Abstract
Induction of mucosal immunity may be important for preventing SARS-CoV infections. For safe and effective delivery of viral antigens to the mucosal immune system, we have developed a novel surface antigen display system for lactic acid bacteria using the poly-γ-glutamic acid synthetase A protein (PgsA) of Bacillus subtilis as an anchoring matrix. Recombinant fusion proteins comprised of PgsA and the Spike (S) protein segments SA (residues 2 to 114) and SB (residues 264 to 596) were stably expressed in Lactobacillus casei. Surface localization of the fusion protein was verified by cellular fractionation analyses, immunofluorescence microscopy, and flow cytometry. Oral and nasal inoculations of recombinant L. casei into mice resulted in high levels of serum immunoglobulin G (IgG) and mucosal IgA, as demonstrated by enzyme-linked immunosorbent assays using S protein peptides. More importantly, these antibodies exhibited potent neutralizing activities against severe acute respiratory syndrome (SARS) pseudoviruses. Orally immunized mice mounted a greater neutralizing-antibody response than those immunized intranasally. Three new neutralizing epitopes were identified on the S protein using a peptide neutralization interference assay (residues 291 to 308, 520 to 529, and 564 to 581). These results indicate that mucosal immunization with recombinant L. casei expressing SARS-associated coronavirus S protein on its surface provides an effective means for eliciting protective immune response against the virus.
Severe acute respiratory syndrome (SARS) recently emerged as a zoonotic infectious disease with high morbidity and mortality. The causative agent, SARS-associated coronavirus (SARS-CoV), belongs to the Coronaviridae, a family of enveloped viruses containing a single-stranded RNA genome ranging from 27 to 32 kb in length (22). SARS-CoV is transmitted through the mucosal surfaces of the upper respiratory or gastrointestinal tracts, leading to pneumonia and enteritis in humans, with a limited occurrence of systemic diseases (14, 18, 19, 27). Recent studies have identified the spike (S) protein as a major target for vaccine development because of its ability to induce neutralizing antibodies (8, 10, 31, 33, 36). Several vaccine strategies have been examined for the prevention of SARS infection, including whole inactivated virus, DNA vaccine, and recombinant viral vectors based on adenovirus, vaccinia virus, and parainfluenza virus (3, 4, 11, 20, 35).
Studies of the development of animal coronavirus vaccines have demonstrated that systemic humoral or cell-mediated immune responses induced by parenteral administration may not be enough to prevent infection in mucosal areas (17). The prominent role of the mucosa in SARS transmission and infection suggests that direct mucosal immunization could be an effective strategy for prophylaxis by induction of systemic and mucosal immune responses. For mucosal immunization, lactic acid bacteria (LAB) are more attractive as delivery vehicles than other live-vaccine vectors (e.g., Shigella, Salmonella, and Listeria) (6, 12, 24, 25) because LAB are considered safe, they exhibit adjuvant properties, and they are weakly immunogenic (16, 21, 23, 26, 34). In addition, extracellularly accessible antigens expressed on the surfaces of bacteria are better recognized by the immune system than those that are intracellular (12).
For surface display of antigens on LAB, we have developed a novel expression vector using the pgsA gene product as an anchoring matrix. PgsA is a transmembrane protein derived from the poly-γ-glutamic acid synthetase complex (the PgsBCA system) of Bacillus subtilis (1, 2). In this study, we describe the expression of two segments of SARS-CoV S protein fused to PgsA that are displayed on the surface of Lactobacillus casei. Intranasal and oral vaccinations of mice with the recombinant L. casei elicited systemic and mucosal immune responses that had potent neutralizing activities against the SARS pseudovirus. The results of this study suggest a potential use for our surface expression system against other pathogens that are transmitted mucosally.
MATERIALS AND METHODS
Bacterial strains, cloning, and construction of surface display plasmids.
Escherichia coli JM83 and L. casei BLS-S8 were used. Plasmid pPGS1, harboring the pgsBCA genes (GenBank accession no. AB016245) of B. subtilis, was used to construct a minimal surface expression vector. The published sequence of SARS-CoV S protein (Tor2 strain; GenBank accession no. AY274119) was used to chemically synthesize codon-optimized DNA fragments encoding the SA (amino acids [aa] 2 to 114) and SB (aa 264 to 596) polypeptides. The DNA fragments were cloned into the pGEM T vector to generate pGT-SA and pGT-SB, respectively.
To generate the surface display vector pHAT:pgsA, which contains pgsA under the control of the high constitutive expression constitutive promoter, the pgsA gene was PCR amplified from pPGS1 using primers 5′-CAT ATG AAA AAA GAA CTG AGC-3′ and 5′-GGA TCC AGA TTT TAG TTT GTC-3′. The amplified DNA fragment was cloned into pHCEIIB (TAKARA) to generate pHCEIIB-pgsA. Subsequently, the fragment containing the HCE promoter, pgsA gene, multicloning site, and transcriptional terminator (rrnBT1T2) was excised from pHCEIIB-pgsA and cloned into pAT19 (29) to generate pHAT:pgsA. To generate constructs fused to the S protein, the SA and SB segments were PCR amplified from pGT-SA and pGT-SB using primer pairs 5′-CGC GGA TCC TTT ATT TTC TTA TTA-3′ plus 5′-CGG GGT ACC TTA CAC AGA CTG TGA CTT-3′ and 5′-CGC GGA TCC CTC AAG TAT GAT GAA AAT-3′ plus 5′-CGG GGT ACC TTA AAC AGC AAC TTC AGA-3′, respectively. The PCR-amplified fragments were inserted into BamHI/KpnI-digested pHAT:pgsA to yield pHAT:pgsA-SA and pHAT:pgsA-SB, respectively.
Cell wall fractionation, immunoblotting, flow cytometry, and immunofluorescence microscopy.
