid line) the presence of BMS-488043. (B) Effect of BMS-488043 concentration on the CDmin wavelength shift.
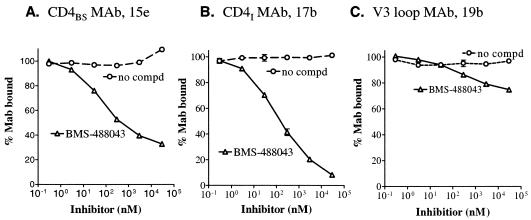
The second approach looked at the gp120 structure with conformation-sensitive gp120 MAbs that recognize the CD4 binding site or the CD4-induced epitopes. Interaction of the CD4 binding site MAb (CD4BS, 15e) (16) with gp120 was greatly reduced with increasing concentrations of BMS-488043 (Fig. 5A). Thus, the compound likely induced a conformational change affecting sCD4 binding. Binding of MAb 17b (targeting the CD4-induced CD4I epitopes) (39) to gp120 was also decreased by BMS-488043 in a dose-dependent fashion (Fig. 5B). The same also holds true for two other CD4I MAbs, 25e and 4.12d (5; data not shown). In contrast, sCD4 stimulates CD4I MAb binding to gp120 (39, 45). These results indicate that binding of BMS-488043 also mediates a gp120 conformational change affecting the CD4I epitopes. Furthermore, BMS-488043 inhibition activity against CD4I MAb binding to gp120 could not be observed if gp120 was preprimed with sCD4, agreeing with the notion that the binding of CD4 precludes the interaction of an AI with gp120.
FIG. 5.
Effect of BMS-488043 on gp120-MAb binding. Binding of a conformation-sensitive MAb to gp120JRFL was measured with a gp120-MAb binding ELISA with various concentrations of BMS-488043 added. (A) Binding of CD4BS to gp120. (B) Antibody binding to CD4-induced epitopes in the absence of sCD4. (C) Binding of V3 loop MAb to gp120. compd, compound.
The final approach probed gp120 conformations by using proteolytic susceptibility. The thrombin susceptibility of gp120 was employed to probe gp120 conformational changes in the V3 loop region (6, 37). A fivefold difference in V3 loop susceptibility to thrombin, between the sCD4- and inhibitor-bound gp120s (t1/2s, 0.5 and 2.5 h, respectively) was observed, indicating differential V3 loop conformations of the two ligand-bound gp120s. The rate of V3 loop cleavage in free gp120 (t1/2, 1.7 h) was between the two different ligand-bound gp120s. The moderate effect exerted by BMS-488043 on the V3 loop region (t1/2, 2.5 h versus 1.7 h for free gp120) agreed well with the weak influence of BMS-488043 on the V3 loop MAb binding. At inhibitor concentrations as high as 30 μM, binding of MAb 19b (28) and other V3 MAbs CO11 and 39F (Fig. 5C and unpublished results), to gp120 was also only marginally reduced.
Cumulatively, results obtained by all three approaches (CD spectral analysis, conformation-dependent MAb binding, and thrombin susceptibility) support the working hypothesis that AIs bind specifically to gp120, causing conformational changes that disrupt its binding to cellular CD4 receptors. The altered conformation is likely to span the CD4 and coreceptor binding regions and to impact downstream events in the viral entry process.
Blocking of virion binding to sCD4.
Since gp120 monomers and viral envelope trimers may interact differently with compounds, AIs were assayed for the ability to inhibit the binding of the envelope trimers to sCD4. HIV-1 LAI virions were treated with up to 3,000 nM BMS-488043, and the amount of virions captured by CD4-IgG was quantitated with a p24 ELISA. Results showed that BMS-488043 blocked virion binding to CD4-IgG with an IC50 of 0.05 μM, supporting the inhibition of envelope trimer-CD4 binding by compounds. Other structural analogs of BMS-488043 exhibited similar dose-dependent inhibition in this assay (data not shown). Comparable results were obtained when BMS-488043 was assayed against LAI, YU2, and JRFL virions captured by the anti-CD4 MAb OKT4 (Table 1) when coincubated with sCD4. In agreement with recombinant monomeric gp120-sCD4 binding studies (Fig. 2A), elevation of sCD4-IgG concentrations (1 → 5 μg/ml) also decreased the inhibitor potency (IC50, 22 → 50 nM) against virion-CD4-IgG binding, again suggesting a competitive mode of inhibition against virion envelope. Furthermore, the relative IC50s of a series of AIs tested in the virion-CD4-IgG binding assay are in alignment with the antiviral potencies of these compounds (data not shown), further supporting interruption of CD4-envelope interaction as the major target for AIs.
TABLE 1.