Recombinant L. casei BLS-S8 cells were grown at 30°C, and cell fractionations and protein extractions were performed as previously described (5). For immunodetection of fusion proteins, rabbit anti-PgsA (1:1,000) or rabbit anti-SARS S (1:1,000; ABGENT, San Diego, CA) polyclonal antibodies were used. Biotin-conjugated anti-rabbit immunoglobulin G (IgG) was used as a secondary antibody, which was visualized with avidin-biotin solution (Vector Laboratories, Burlingame, CA), substrate solution (containing 30 mg diaminobenzidine, 100 mM Tris-HCl, 4 mM NiCl, pH 7.5), and 30% hydrogen peroxide (H2O2). For immunofluorescence staining, L. casei BLS-S8 cells were cultured in MRS broth (Difco) overnight at 30°C. The cell pellets were sequentially incubated with rabbit anti-SARS S polyclonal antibodies (1:1,000), biotin-conjugated anti-rabbit IgG secondary antibodies (1:1,000; Sigma, St. Louis, MO), and fluorescein-conjugated streptavidin (Vector Laboratories, Burlingame, CA). Finally, 30,000 cells were analyzed with FACSCalibur (Becton Dickinson, Oxnard, CA) equipped with CellQuest software. For immunofluorescence microscopy, cells labeled with anti-SARS S polyclonal antibodies and fluorescein isothiocyanate-conjugated anti-rabbit antibodies were examined using a Carl Zeiss Axioskop 2 fluorescence microscope. Photographs were taken with an Axiocam high-resolution camera.
Immunizations and sample collection.
Groups of 12 C57BL/6 mice were immunized either orally or intranasally with equal mixtures of live L. casei BLS-S8 that express recombinant SARS proteins from plasmids pHAT:pgsA-SA and pHAT:pgsA-SB. L. casei BLS-S8 harboring the parental plasmid, pHAT:pgsA, was used as a negative control. For the oral route, 5 × 109 L. casei BLS-S8 cells in 100 μl suspension were administered daily via intragastric lavage on days 0 to 4, 7 to 11, 21 to 25, and 49 to 53. For the intranasal route, 2 × 109 L. casei BLS-S8 cells in 20 μl suspension were administered into the nostrils of lightly anesthetized mice on days 0 to 2, 7 to 9, 21, and 49. Blood samples were collected from the tail vein on days 0 (preimmune), 14, 28, 42, 56, 70, and 84. Sera were prepared from the blood and stored at −20°C until they were analyzed.
To purify IgG from serum samples, a NAb Protein G Spin Purification Kit (PIERCE) was used according to the manufacturer's protocol. Briefly, 100 to 200 μl of serum sample was added to a Handee Spin Cup Column containing ImmunoPure Immobilized Protein G Plus gel slurry. After 30 min of incubation at room temperature with shaking, the column was washed and IgG was eluted. The purified IgG was stored at −20°C until it was analyzed. To obtain bronchoalveolar and intestinal lavage samples, mice were killed on day 70 or 84. Bronchoalveolar and intestinal lavage fluids were obtained by washing the respective organs three times with 0.5 ml of ice-cold saline containing protease inhibitors. Samples were centrifuged at 2,500 × g for 20 min at 4°C, and the supernatants were stored at −20°C until they were analyzed.
ELISA.
Enzyme-linked immunosorbent assay (ELISA) plates were coated overnight at 4°C with 600 ng/well of pooled synthetic peptides (Table 1) in carbonate buffer (pH 9.6). After the wells were blocked for 1 h at room temperature with 2% bovine serum albumin (Sigma), serially diluted serum samples were added in duplicate and incubated for 1 h at 37°C. Preimmune sera (1:50 dilution) were used as negative controls. After the plates were washed five times with phosphate-buffered saline (PBS) containing 0.05% Tween 20, peroxidase-conjugated rabbit anti-mouse IgG antibody (Sigma) was added to each well (1:5,000) and incubated for an additional 1 h at 37°C. Following another round of washing, substrate solution containing tetramethylbenzidine and H2O2 was added. The reaction was allowed to proceed for 30 min at room temperature before it was terminated by adding stop solution (H2SO4). The optical density was measured at 450 nm using an ELISA autoreader (Molecular Devices). ELISA was performed three times for each serum sample. End point titers were defined as the maximum dilutions giving an A450 measurement of 0.1. This cutoff value represents the mean optical density plus 2 standard deviations of 10 normal mouse serum samples tested at 1:50 dilution. Statistical comparison was made by the Mann-Whitney U test.
TABLE 1.
SARS spike peptide pool used for antigen coating mixture
| Peptide | Residues | Sequence |
|---|---|---|
| SARS-SA/P1 | 17-36 | DRCTTFDDVQAPNYTQHTSS |
| SARS-SA/P2 | 33-52 | HTSSMRGVYYPDEIFRSDTL |
| SARS-SA/P3 | 91-114 | ATEKSNVVRGSTMNNKSQSV |
| SARS-SB/P4 | 285-302 | ELKCSVKSFEIDKGIYQT |
| SARS-SB/P5 | 510-529 | VCGPKLSTDLIKNQCVNFNF |
| SARS-SB/P6 | 550-569 | QFGRDVSDFTDSVRDPKTSE |
To determine the isotype profiles of antigen-specific antibodies, an ImmunoPure Isotyping Kit I was used according to the manufacturer's protocol {horseradish peroxidase-[2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid)]; Pierce}. Briefly, the general procedure described above was used, except that subclass-specific rabbit anti-mouse immunoglobulins (IgG1, IgG2a, IgG2b, IgG3, IgA, and IgM) were used as secondary antibodies (1:3,000). Subsequently, peroxidase-conjugated goat anti-rabbit IgG antibody was used (1 h at 37°C). For determining antigen-specific IgA levels in the mucosa, bronchoalveolar and intestinal lavage fluids (diluted 1:5 in PBS containing 1% bovine serum albumin) were added to peptide-coated wells in duplicate. Following 1 h of incubation at 37°C, the plates were washed with PBS containing 0.05% Tween 20 and IgA was detected using peroxidase-conjugated goat anti-mouse IgA antibody (Sigma; 1:5,000).
SARS pseudovirus neutralization assay.
SARS pseudovirus production, infection, and neutralization assays were performed as previously described (9). Briefly, pHCMV-S plasmid encoding SARS-CoV S protein was transfected into TELCeB6 cells, which constitutively produce Moloney murine leukemia virus (Mo-MuLV) particles. Lipofectin (Invitrogen; 30 μl) was used to transfect 30 μg of DNA into cells in a T25 flask. After 3 days, the culture medium was collected and cell debris was removed by centrifugation. The resulting supernatant was aliquoted and stored at −80°C. For SARS pseudovirus infection and neutralization assays, 100 infectious units of virus was incubated in the presence or in the absence of antibodies for 1 h at 37°C. Subsequently, antibody-virus mixtures were added to Vero E6 cells in 96-well plates, and the infection was allowed to proceed for about 1.5 days. The cells were fixed with 1% formaldehyde and 0.05% glutaraldehyde in PBS. Infected cells were stained for β-galactosidase activity (5 mM potassium ferricyanide, 5 mM potassium ferrocyanide, 2 mM magnesium chloride, and 1 mg/ml X-Gal [5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside] in PBS) and quantified.