Inhibition of virion (WT versus ΔCT)-sCD4 binding by attachment inhibitors
| Ligand and virusb | IC50 (μM)a of:
|
|||
|---|---|---|---|---|
| BMS-488043
|
BMS-378806
|
|||
| WT | ΔCT | WT | ΔCT | |
| sCD4 | ||||
| LAI | 0.03 | NAc | NDd | NA |
| JRFL | 0.03 | >3.0 | 0.04 | >3.0 |
| YU2 | 0.3 | >3.0 | 0.3 | >3.0 |
| 17b/sCD4 | ||||
| LAI | 0.03 | NA | ND | NA |
| YU2 | 0.07 | >3.0 | 0.18 | >3.0 |
| C34/sCD4 | ||||
| LAI | 0.01 | NA | ND | NA |
| JRFL | NA | 0.3 | NA | ND |
| YU2 | 0.04 | 0.06 | ND | ND |
Virion binding to sCD4, MAb 17b (with sCD4 priming), and C34 (in the presence of sCD4) was assayed in the presence of various concentrations of inhibitors. Ligand-bound virions were captured and lysed, and p24 levels were determined with a p24 ELISA. Concentrations of inhibitors required to inhibit virion-ligand binding by 50% are reported.
LAI was a full-length molecular clone. JRFL and YU2 were virions pseudotyped with respective envelopes.
NA, not available due to lack of viable virion or lack of binding.
ND, not done.
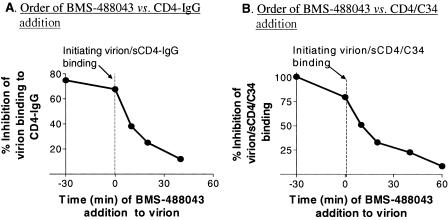
To prove that inhibition occurs prior to productive envelope-CD4 engagement, a time-of-addition study was performed. While maintaining a consistent time of virion exposure to sCD4-IgG (40 min), BMS-488043 (200 nM) was added at various times, either prior to or after initiation of virion incubation with sCD4-IgG. This experiment was possible due to the fast binding of the compound to gp120 (t1/2, <5 min). The results obtained showed that BMS-488043 was most effective at preventing virus binding to CD4-IgG when added prior to or at the beginning of the CD4-IgG exposure period (Fig. 6A). Adding BMS-488043 after the virion-sCD4-IgG incubation period reduced its inhibition activity in a time-dependent manner (Fig. 6A). These results, together with observations from the [125I]sCD4-gp120 binding study (Fig. 3A), indicate that BMS-488043 acts on the viral envelope protein prior to its productive CD4 engagement.
FIG. 6.
Dependence of BMS-488043 activity on the order in which ligands are added to virions. (A) Inhibition of HIV LAI virion binding to sCD4-IgG by BMS-488043. The inhibitor was added to the assay mixture at the time indicated, and the level of virion (p24) bound to sCD4-IgG was measured. Bound-virion levels were normalized to virion levels detected in the absence of the inhibitor. Initiation of virion sCD4-IgG binding is marked as time zero. (B) Inhibition of sCD4-induced virion-C34 binding. Virion was coincubated with sCD4 and C34 peptide (from time zero to 60 min) while the BMS-488043 addition time was varied. C34 peptide-bound virions were captured with MAb NC-1, and the p24 level was determined.
We next examined whether BMS-488043 binding also affects events that occur after CD4 binding, i.e., CD4-induced exposure of the gp41 region, as suggested by Si et al. CD4-induced exposure of the virion N-terminal heptad repeat groove was probed by C34 (gp41-CHR [C-terminal heptad repeat]) peptide binding, followed by virion capturing to the immobilized NC-1 MAb (recognizes the gp41 peptide N36-C34 complex) (18). The step at which the compound functions was investigated by changing the order of addition of the compound and sCD4. As shown in a representative result from multiple experiments (Fig. 6B), a time-dependent reduction of BMS-488043 inhibition of sCD4-induced HIV LAI virion binding to C34 peptide was evident. Preexposure of virions to inhibitor, relative to sCD4, gave the highest inhibition level. The efficacy of the inhibitor continued to drop as the sCD4-virion contact time was extended, prior to compound addition. This time-dependent reduction in compound activity is similar to that observed in the virion-CD4-IgG binding study (Fig. 6A) and clearly indicates that BMS-488043 acts on the pre-CD4 bound envelope to prevent subsequent CD4-gp120 interactions and the downstream events.
In a similar assay, under equilibrium binding conditions that allow the sCD4-mediated gp41 conformational change to take place (10, 11), inhibitor potency decreased with increasing sCD4 concentrations (IC50s were 10 and 22 nM for 5- and 20-μg/ml sCD4, respectively). Together with the virion-CD4-IgG binding results, these observations confirm that BMS-488043 competes with sCD4 for virion envelope binding.