To identify neutralization epitopes, 45 overlapping peptides derived from the SARS-CoV S glycoprotein (aa 257 to 598) were used (Table 2). The peptides were obtained from the NIH Biodefense and Emerging Infections Research Resources Repository (Bethesda, MD). They ranged in size from 15 to 18 amino acids, with a 10-amino-acid overlap. The peptides were initially dissolved (5 mM stock) in PBS containing either 5 or 10% dimethyl sulfoxide. Antibodies were incubated in the presence or in the absence of peptides (100 μM) for 1 h at 37°C. Subsequently, 100 infectious units of SARS pseudovirus was added to each antibody-peptide mixture and incubated for 1 h. The antibody-peptide-pseudovirus mixtures were then added to Vero E6 cells, and infections were monitored as described above.
TABLE 2.
SARS spike protein peptides useda
| Mixture 1
|
Mixture 2
|
Mixture 3
|
Mixture 4
|
||||
|---|---|---|---|---|---|---|---|
| Residues | Sequence | Residues | Sequence | Residues | Sequence | Residues | Sequence |
| 257-272 | LKPTTFMLKYDENGTI | 343-360 | KKISNCVADYSVLYNSTF | 427-444 | NIDATSTGNYNYKYRYLR | 505-521 | NAPATVCGPKLSTDLIK |
| 263-280 | MLKYDENGTTTDAVDCSQ | 351-367 | DYSVLYNSTFFSTFKCY | 435-451 | NYNYKYRYLRHGKLRPF | 512-529 | GPKLSTDLIKNQCVNFNF |
| 271-287 | TITDAVDCSQNPLAELK | 358-374 | STFFSTFKCYGVSATKL | 442-458 | YLRHGKLRPFERDISNV | 520-537 | IKNQCVNFNFNGLTGTGV |
| 278-295 | CSQNPLAELKCSVKSFEI | 365-382 | KCYGVSATKLNDLCFSNV | 449-465 | RPFERDISNVPFSPDGK | 528-545 | NFNGLTGTGVLTPSSKRF |
| 286-300 | LKCSVKSFEIDKGIY | 373-390 | KLNDLCFSNVYADSFVVK | 456-472 | SNVPFSPDGKPCTPPAL | 536-553 | GVLTPSSKRFQPFQQFGR |
| 291-308 | KSFEIDKGIYQTSNFRVV | 381-398 | NVYADSFVVKGDDVRQI | 463-478 | DGKPCTPPALNCYWPL | 544-558 | RFQPFQQFGRDVSDF |
| 299-316 | IYQTSNFRVVPSGDVVRF | 389-406 | VKGDDVRQIAPGQTGVIA | 469-484 | PPALNCYWPLNDYGFY | 549-566 | QQFGRDVSDFTDSVRDPK |
| 307-322 | VVPSGDVVRFPNITNL | 397-412 | IAPGQTGVIADYNYKL | 476-491 | WPLNDYGFYTTTGIGY | 557-573 | DFTDSVRDPKTSEILDI |
| 313-329 | VVRFPNITNLCPFGEVF | 403-420 | GVIADYNYKLPDDFMGCV | 482-499 | GFYTTTGIGYQPYRVVVL | 564-581 | DPKTSEILDISPCSFGGV |
| 320-337 | TNLCPFGEVFNATKFPSV | 411-428 | KLPDDFMGCVLAWNTRNI | 490-506 | GYQPYRVVVLSFELLNA | 572-590 | DISPCSFGGVSVITPGTNA |
| 328-345 | VFNATKFPSVYAWERKKI | 419-436 | CVLAWNTRNIDATSTGNY | 497-514 | VVLSFELLNAPATVCGPK | 581-598 | VSVITPGTNASSEVAVLY |
| 336-352 | SVYAWERKKISNGVADY | ||||||
Boldface type indicates peptides which completely inhibited neutralizing activity.
RESULTS
Construction of a novel surface display vector and expression of SARS viral-antigen hybrids on L. casei.
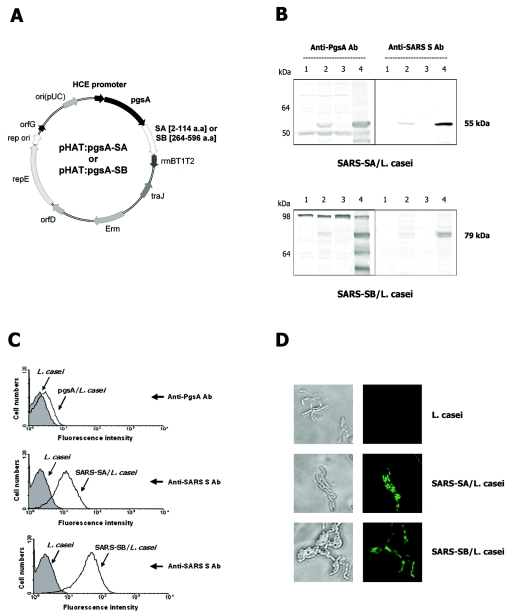
We generated expression vectors encoding PgsA fused to two fragments of the SARS-CoV S protein, SA and SB, containing amino acid residues 2 to 114 and 264 to 596, respectively (pHAT:pgsA-SA and pHAT:pgsA-SB) (Fig. 1A). These sequences include the B-cell epitopes and the receptor-binding domain (RBD) located at the N terminus of the S protein (33). E. coli harboring the plasmids was grown overnight at 37°C. The cells were harvested, and expression of the expected chimeric proteins was confirmed by immunoblotting using anti-PgsA and anti-SARS S polyclonal antibodies (data not shown). L. casei cells were then transformed with the plasmids and cultured at 30°C. Expression of the PgsA-SA and PgsA-SB fusion proteins was monitored by immunoblotting whole-cell lysates of serially passaged recombinant L. casei (Fig. 1B, lane 2). Both PgsA-SA and PgsA-SB were stably expressed through more than 10 serial passages and maintained their predicted molecular masses (55 kDa and 79 kDa, respectively).
FIG. 1.
Construction and expression of chimeric S proteins. (A) A schematic diagram of pHAT:pgsA-SA and pHAT:pgsA-SB. (B) Western blot analyses of PgsA-SA (top) and PgsA-SB (bottom) expression in L. casei using anti-pgsA (left) and anti-SARS S (right) polyclonal antibodies. Lanes 1 and 2 show whole-cell lysates of wild-type (parental vector) and recombinant L. casei, respectively. Lanes 3 and 4 show cytoplasmic and membrane fractions of recombinant L. casei, respectively. Protein bands of ∼55 and 79 kDa, corresponding to the expected sizes of PgsA-SA and PgsA-SB, respectively, were detected in lanes 2 and 4. (C) Fluorescence-activated cell sorter histograms of wild-type (filled) and recombinant (open) L. casei cells. The cells were probed with either polyclonal rabbit anti-PgsA (top) or anti-SARS S (middle and bottom) polyclonal antibodies, followed by biotin-conjugated anti-rabbit IgG antibody and fluorescein isothiocyanate-conjugated streptavidin. (D) Representative immunofluorescence images of wild-type (control) and recombinant L. casei cells expressing PgsA-SA and PgsA-SB. Bright-field images are shown on the left.
To determine cellular localization of the proteins, membrane and cytoplasmic fractions of L. casei cells were subjected to Western immunoblotting. As expected, both PgsA-SA and PgsA-SB fusion proteins were detected in the membrane, but not the cytoplasmic fraction (Fig. 1B, lanes 4 and 3, respectively). Immunoreactive bands of lower molecular mass were also detected using anti-PgsA antibodies, but not with anti-SARS S antibodies (Fig. 1B, lane 4). Although the exact identities of these bands have not been determined, they are presumed to be degradation products of chimeric proteins (e.g., N-terminal protein fragments that do not contain SA or SB regions), which might have been generated during the membrane fractionation procedure.
The localization of the fusion S proteins on the surface of L. casei was further verified by flow cytometry and fluorescence microscopy (Fig. 1C and D, respectively). Flow cytometry analyses using rabbit anti-PgsA and anti-SARS S polyclonal antibodies showed significant increase of fluorescence in cells harboring pHAT:pgsA-SA and pHAT:pgsA-SB compared to that in control cells harboring the parental pHAT:pgsA plasmid (Fig. 1C). Analyses by immunofluorescence microscopy also revealed fluorescence only in the recombinant bacteria harboring pHAT:pgsA-SA and pHAT:pgsA-SB, but not in control cells harboring pHAT:pgsA (Fig. 1D). These results showed that SARS S protein fragments were properly and efficiently displayed on the cell surface of L. casei using PgsA as a membrane-anchored protein display motif.
Systemic and mucosal immunogenicities of hybrid proteins expressed on L. casei.
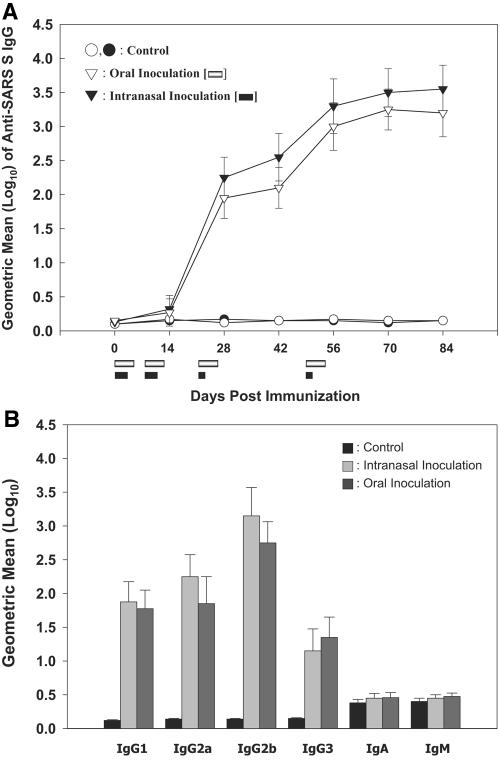
C57BL/6 mice were immunized nasally or orally with L. casei expressing the SA or SB protein fragment on the cell surface. As negative controls, mice were immunized with isogenic cells harboring the pHAT:pgsA plasmid. For oral immunization, the animals were inoculated daily on days 0 to 4, 7 to 11, 21 to 25, and 49 to 53. A lighter schedule was employed for nasal immunization (days 0 to 2, 7 to 9, 21, and 49). Serum samples were collected at 2-week intervals and analyzed by ELISA using six peptides derived from the SA and SB fragments of the S protein (Table 1). During the first two series of immunizations, very low levels of antibodies were detected (Fig. 2, left). However, high antibody levels were detected shortly after the third immunization (day 28; P < 0.01). Further increase in antibody titer was observed after the fourth immunization (day 56). A similar antibody response pattern was observed for intranasal and oral immunizations. At the end of immunization, the mean serum IgG titers in both experimental groups were over 1,000 times higher than those in the control groups. There were no significant differences in the antibody titers between the oral- and intranasal-inoculation groups.
FIG. 2.
Systemic anti-SARS S protein response following mucosal immunization. Groups of 12 mice were immunized orally (open) or intranasally (closed) with either wild-type (circles) or a mixture of recombinant (inverted triangles) L. casei. (Left) Kinetics of anti-SARS S serum IgG response. ELISA was performed in triplicate using pooled peptides shown in Table 1, and titers are defined as the reciprocal of the maximum dilution of sera that yielded an absorbance equal to that of preimmune samples. Immunization periods are indicated by bars below the x axis. (Right) Isotype profiles of anti-SARS S serum antibodies. Serum samples collected on day 84 postimmunization were used. End point titers were calculated as the reciprocals of serum dilutions yielding the same optical density as a 1/50 dilution of a pooled preimmune serum. The data are presented as means ± standard deviations. Statistical comparisons between groups were made by the Mann-Whitney U test.
To better characterize antibody responses against S protein fragments, the levels of antigen-specific IgG subclasses (IgG1, IgG2a, IgG2b, and IgG3) and other antibody isotypes (IgA and IgM) were assessed by ELISA. Pooled immune sera collected 84 days after the first inoculation were used. Both intranasally and orally immunized mice developed S protein-specific antibodies that were predominantly IgG2b, with moderate levels of IgG2a and IgG1 (Fig. 2, right). The subclass ratio, calculated as IgG1/(IgG2a + IgG2b), was less than 0.4, indicating a Th1-type response. The mean titers of these subtypes were significantly different from the baseline titers in the control group (P < 0.01). In contrast, no significant differences were observed for IgA and IgM isotypes.