Inhibition of mutant virions carrying a C-terminally deleted envelope.
Si et al. did not detect BMS-378806 inhibition of fluorescein isothiocyanate (FITC)-sCD4 binding to a cell surface-expressed envelope variant (ΔCT), containing a C-terminal deletion at amino acid 712 (38) but did observe the inhibition of HR2 (C34) binding to gp41. To reconcile this difference with our observations, we further investigated how this particular HIV variant with a significantly altered gp120 conformation would interact with AIs and impact their action.
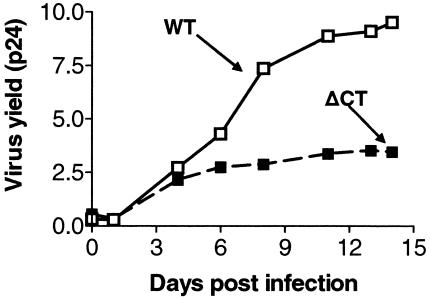
ΔCT HIV-1 YU-2 virions exhibit apparent growth defects (Fig. 7), suggesting that they may not be widely observed in patients. Nonetheless, pseudotyped viruses with the ΔCT envelope displayed viral entry activity and susceptibility to BMS-488043 with a 50% effective concentration in the low nanomolar range. Similar results were observed with another variant, ΔCT HIV JRFL. The fact that infection could not be detected with pseudotyped ΔCT HIV LAI further illustrates the detrimental effect of a C-terminal deletion on envelope function.
FIG. 7.
Growth kinetics of WT HIV-1 YU-2 versus those of the ΔCT envelope variant. After a 3-day stimulation with phytohemagglutinin, peripheral blood mononuclear cells were infected with WT or ΔCT virions at a multiplicity of infection of 0.005 and cultured for 10 days. Aliquots (200 μl) were collected daily, and the p24 level was determined to monitor the virus yield.
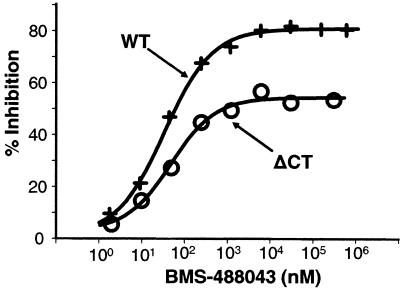
To gain insight into how AIs interact with these ΔCT envelope variants, we compared compound activities against virions pseudotyped with ΔCT envelope to those with wild-type (WT) envelopes. By using the sCD4-virion binding, sCD4-induced 17b-virion binding, and sCD4-induced C34-virion binding assays, we evaluated BMS-488043 inhibition of virion binding to sCD4, 17b (sCD4 induced), or C34 (gp41-CHR) peptide, respectively. Results showed that BMS-488043 was highly effective in preventing sCD4 binding to virions carrying WT envelope (LAI, JRFL, and YU2), with IC50s in the 30 to 300 nM range (Table 1). Much lower activity was detected with the ΔCT YU2 and ΔCT JRFL mutant viruses (Table 1), with <50% of virion binding inhibited even at 3 μM inhibitor. Similar results were obtained for BMS-378806. Decreased AI activities were also evident when examining sCD4-dependent MAb 17b binding to mutant YU2 virions (Table 1). The reduced compound inhibition of the gp120-CD4 interaction indicated that these conformationally altered gp120 variants interacted with AIs differently from full-length envelope. Nonetheless, BMS-488043 retained its potent inhibition of sCD4-induced C34 peptide binding to gp41 of ΔCT YU2 virions, yielding similar IC50s (0.04 and 0.06 μM, respectively) for both WT and ΔCT virions (Table 1). BMS-488043 also inhibited CD4-induced C34 binding to ΔCT JRFL. These data revealed an alternative mechanism to that of WT envelopes and suggested that upon inhibitor binding, the ΔCT virion envelope displays a conformation distinct from that of WT envelope.
To determine the basis of ΔCT virion inhibition, we studied the inhibition pattern of virion-sCD4 binding at high compound concentrations. Incomplete inhibition of ΔCT JRFL virion binding to sCD4 (plateau at ∼50% inhibition, Fig. 7) was evident even at 300 μM BMS-488043, unlike the nearly complete inhibition of WT virion binding. It appears that BMS-488043 may noncompetitively inhibit ΔCT envelope binding to sCD4 via an altered gp120 conformation. This distinct pattern of inhibition suggests that truncation of the cytoplasmic domain of the envelope protein results in a conformational structure with unique interactions among gp120, CD4, and the AIs. Together with the potent inhibition of C34 binding to sCD4-primed mutant virions, these results suggest that BMS-488043 binding to an envelope with a more triggered structure may actually result in conformational changes capable of simultaneously accommodating both compound and CD4 yet still prohibiting further conformational changes necessary for successful N-terminal heptad repeat-CHR interaction.