To assess mucosal immune responses, S protein-specific IgA levels in intestinal and bronchoalveolar lavage fluids were determined by ELISA. Fluids collected on days 70 and 84 after the first inoculation were examined using a mixture of six peptides as coating antigens (Table 1). Both intranasal and oral immunizations elicited S protein-specific mucosal IgA responses at the site of inoculation, as well as the remote mucosal site (Fig. 3). Not surprisingly, greater antibody titers were detected at the locality of immunization. In contrast, only background levels of antibodies were detected in control animals.
FIG. 3.
Anti-SARS mucosal IgA antibody responses. Intestinal and bronchoalveolar lavage fluids, harvested from mice sacrificed 70 or 84 days postimmunization, were analyzed by ELISA in triplicate. Optical densities (OD) of samples from animals immunized orally (A) or intranasally (B) are shown. Fluids from control animals are shown as black bars. The error bars represent standard deviations.
SARS-CoV-neutralizing activity.
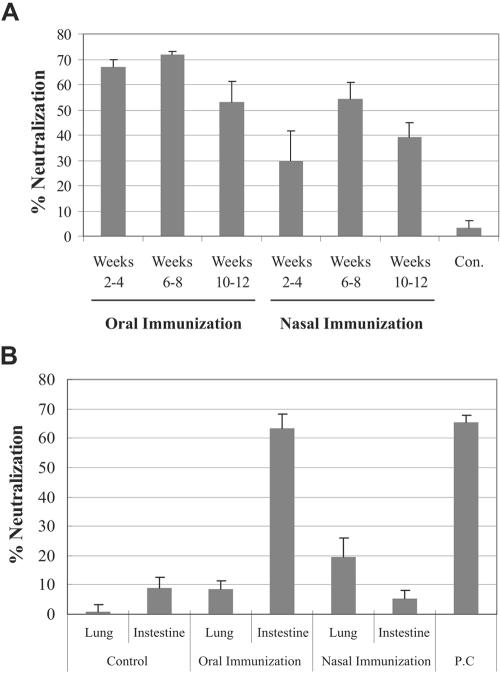
To determine whether anti-S protein antibodies exhibited antiviral activity, SARS pseudovirus neutralization assays were performed as we have previously described (9). This assay uses Mo-MuLV pseudotyped with the SARS-CoV S glycoprotein. Since Mo-MuLV encodes β-galactosidase, virus-infected cells can be quantified after staining them with X-Gal. First, we examined the neutralizing activity of purified serum IgG. Both intranasally and orally immunized mouse groups mounted potent SARS pseudovirus-neutralizing activities (Fig. 4A). In contrast, no antiviral activity was observed with IgG purified from control animals. For both immunization routes, the neutralizing-antibody levels peaked 6 to 8 weeks after the first immunization. No neutralizing activity was observed against control Mo-MuLVs pseudotyped with vesicular stomatitis virus G glycoproteins (data not shown).
FIG. 4.
SARS-CoV pseudovirus-neutralizing activity. Purified IgG from sera (A) and IgA from either intestinal or bronchoalveolar lavage fluids (B) were evaluated for SARS pseudovirus-neutralizing activities. Mice immunized either orally or intranasally were sampled at the indicated number of weeks after the first immunization. Approximately 100 infectious units of pseudovirus were used. Con., control; P.C., positive control. The assays were performed in duplicate, and the graphs are representative of two independent experiments. The error bars represent standard deviations.
Next, the neutralizing activities of anti-SARS IgA in intestinal and bronchoalveolar lavage fluids were determined to assess mucosal immune responses. Not surprisingly, potent neutralizing activity was observed in intestinal lavage fluids from mice immunized orally (Fig. 4B). In contrast, only a low level of neutralizing activity was detected in bronchoalveolar lavage fluids. In intranasally immunized mice, a moderate level of neutralizing activity was observed in bronchoalveolar lavage fluids. However, very little, if any, was detected in intestinal lavage fluids. The higher level of neutralizing mucosal IgA response in mice immunized orally is consistent with greater neutralizing activity of serum IgG from the same animals (Fig. 4A). Similar to IgA levels determined by peptide ELISA (Fig. 3), greater neutralizing activity was observed at the locality of immunization. Although S protein-specific IgA was detected in intestinal lavage fluids of intranasally immunized mice (Fig. 3B), we failed to detect neutralizing activity (Fig. 4B). These results are not necessarily contradictory; they may simply indicate that the fraction of antibodies that has neutralizing activity is below the level of detection. This may be further complicated by a higher background activity for fluids collected from the intestine than from the lung.
Identification of neutralizing epitopes.
Identification of neutralizing epitopes could facilitate not only the development of an effective vaccine against SARS-CoV, but also our understanding of protein structure and function. To identify epitopes targeted by neutralizing antibodies generated with our recombinant L. casei, we employed a peptide neutralization interference assay. The principle behind this assay is that peptides should interfere with neutralizing activity if they are structurally similar to epitopes targeted by neutralizing antibodies. It should be noted that this assay is capable of identifying linear epitopes, but not discontinuous or highly conformation-dependent epitopes. Since our recombinant L. casei strains expressed two segments of the S glycoprotein (amino acids 2 to 114 and 264 to 595) and since the receptor-binding domain has been shown to reside between amino acids 318 and 510 (36), we looked for potential neutralization epitopes between amino acids 264 and 595. Forty-five overlapping peptides between amino acids 257 and 598 were evaluated (Table 2). To conserve antibodies, we initially examined the neutralization interference activities of four different pools of peptides (mixtures 1 to 4). Each mixture contained 11 peptides, except for mixture 1, which contained 12. As shown in Fig. 5A, peptide mixtures 1 and 4 strongly inhibited the neutralizing activity of purified serum IgG, indicating that potential linear neutralization epitopes existed between amino acids 257 and 352 and between 505 and 598. To further delineate neutralizing epitopes, we evaluated individual peptides within mixture 1 and mixture 4. As shown in Fig. 5B, four peptides were able to completely inhibit neutralizing activity (peptides 291 to 308, 512 to 529, 520 to 537, and 564 to 581). Since the peptides 512 to 529 and 520 to 537 overlap, the critical antibody recognition site most likely resides between 520 and 529. Two other peptides also exhibited inhibitory effects, albeit less potently (peptides 278 to 295 and 528 to 545). These results indicate that there are at least three distinct epitopes that are the principal determinants of neutralizing-antibody responses mounted against our recombinant L. casei (residues 291 to 308, 520 to 529, and 564 to 581).
FIG. 5.