DISCUSSION
We have previously shown that AIs are potent and effective inhibitors of viral replication and that BMS-378806 blocks HIV entry by binding specifically to viral gp120 protein and preventing its interaction with cellular CD4 receptors (13, 23). In the present study, the AI BMS-488043 (an analog of BMS-378806) was further characterized to define its inhibition mechanism and to investigate the cause(s) for lack of CD4 inhibition observed by Si et. al (38).
Like BMS-378806, BMS-488043 binds to gp120 and potently inhibits the binding of monomeric gp120 to sCD4. This inhibition mechanism likely applies to the majority of susceptible HIV envelopes, since sCD4 binding to gp120 proteins of 11 independent HIV origins was susceptible to compound inhibition. Binding of AIs and sCD4 to gp120 is competitive, since under the equilibrium binding conditions employed in our assays, increasing inhibitor concentrations resulted in a decrease in the apparent binding affinity (KD) of sCD4 for gp120 without affecting the sCD4 binding capacity (Bmax) of gp120 (Fig. 2). These results demonstrate a classical pattern of competitive inhibition between an inhibitor and a ligand. Mutually exclusive binding of AIs and sCD4 to gp120 (Fig. 3) further supports the notion that the inhibition of sCD4 binding is competitive in nature, and order-of-addition studies showed that prior inhibitor-gp120 binding is required for sCD4 exclusion. However, the nonsymmetrical changes in the sCD4 KD and inhibitor IC50s are worth noting. A fourfold increase in the inhibitor concentration yielded an about twofold elevation in the KD of sCD4 binding to gp120 (Fig. 2B). On the contrary, a fivefold increase in the sCD4 concentration (0.1 to 0.5 μg/ml) resulted in an ∼20-fold increase in the IC50 for sCD4-gp120 binding (Fig. 2A). This differential effect on sCD4 and compound binding is likely due to the nonequivalent kinetics and dynamics of binding of the two ligands to gp120. Binding of sCD4 causes substantial gp120 conformational rearrangements, resulting in a large magnitude of binding thermodynamics (22, 30), while formation of a gp120-inhibitor complex involves relatively small conformational changes (reference 38 and this paper). Also in agreement, our studies showed that [125I]sCD4-gp120 binding is less readily reversible than [3H]BMS-488043-gp120 binding.
Due to the competitive nature of AI inhibition (Fig. 2 and 3), the small conformational impact of AIs, and the very high energetic change in gp120 upon sCD4 binding (30), excess sCD4 in an ELISA can be expected to mask the ability of AIs to block sCD4-gp120 or sCD4-induced CD4I-MAb-gp120 binding. The lack of sCD4 inhibition in the gp120-sCD4 binding assay observed by Si et al. (38) is likely due to the high concentration of sCD4 (N. Madani, personal communication, and reference 38) utilized in the assay. Importantly, BMS-488043 and its analogs also competitively blocked the binding of virions to sCD4 and prior BMS-488043-virion interactions were required for sCD4 exclusion (Fig. 6). Moreover, BMS-488043 blocked the sCD4-induced exposure of the gp41 groove in virions, but only when added prior to sCD4 (Fig. 6), indicating that sCD4 exclusion is the key step of inhibition in this assay. Collectively, experiments with both gp120 monomer and virion trimeric glycoproteins support the concept that the primary inhibitory mechanism of AIs is blocking the gp120-CD4 interaction and requires prior compound engagement with envelope. However, it seems unlikely that steric hindrance from AI binding alone could account for inhibition of gp120-CD4 binding, since the docking of envelope to CD4 was reported to culminate in a cascade of conformational changes in gp120 (4, 22, 30) which encompasses an 802-Å2 surface area of gp120 for receptor binding (22). By three independent approaches, the present study demonstrates that AI induced gp120 conformation changes, spanning both CD4 and coreceptor binding regions and beyond. These changes in the viral envelope protein likely inhibit acquisition of the key structure(s) required for CD4 binding and other entry functions.