Identification of neutralization epitopes using a peptide neutralization interference assay. (A) Four different mixtures of overlapping peptides derived from S glycoprotein (Table 2) were evaluated for the ability to interfere with the neutralizing activity of IgG purified from sera collected 6 to 8 weeks after the first oral immunization. (B) Neutralization interference by individual peptides contained in mixtures 1 and 4 using serum IgG. The numbers indicate the amino acid residues of the S glycoprotein. The assays were performed in duplicate, and the graphs are representative of two independent experiments. The error bars represent standard deviations.
DISCUSSION
Mucosal immunization offers a number of advantages over other routes of antigen delivery, including convenience, cost effectiveness, and induction of both local and systemic immune responses (16, 21, 23, 26, 34). The goal is to provide the first line of defense by effectively eliminating pathogens at the mucosal surface. Surface display of antigens on the bacterial surface has been problematic because large antigens perturb membrane topology. In addition, true surface exposure of antigens requires a transmembrane anchor that is long enough to cross the cell wall. The average thickness of the peptidoglycan layer in the genus Lactobacillus has been reported to be approximately 15 to 30 nm (30), and at least 100 amino acids are needed to properly cross the cell wall (13).
In this report, we have described the development of a novel surface display system using the PgsA protein as the transmembrane anchor to present heterologous proteins on L. casei. We adopted PgsA protein derived from the PgsBCA enzyme complex of B. subtilis isolated from chung-kook-jang (a traditional Korean soybean paste used as a food additive) because it has one transmembrane domain near its N terminus (amino acids ∼26 to 42) with the bulk of the protein (about 336 amino acids) located on the outside of the cell membrane (1). The exact conformation of the protein in the peptidoglycan layer cannot be easily predicted. Nevertheless, the immunofluorescence and fluorescence-activated cell sorter data indicated that it was able to cross the cell wall even though much of the extracellular domain of PgsA is likely located in the peptidoglycan layer. The ability to display antigens of high molecular mass would be extremely valuable. In this regard, we have recently used PgsA to display α-amylase (∼81.5 kDa) of Streptococcus bovis with intact activity on the cell surface layer of LAB (unpublished data). The ability to display heterologous proteins of a wide range of molecular masses (from ∼10 to 80 kDa) on gram-positive bacteria confirms that our PgsA display system is highly versatile.
Oral and nasal administration of recombinant L. casei displaying SARS-CoV S protein antigens on the surface induced both systemic and mucosal immune responses against the protein. More importantly, the antibodies present both in the serum and on the mucosal surface exhibited neutralizing activities. Oral immunization was more effective in eliciting neutralizing antibodies than the intranasal route, albeit not without some caveats. First of all, a higher dose of L. casei was administered for oral immunization (5 × 109 versus 2 × 109 organisms). Secondly, the immunization schedules, which were optimized for each route, were slightly different. For oral immunization, animals were immunized for five consecutive days during the third and fourth immunization periods (days 21 to 25 and 49 to 53). In contrast, animals were immunized nasally only once on days 21 and 49. However, these differences in the immunization regimen do not fully explain the disparity in the neutralizing-antibody levels, since similar, if not greater, levels of antibodies were detected using ELISA in intranasally immunized animals than in those immunized orally (Fig. 2). An alternative explanation could be that the antigens are processed and/or presented differently to immune cells in the two mucosal compartments. For example, efficient phagocytosis of recombinant L. casei by alveolar macrophages could result in presentation of primarily degraded S protein peptides to B cells. Consequently, antibodies that target peptide fragments, which may exhibit neutralizing activities weaker than those against intact proteins, would be preferentially generated.
To date, only four neutralizing epitopes of 20 amino acids or less have been identified for SARS-CoV (548 to 567, 607 to 627, 130 to 150, and 490 to 510) (7-8). These epitopes were identified by evaluating the immunoreactivities of serially truncated S protein fragments. This methodology, however, could not be employed in our study because we were characterizing polyclonal antibodies rather than monoclonal antibodies, as in other studies. Consequently, we had to resort to a more complicated peptide neutralization interference assay. Nevertheless, we were able to identify three novel epitopes that are principally targeted by neutralizing antibodies induced using our recombinant L. casei (291 to 308, 520 to 529, and 564 to 581).
Given that we were working with polyclonal antibodies, we were somewhat surprised by two results. The first surprise was that none of the epitopes we identified are situated within the RBD (amino acids 318 to 510) (32). Our results are not due to experimental artifacts, since only one of four previously identified neutralization epitopes lies within the proposed RBD (490 to 510) (7). Although the precise mechanism needs to be further elucidated, one possible explanation is that neutralizing antibodies that bind outside of the 318-to-510 fragment may interfere with one of the post-receptor-binding events of virus entry (e.g., conformational changes). The second surprise was that we observed nearly complete inhibition of neutralizing activity by peptides from three nonoverlapping regions (Fig. 5B). That is, we predicted that if there were only one principal neutralizing antibody/epitope, then we would identify only one peptide that would interfere with the neutralizing activity. In contrast, if there were multiple neutralizing antibodies/epitopes, then we expected to identify multiple peptides that would only partially inhibit neutralizing activity; blocking neutralizing antibodies from binding to one epitope should not prevent the binding of other antibodies that target a different epitope.
To begin ascertaining the significance of our results, we examined neutralizing epitopes in the context of a previously reported three-dimensional model of the S1 domain (28). The model was constructed based on Clostridium botulinum toxin, which shows good structural homology to predicted secondary structures of the S glycoprotein. As shown in Fig. 6, two of the epitopes we identified (520 to 529 and 564 to 581) are situated adjacent to each other and lie between two previously identified epitopes (548 to 567 and 607 to 627). Given the close proximity between the 520-to-529 and 564-to-581 epitopes, it is plausible that a single antibody might be binding to both of these peptide segments. On the other hand, the 291-to-308 epitope appears to be well separated from the other two. However, the conformation of amino acid residues 323 to 502 in the context of this predicted model of the S1 domain is vastly different from that of a recently solved receptor-binding domain of S protein cocrystalized with soluble ACE2 (15). Therefore, the proposed model of the S1 domain could be largely inaccurate, and there is still a possibility that the 291-to-308 neutralizing epitope could be positioned close to epitopes 520 to 529 and 564 to 581. Alternatively, two different antibody molecules might be binding to the 291-to-308 and 520-to-529/564-to-581 regions separately. However, their ability to inhibit infection depends on cooperative interactions between the two antibodies so that the virus is not neutralized unless both antibodies are bound. Additional studies will be needed to test these hypotheses.