Consistent with this hypothesis, the most relevant resistance substitutions (23, 25) selected by AIs also mapped to gp120 amino acids within the CD4 binding pocket (22). Furthermore, several secondary substitutions were located in CCR5 binding sites or the gp41 region, suggesting that resistance may result from interactions of various regions of the HIV envelope and implying that compound binding may have a wide range of allosteric effects. This latter result is not surprising if AIs confer their activity by structural modulations of the envelope. Importantly, a potential BMS-378806 binding location was recently suggested by mapping BMS-378806 resistance substitutions to a hydrophobic cavity on an X-ray crystal model of unliganded SIV gp120 (4). It is notable that many CD4 contact sites (22) were also mapped to the same region. These observations are consistent with the idea of AIs as competitive inhibitors of CD4 in gp120 binding. Together, resistance mapping, SIV gp120 structure modeling, and our biochemical data indicate that AIs and sCD4 likely share some binding determinants on the unliganded form of gp120, reinforcing the primary CD4 inhibition mechanism of this series of AIs.
Since AIs function through gp120 conformation changes, and cytoplasmic-tail-truncated (ΔCT) envelope variants, like that used by Si et al. (38), exhibited changed envelope conformations with enhanced exposure of CD4 and coreceptor binding regions (8, 9, 38), we examine if the lack of compound inhibition of FITC-CD4 binding to cell surface envelope (38) was due to the use of this variant envelope in their study. These altered envelope properties have also been linked to viruses displaying CD4 independence (8, 17). The higher cell surface expression of ΔCT envelopes (than the WT) seems necessary for the detection of FITC-CD4 binding (38) and makes a WT comparison study technically very challenging. Therefore, we relied on the virion binding assay for a direct comparison of AI function against full-length and ΔCT envelopes.
In studying the mechanism of inhibition of HIV-1 variants, virions pseudotyped with ΔCT envelope variants (including the one studied by Si et al. [38]) displayed a significantly diminished susceptibility to AI inhibition of sCD4-virion binding or sCD4-induced 17b-virion binding (Table 1). However, inhibition of C34 peptide binding to the sCD4-primed ΔCT virions was not affected (Table 1). These results are in agreement with the lack of BMS-378806 activity in blocking FITC-CD4 binding or FITC-CD4-induced 17b binding to the cell surface-expressed ΔCT YU2 envelope variant reported by Si et al. (38). The data are also consistent with the finding of Si et al. that AI inhibits C34-phycoerythrin binding to FITC-sCD4-treated ΔCT YU2 envelope (38). Ultimately, BMS-488043 appears to inhibit sCD4 binding to the ΔCT variants via a noncompetitive inhibition mechanism (Fig. 8), in contrast to the competitive inhibition observed for WT virions. It is conceivable that envelope variants with this unusual conformation can accommodate sCD4 after binding to AIs, which remain effective in disrupting the subsequent viral entry event(s). Moreover, we observed compound inhibition of CD4-independent HIV-1 infection when using the same construct (ADA S190R N197) as Si et al. (38). Since this CD4-independent HIV-1 also exhibited an altered envelope conformation with a more exposed coreceptor binding area (21), presumably compound inhibition occurred via alternative conformational changes.
FIG. 8.
BMS-488043 inhibition of WT versus ΔCT virion binding to sCD4. Virions (JRFL) were incubated with sCD4 (1 μg/ml) in the presence of various inhibitor concentrations. sCD4-bound virions were captured (anti-CD4 OKT4). The p24 level was determined and normalized to that of the no-inhibitor control.
In summary, our cumulative data support the idea that AIs prohibit HIV-1 infection via envelope conformational alterations and that they primarily interfere with cellular CD4 binding. Factors such as the concentration of sCD4 used in the gp120-sCD4 binding assay and the envelope conformation used in the study can clearly impact the experimental outcome. Under special circumstances, like those reported by Si et al., inhibition of viral entry may be accomplished without completely disrupting envelope-CD4 binding. Considering the heterogeneous nature of HIV envelopes, the extended effects exerted by the compound on various gp120 conformations may enhance the utility of these novel inhibitors in clinical settings. Investigation of this series of inhibitors may also further our understanding of the HIV-1 envelope structure and the viral entry process.
Acknowledgments
We sincerely thank Hwei-Gene Wang for critical reading, valuable suggestions, and contributions to manuscript preparation. We are also grateful to Shibo Jiang for providing the monoclonal antibody NC-1 and to Betsy Egger for her timely technical assistance.
REFERENCES
- 1.Bisson, G., R. Gross, V. Miller, I. Weller, A. Walker, P. Arlett, A. Carr, S. Evans, D. Graham, A. Justice, C. Kreft-Jais, J. D. Lundgren, B. Munk, J. Murray, M. Pirmohamed, D. Pizzuti, and A. Szarfman. 2003. Monitoring of long-term toxicities of HIV treatments: an international perspective. AIDS 17:2407-2417. [DOI] [PubMed] [Google Scholar]
- 2.Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93:681-684. [DOI] [PubMed] [Google Scholar]
- 3.Chang, D.-K., and S.-F. Cheng. 1996. Circular dichroism study on the secondary structural change induced by complex formation of a peptide derived from a CD4 binding site of HIV-1 envelope glycoprotein gp120 and a peptide from the N-terminal domain of CD4. Lett. Peptide Sci. 3:293-300. [Google Scholar]
- 4.Chen, B., E. M. Vogan, H. Gong, J. J. Skehel, D. C. Wiley, and S. C. Harrison. 2005. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature 433:834-841. [DOI] [PubMed] [Google Scholar]
- 5.Choe, H., W. Li, P. L. Wright, N. Vasilieva, M. Venturi, C. C. Huang, C. Grundner, T. Dorfman, M. B. Zwick, L. Wang, E. S. Rosenberg, P. D. Kwong, D. R. Burton, J. E. Robinson, J. G. Sodroski, and M. Farzan. 2003. Tyrosine sulfation of human antibodies contributes to recognition of the CCR5 binding region of HIV-1 gp120. Cell 114:161-170. [DOI] [PubMed] [Google Scholar]
- 6.Clements, G., M. Price-Jones, P. Stephens, C. Sutton, T. Schulz, P. Clapham, J. McKeating, M. McClure, S. Thomson, M. Marsh, J. Kay, R. Weiss, and J. Moore. 1991. The V3 loops of the HIV-1 and HIV-2 surface glycoproteins contain proteolytic cleavage sites: a possible function in viral fusion? AIDS Res. Hum. Retrovir. 7:3-16. [DOI] [PubMed] [Google Scholar]
- 7.Cooley, L. A., and S. R. Lewin. 2003. HIV-1 cell entry and advances in viral entry inhibitor therapy. J. Clin. Virol. 26:121-132. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, T. G., T. L. Hoffman, F. Baribaud, S. Wyss, C. C. LaBranche, J. Romano, J. Adkinson, M. Sharron, J. A. Hoxie, and R. W. Doms. 2001. Relationships between CD4 independence, neutralization sensitivity, and exposure of a CD4-induced epitope in a human immunodeficiency virus type 1 envelope protein. J. Virol. 75:5230-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards, T. G., S. Wyss, J. D. Reeves, S. Zolla-Pazner, J. A. Hoxie, R. W. Doms, and F. Baribaud. 2002. Truncation of the cytoplasmic domain induces exposure of conserved regions in the ectodomain of human immunodeficiency virus type 1 envelope protein. J. Virol. 76:2683-2691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gallo, S., A. Puri, and R. Blumenthal. 2001. HIV-1 gp41 six-helix bundle formation occurs rapidly after the engagement of gp120 by CXCR4 in the HIV-1 Env-mediated fusion process. Biochemistry 40:12231-12236. [DOI] [PubMed] [Google Scholar]
- 11.Gallo, S. A., G. M. Clore, J. M. Louis, C. A. Bewley, and R. Blumenthal. 2004. Temperature-dependent intermediates in HIV-1 envelope glycoprotein-Mediated fusion revealed by inhibitors that target N- and C-terminal helical regions of HIV-1 gp41. Biochemistry 43:8230-8233. [DOI] [PubMed] [Google Scholar]
- 12.Gulick, R. M. 2003. New antiviral drugs. Clin. Microbiol. Infect. 9:186-193. [DOI] [PubMed] [Google Scholar]
- 13.Guo, Q., H.-T. Ho, I. Dicker, L. Fan, N. Zhou, J. Friborg, T. Wang, B. V. McAuliffe, H.-G. H. Wang, R. E. Rose, H. Fang, H. T. Scarnati, D. R. Langley, N. A. Meanwell, R. Abraham, R. J. Colonno, and P.-F. Lin. 2003. Biochemical and genetic characterizations of a novel human immunodeficiency virus type 1 inhibitor that blocks gp120-CD4 interactions. J. Virol. 77:10528-10536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanna, G., J. Lalezari, J. Hellinger, D. Wohl, T. Masterson, W. Fiske, J. Kadow, P. Lin, M. Giordano, R. Colonno, and D. Grasela. 2004. Antiviral activity, safety, and tolerability of a novel, oral small-molecule HIV-1 attachment inhibitor, BMS-488043, in HIV-1-infected subjects, abstr. 141. 11th Conference on Retroviruses and Opportunistic Infections. http://www.retroconference.org/2004/home.htm.