FIG. 6.
Locations of SARS neutralization epitopes identified to date. A proposed three-dimensional model of the S1 domain (aa 17 to 680; Protein Data Bank identifier, 1Q4Z) was used to plot neutralization epitopes identified by this and previously reported studies. The orientation of the molecule was arbitrarily chosen for optimal display of the epitopes. The aspartic acid residue at 454 (indicated in red), which has been shown to be critical for binding to ACE2, is shown as a reference point. Panels A and B show the opposite sides of the protein. The three epitopes identified in this study are indicated in yellow (520 to 529), purple (564 to 581), and blue (291 to 308).
Considering that SARS-CoV can be transmitted via inhalation or the fecal-oral route, a vaccine that can induce mucosal immunity might be more protective than those that elicit only a systemic immune response. In this regard, the results of our study indicate that recombinant L. casei expressing S protein antigens may be a promising mucosal vaccine candidate against SARS-CoV infection. Furthermore, our surface display system could be used to design vaccines against other pathogens that are transmitted mucosally (e.g., human immunodeficiency virus type 1).
Acknowledgments
This work was supported by a National Research Laboratory Program grant (2000-N-NL-01-C-171) from the Korea Ministry of Science and Technology to C.-J. K., and the work of M.-H. S. was supported by the Biochallenger program (M1-0310-82-0000) from the Korea Ministry of Commerce, Industry and Energy. The work of M.W.C was supported by U.S. National Institutes of Health grant U54 AI057160 to the Midwest Regional Center of Excellence for Biodefense and Emerging Infectious Diseases Research (MRCE) and by R21 AI059217.
We thank Seiki Kuramitsu and Noriko Nakagawa at Osaka University for technical help in understanding of PgsA topology and the NIH Biodefense and Emerging Infections Research Resources Repository for providing SARS S protein peptides.
REFERENCES
- 1.Ashiuchi, M., C. Nawa., T. Kamei, J. J. Song, S. P. Hong, M. H. Sung, K. Soda, T. Yagi, and H. Misono. 2001. Physiological and biochemical characteristics of poly-γ-glutamate synthetase complex of Bacillus subtilis. Eur. J. Biochem. 268:5321-5328. [DOI] [PubMed] [Google Scholar]
- 2.Ashiuchi, M., K. Soda, and H. Misono. 1999. A poly-γ-glutamate synthetic system of Bacillus subtilis IFO 3336: gene cloning and biochemical analysis of poly-γ-glutamate produced by Escherichia coli clone cells. Biochem. Biophys. Res. Commun. 263:6-12. [DOI] [PubMed] [Google Scholar]
- 3.Bisht, H., A. Roberts, L. Vogel, A. Bukreyev, P. L. Collins, B. R. Murphy, K. Subbarao, and B. Moss. 2004. Severe acute respiratory syndrome coronavirus spike protein expressed by attenuated vaccinia virus protectively immunizes mice. Proc. Natl. Acad. Sci. USA 101:6641-6646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bukreyev, A., E. W. Lamirande, U. J. Buchholz, L. N. Vogel, W. R. Elkins, M. St. Claire, B. R. Murphy, K. Subbarao, and P. L. Collins. 2004. Mucosal immunization of African green monkeys (Cercopithecus aethlops) with an attenuated parainfluenza virus expressing the SARS coronavirus spike protein for the prevention of SARS. Lancet 363:2122-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dieye, Y., S. Usai, F. Clier, A. Gruss, and J. C. Piard. 2001. Design of protein-targeting system for lactic acid bacteria. J. Bacteriol. 183:4157-4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman, R. S., F. R. Frankel, Z. Xu, and J. Lieberman. 2000. Induction of human immunodeficiency virus (HIV)-specific CD8 T-cell responses by Listeria monocytogenes and a hyperattenuated Listeria strain engineered to express HIV antigens. J. Virol. 74:9987-9993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greenough, T. C., G. J. Babcock, A. Roberts, H. J. Hernandez, W. D. J. Thomas, J. A. Coccia, R. F. Graziano, M. Srinivasan, L. Lowy, R. W. Finberg, K. Subbarao, L. Vogel, M. Somasundaran, K. Luzuriaga, J. L. Sullivan, and D. M. Ambrosino. 2005. Development and characterization of a severe acute respiratory syndrome-associated coronavirus-neutralizing human monoclonal antibody that provides effective immunoprophylaxis in mice. J. Infect. Dis. 191:507-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guillen, J., A. J. Perez-Berna, M. R. Moreno, and J. Villalain. 2005. Identification of the membrane-active regions of the severe acute respiratory syndrome coronavirus spike membrane glycoprotein using a 16/18-mer peptide scan: implications for the viral fusion mechanism. J. Virol. 79:1743-1752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han, D. P., H. G. Kim, Y. B. Kim, L. L. M. Poon, and M. W. Cho. 2004. Development of a safe neutralization assay for SARS-CoV and characterization of S-glycoprotein. Virology 326:140-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.He, Y., Y. Zhou, S. Liu, Z. Kou, W. Li, M. Farzan, and S. Jiang. 2004. Receptor-binding domain of SARS-CoV spike protein induces highly potent neutralizing antibodies: implication for developing subunit vaccine. Biochem. Biophys. Res. Commun. 324:773-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He, Y., Y. Zhou, P. Siddiqui, and S. Jiang. 2004. Inactivated SARS-CoV vaccine elicits high titers of spike protein-specific antibodies that block receptor binding and virus entry. Biochem. Biophys. Res. Commun. 325:445-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J. S., K. S. Shin, J. G. Pan, and C. J. Kim. 2000. Surface-displayed viral antigens on Salmonella carrier vaccine. Nat. Biotechnol. 18:645-648. [DOI] [PubMed] [Google Scholar]
- 13.Leenhouts, K., G. Buist, and J. Kok. 1999. Anchoring of proteins to lactic acid bacteria. Antonie Leeuwenhoek 76:367-376. [PubMed] [Google Scholar]
- 14.Leung, W. K., K. F. To, P. K. S. Chan, H. L. Y. Chan, A. K. L. Wu, N. Lee, K. Y. Yuen, and J. J. Y. Sung. 2003. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. Gastroenterology 125:1011-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li, F., W. Li, M. Farzan, and S. C. Harrison. 2005. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 309:1864-1868. [DOI] [PubMed] [Google Scholar]