- 15.Hanna, G., J.-H. Yan, W. Fiske, T. Masterson, D. Zhang, and D. Grasela. 2004. Safety, tolerability, and pharmacokinetics of a novel, small-molecule HIV-1 attachment inhibitor, BMS-488043, after single and multiple oral doses in healthy subjects, abstr. 535. 11th Conference on Retroviruses and Opportunistic Infections. http://www.retroconference.org/2004/home.htm.
- 16.Ho, D. D., J. A. McKeating, X. L. Li, T. Moudgil, E. S. Daar, N. C. Sun, and J. E. Robinson. 1991. Conformational epitope on gp120 important in CD4 binding and human immunodeficiency virus type 1 neutralization identified by a human monoclonal antibody. J. Virol. 65:489-493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffman, T. L., C. C. LaBranche, W. Zhang, G. Canziani, J. Robinson, I. Chaiken, J. A. Hoxie, and R. W. Doms. 1999. Stable exposure of the coreceptor-binding site in a CD4-independent HIV-1 envelope protein. Proc. Natl. Acad. Sci. USA 96:6359-6364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jiang, S., K. Lin, and M. Lu. 1998. A conformation-specific monoclonal antibody reacting with fusion-active gp41 from the human immunodeficiency virus type 1 envelope glycoprotein. J. Virol. 72:10213-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kelly, S. M., and N. C. Price. 2000. The use of circular dichroism in the investigation of protein structure and function. Curr. Protein Pept. Sci. 1:349-384. [DOI] [PubMed] [Google Scholar]
- 20.Kilby, J. M., and J. J. Eron. 2003. Novel therapies based on mechanisms of HIV-1 cell entry. N. Engl. J. Med. 348:2228-2238. [DOI] [PubMed] [Google Scholar]
- 21.Kolchinsky, P., E. Kiprilov, P. Bartley, R. Rubinstein, and J. Sodroski. 2001. Loss of a single N-linked glycan allows CD4-independent human immunodeficiency virus type 1 infection by altering the position of the gp120 V1/V2 variable loops. J. Virol. 75:3435-3443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kwong, P. D., R. Wyatt, J. Robinson, R. W. Sweet, J. Sodroski, and W. A. Hendrickson. 1998. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature 393:648-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin, P.-F., W. Blair, T. Wang, T. Spicer, Q. Guo, N. Zhou, Y.-F. Gong, H.-G. H. Wang, R. Rose, G. Yamanaka, B. Robinson, C.-B. Li, R. Fridell, C. Deminie, G. Demers, Z. Yang, L. Zadjura, N. Meanwell, and R. Colonno. 2003. A small molecule HIV-1 inhibitor that targets the HIV-1 envelope and inhibits CD4 receptor binding. Proc. Natl. Acad. Sci. USA 100:11013-11018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little, S. J., S. Holte, J.-P. Routy, E. Daar, M. Markowitz, A. C. Collier, R. A. Koup, J. W. Mellors, E. Connick, B. Conway, M. Kilby, L. Wang, J. M. Whitcomb, N. S. Hellmann, and D. D. Richman. 2002. Antiretroviral-drug resistance among patients recently infected with HIV. N. Engl. J. Med. 347:385-394. [DOI] [PubMed] [Google Scholar]
- 25.Madani, N., A. L. Perdigoto, K. Srinivasan, J. M. Cox, J. J. Chruma, J. LaLonde, M. Head, A. B. Smith III, and J. G. Sodroski. 2004. Localized changes in the gp120 envelope glycoprotein confer resistance to human immunodeficiency virus entry inhibitors BMS-806 and #155. J. Virol. 78:3742-3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marks, K., and R. M. Gulick. 2004. New antiretroviral agents for the treatment of HIV infection. Curr. Infect. Dis. Rep. 6:333-339. [DOI] [PubMed] [Google Scholar]
- 27.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moore, J. P., F. E. McCutchan, S. W. Poon, J. Mascola, J. Liu, Y. Cao, and D. D. Ho. 1994. Exploration of antigenic variation in gp120 from clades A through F of human immunodeficiency virus type 1 by using monoclonal antibodies. J. Virol. 68:8350-8364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moore, J. P., and R. J. Shattock. 2003. Preventing HIV-1 sexual transmission—not sexy enough science, or no benefit to the bottom line? J. Antimicrob. Chemother. 52:890-892. [DOI] [PubMed] [Google Scholar]