- 16.Mannam, P., K. F. Jones, and B. L. Geller. 2004. Mucosal vaccine made from live, recombinant Lactococcus lactis protects mice against pharyngeal infection with Streptococcus pyogenes. Infect. Immun. 72:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park, S., K. Sestak, D. C. Hodgins, D. I. Shoup, L. A. Ward, D. J. Jackwood, and L. J. Saif. 1998. Immune response of sows vaccinated with attenuated transmissible gastroenteritis virus (TGEV) and recombinant TGEV spike protein vaccine and protection of their suckling piglets against TGEV challenge. Am. J. Vet. Res. 59:1002-1008. [PubMed] [Google Scholar]
- 18.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, K. Y. Yeun, and HKU/UCH SARS Study Group. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peiris, J. S., K. Y. Yuen, A. D. Osterhaus, and K. Stohr. 2003. The severe acute respiratory syndrome. N. Engl. J. Med. 349:2431-2441. [DOI] [PubMed] [Google Scholar]
- 20.Qu, D., B. Zheng, X. Yao, Y. Guan, Z. H. Yuan, N. S. Zhong, L. W. Lu, J. P. Xie, and Y. M. Wen. 2005. Intranasal immunization with inactivated SARS-CoV (SARS-associated coronavirus) induced local and serum antibodies in mice. Vaccine 23:924-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson, K., L. M. Chamberlain, K. M. Schofield, J. M. Wells, and R. W. F. Le Page. 1997. Oral vaccination of mice against tetanus with recombinant Lactococcus lactis. Nat. Biotechnol. 15:653-657. [DOI] [PubMed] [Google Scholar]
- 22.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. T. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rassmussen, R. Fouchier, S. Gunther, A. D. M. E. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 23.Seegers, J. F. M. L. 2002. Lactobacilli as live vaccine delivery vectors: progress and prospects. Trends Biotechnol. 20:508-515. [DOI] [PubMed] [Google Scholar]
- 24.Shata, M. T., and D. M. Hone. 2001. Vaccination with a Shigella DNA vaccine vector induces antigen-specific CD8+ T cells and antiviral protective immunity. J. Virol. 75:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shata, M. T., M. S. Reitz, Jr., A. L. DeVico, G. K. Lewis, and D. M. Hone. 2001. Mucosal and systemic HIV-1 Env-specific CD8+ T-cells develop after intragastric vaccination with a Salmonella Env DNA vaccine vector. Vaccine 20:623-629. [DOI] [PubMed] [Google Scholar]
- 26.Shaw, D. M., B. Gaerthe, R. J. Leer, J. G. M. M. Van der Stap, C. Smittenaar, M. J. Heijne den Bak-Glashouwer, J. E. R. Thole, F. J. Tielen, P. H. Pouwels, and C. E. G. Havenith. 2000. Engineering the microflora to vaccinate the mucosa: serum immunoglobulin G responses and activated draining cervical lymph nodes following mucosal application of tetanus toxin fragment C-expressing lactobacilli. Immunology 100:510-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi, X., E. Gong, D. Gao, B. Zhang, J. Zheng, Z. Gao, Y. Zhong, W. Zou, B. Wu, W. Fang, S. Liao, S. Wang, Z. Xie, M. Lu, L. Hou, H. Zhong, H. Shao, N. Li, C. Liu, F. Pei, J. Yang, Y. Wang, Z. Han, X. Shi, Q. Zhang, J. You, X. Zhu, and J. Gu. 2005. Severe acute respiratory syndrome associated coronavirus is detected in intestinal tissues of fatal cases. Am. J. Gastroenterol. 100:169-176. [DOI] [PubMed] [Google Scholar]
- 28.Spiga, O., A. Bernini, A. Ciutti, S. Chiellini, N. Menciassi, F. Finetti, V. Causarono, F. Anselmi, F. Prischi, and N. Niccolai. 2003. Molecular modelling of S1 and S2 subunits of SARS coronavirus spike glycoprotein. Biochem. Biophys. Res. Commun. 310:78-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Trieu-Cuot, P., C. Carlier, C. Poyart-Salmeron, and P. Courvalin. 1991. Shuttle vectors containing a multiple cloning site and a lacZα gene for conjugal transfer of DNA from Escherichia coli to gram-positive bacteria. Gene 102:99-104. [DOI] [PubMed] [Google Scholar]
- 30.Ventura, M., I. Jankovic, D. C. Walker, R. D. Pridmore, and R. Zink. 2002. Identification and characterization of novel surface proteins in Lactobacillus johnsonii and Lactobacillus gasseri. Appl. Environ. Microbiol. 68:6172-6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, S., T. W. Chou, P. V. Sakhatskyy, S. Huang, J. M. Lawrence, H. Cao, X. Huang, and S. Lu. 2005. Identification of two neutralizing regions on the severe acute respiratory syndrome coronavirus spike glycoprotein produced from the mammalian expression system. J. Virol. 79:1906-1910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wong, S. K., W. Li, M. J. Moore, H. Choe, and M. Farzan. 2004. A 193-amino acid fragment of the SARS coronavirus S protein efficiently binds angiotensin-converting enzyme 2. J. Biol. Chem. 279:3197-3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiao, X., S. Chakraborti, A. S. Dimitrov, K. Gramatikoff, and D. S. Dimitrov. 2003. The SARS-CoV S glycoprotein: expression and functional characterization. Biochem. Biophys. Res. Commun. 312:1159-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xin, K. Q., Y. Hoshino, Y. Toda, S. Igimi, Y. Kojima, N. Jounai, K. Ohba, A. Kushiro, M. Kiwaki, K. Hamajima, D. Klinman, and K. Okuda. 2003. Immunogenicity and protective efficacy of orally administered recombinant Lactococcus lactis expressing surface-bound HIV Env. Blood 102:223-228. [DOI] [PubMed] [Google Scholar]
- 35.Yang, Z. Y., W. P. Kong, Y. Huang, A. Roberts, B. R. Murphy, K. Subbarao, and G. J. Nabel. 2004. A DNA vaccine induces SARS coronavirus neutralization and protective immunity in mice. Nature 428:561-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhou, T., H. Wang, D. Lou, T. Rowe, Z. Wang, R. J. Hogan, S. Qiu, R. J. Bunzel, G. Huang, V. Mishra, T. G. Voss, R. Kimberly, and M. Lou. 2004. An exposed domain in the severe acute respiratory syndrome coronavirus spike protein induces neutralizing antibodies. J. Virol. 78:7217-7226. [DOI] [PMC free article] [PubMed] [Google Scholar]