- 30.Myszka, D. G., R. W. Sweet, P. Hensley, M. Brigham-Burke, P. D. Kwong, W. A. Hendrickson, R. Wyatt, J. Sodroski, and M. L. Doyle. 2000. Energetics of the HIV gp120-CD4 binding reaction. Proc. Natl. Acad. Sci. USA 97:9026-9031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomerantz, R. J., and D. L. Horn. 2003. Twenty years of therapy for HIV-1 infection. Nat. Med. 9:867-873. [DOI] [PubMed] [Google Scholar]
- 32.Reiss, P. 2003. How bad is HAART for the HEART? AIDS 17:2529-2531. [DOI] [PubMed] [Google Scholar]
- 33.Richman, D. D. 2001. HIV chemotherapy. Nature 410:995-1001. [DOI] [PubMed] [Google Scholar]
- 34.Sattentau, Q. J., and J. P. Moore. 1993. The role of CD4 in HIV binding and entry. Philos. Trans. R. Soc. Lond. B Biol. Sci. 342:59-66. [DOI] [PubMed] [Google Scholar]
- 35.Schagger, H., W. A. Cramer, and G. von Jagow. 1994. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal. Biochem. 217:220-230. [DOI] [PubMed] [Google Scholar]
- 36.Schulke, N., M. S. Vesanen, R. W. Sanders, P. Zhu, M. Lu, D. J. Anselma, A. R. Villa, P. W. Parren, J. M. Binley, K. H. Roux, P. J. Maddon, J. P. Moore, and W. C. Olson. 2002. Oligomeric and conformational properties of a proteolytically mature, disulfide-stabilized human immunodeficiency virus type 1 gp140 envelope glycoprotein. J. Virol. 76:7760-7776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz, T. F., J. D. Reeves, J. G. Hoad, C. Tailor, P. Stephens, G. Clements, S. Ortlepp, K. A. Page, J. P. Moore, and R. A. Weiss. 1993. Effect of mutations in the V3 loop of HIV-1 gp120 on infectivity and susceptibility to proteolytic cleavage. AIDS Res. Hum. Retrovir. 9:159-166. [DOI] [PubMed] [Google Scholar]
- 38.Si, Z., N. Madani, J. M. Cox, J. J. Chruma, J. C. Klein, A. Schon, N. Phan, L. Wang, A. C. Biorn, S. Cocklin, I. Chaiken, E. Freire, A. B. Smith III, and J. G. Sodroski. 2004. Small-molecule inhibitors of HIV-1 entry block receptor-induced conformational changes in the viral envelope glycoproteins. Proc. Natl. Acad. Sci. USA 101:5036-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thali, M., J. P. Moore, C. Furman, M. Charles, D. D. Ho, J. Robinson, and J. Sodroski. 1993. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J. Virol. 67:3978-3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ugolini, S., I. Mondor, and Q. J. Sattentau. 1999. HIV-1 attachment: another look. Trends Microbiol. 7:144-149. [DOI] [PubMed] [Google Scholar]
- 41.Volberding, P. 1999. Advances in the medical management of patients with HIV-1 infection: an overview. AIDS 13:S1-S9. [PubMed] [Google Scholar]
- 42.Wang, T., O. B. Wallace, Z. Zhang, N. A. Meanwell, and J. A. Bender. May2005. Antiviral azaindole derivatives. U.S. patent 6, 900,323 B2.
- 43.Wang, T., Z. Zhang, O. B. Wallace, M. Deshpande, H. Fang, Z. Yang, L. M. Zadjura, D. L. Tweedie, S. Huang, F. Zhao, S. Ranadive, B. S. Robinson, Y.-F. Gong, K. Ricarrdi, T. P. Spicer, C. Deminie, R. Rose, H. G. H. Wang, W. S. Blair, P.-Y. Shi, P. F. Lin, R. J. Colonno, and N. A. Meanwell. 2003. Discovery of 4-benzoyl-1-[(4-methoxy-1H-pyrrolo[2,3-b]pyridin-3-yl)oxoacetyl]-2-(R)-methylpiperazine (BMS-378806): a novel HIV-1 attachment inhibitor that interferes with CD4-gp120 interactions. J. Med. Chem. 46:4236-4239. [DOI] [PubMed] [Google Scholar]
- 44.Wegner, S. A., S. K. Brodine, J. R. Mascola, S. A. Tasker, R. A. Shaffer, M. J. Starkey, A. Barile, G. J. Martin, N. Aronson, W. W. Emmons, K. Stephan, S. Bloor, J. Vingerhoets, K. Hertogs, and B. Larder. 2000. Prevalence of genotypic and phenotypic resistance to anti-retroviral drugs in a cohort of therapy-naive HIV-1 infected US military personnel. AIDS 14:1009-1015. [DOI] [PubMed] [Google Scholar]
- 45.Wyatt, R., J. Moore, M. Accola, E. Desjardin, J. Robinson, and J. Sodroski. 1995. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J. Virol. 69:5723-5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wyatt, R., and J. Sodroski. 1998. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science 280:1884-1888. [DOI] [PubMed] [Google Scholar]