Abstract
Baculovirus lef-4 encodes one subunit of the viral RNA polymerase. Here, we demonstrate the essential nature of LEF-4 by RNA interference and bacmid knockout technology. Silencing of LEF-4 in wild-type virus-infected cells suppressed expression of structural genes, while early expression was unaffected, demonstrating its essential role in late gene expression. After transfection of insect cells with lef-4 mutant bacmid, no viral progeny was produced, further defining its central role in infection. Cotransfection with wild-type lef-4 plasmid restored normal replication, but plasmid encoding a guanyltransferase-deficient version failed to rescue. These results emphasize the importance of the mRNA capping function of LEF-4.
Baculoviruses are unique among eukaryotic DNA viruses because early genes are transcribed by host RNA polymerase II (5, 13), while late genes are transcribed by a virus-encoded RNA polymerase (6, 8, 10). In Autographa californica multiple nucleopolyhedrovirus (AcMNPV), the core viral polymerase is composed of four viral proteins, LEF-4, LEF-8, LEF-9, and p47 (10). LEF-8 and LEF-9, which contain motifs common to β and β′ RNA polymerases, are believed to form the catalytic site (19, 26). LEF-4 is an mRNA capping enzyme (7, 9, 15), and the function of p47 is unknown.
The LEF proteins were originally mapped as late expression factors by transient assays that used a late reporter construct and fragments of viral DNA (25). The lef-4 gene was also identified as the site of a temperature-sensitive (ts) mutation with a late expression phenotype (2, 24). To address the essential function of LEF-4, we performed RNA silencing experiments and gene knockout experiments using bacmid technology. Previous studies have demonstrated the value of these techniques in the study of essential viral genes in Spodoptera frugiperda cells (4, 14, 17, 18, 20, 28).
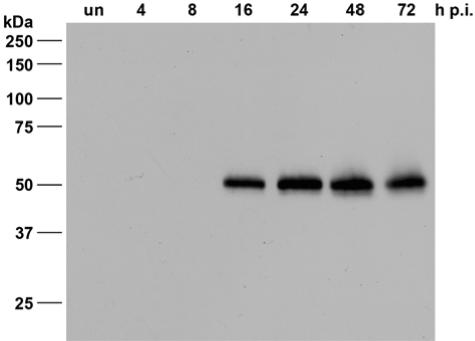
First, a high quality LEF-4 antiserum was produced in rabbits using LEF-4 protein expressed in bacteria (9). S. frugiperda cells were infected with AcMNPV at 10 PFU/cell, and detergent-based nuclear extracts were prepared from infected cells 4, 8, 16, 24, 48, and 72 h postinfection (p.i.). As previously described, the cell pellet was lysed in 0.5% NP-40 and centrifuged and the supernatant (cytoplasmic fraction) was adjusted to 0.1 N NaOH (21). The pelleted nuclei were resuspended in 1% NP-40 and adjusted to 0.1 N NaOH. Immunoblot experiments showed that LEF-4 antiserum recognized a polypeptide of about 50 kDa from 16 through 72 h p.i., without cross-reactivity (Fig. 1).
FIG. 1.
Expression of LEF-4 in AcMNPV-infected S. frugiperda cells. Detergent-based nuclear extracts were prepared from uninfected S. frugiperda cells (“un”) or from infected cells at 4, 8, 16, 24, 48, and 72 h p.i. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and stained with the rabbit anti-LEF-4 antiserum. Protein size markers are given on the left.
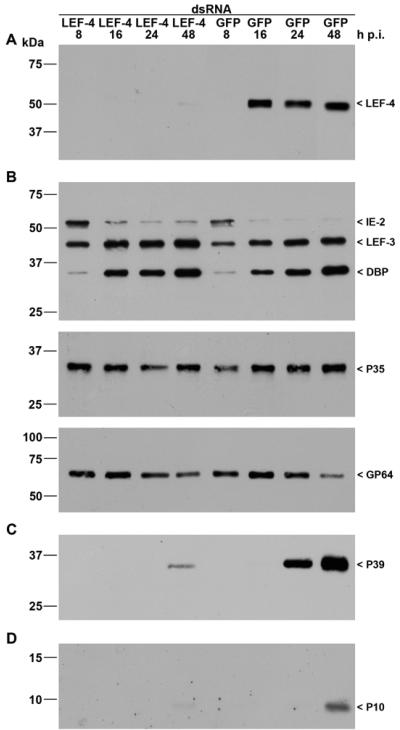
To conduct the RNA silencing experiments, specific double-stranded RNA (dsRNA) was prepared using the T7 RiboMax Express RNAi system (Promega). The cloned HindIII-C fragment of the AcMNPV genome served as DNA template to generate dsRNA corresponding to 543 bp at the 5′ terminus of lef-4 (primer 1a, 5′-GGGTAATACGACTCACTATAGGGGCGATTTTGTGATTGAG-3′; primer 1b, 5′-GGCAAATTCATATTCGAGACG-3′; primer 2a, 5′-GGGTAATACGACTCACTATAGGCAAATTCATATTCGAGACG-3′; primer 2b, 5′-GGGGCGATTTTGTGATTGAG-3′). As a control we used dsRNA corresponding to a 600-bp region of the green fluorescent protein (GFP) gene. S. frugiperda cells were transfected with 5 μg of LEF-4 dsRNA or GFP dsRNA as calcium phosphate precipitates (BD BaculoGold). Cells were subsequently infected with AcMNPV (10 PFU/cell) at 20 h posttransfection, followed by detergent-based nuclear extraction (21). Immunoblot analysis showed that transfection of LEF-4 dsRNA silenced expression of LEF-4. Transfection of the control GFP dsRNA, however, did not affect LEF-4 expression, thus demonstrating specificity (Fig. 2A). The efficiency of lef-4 silencing indicates that a high percentage of cells must have taken up the dsRNA.
FIG. 2.
Inhibition of viral gene expression by lef-4 silencing. S. frugiperda cells were transfected with either LEF-4 or GFP dsRNA. Cells were subsequently infected with AcMNPV (10 PFU/cell) at 20 h posttransfection, and detergent-based nuclear and cytoplasmic extracts were prepared from infected cells 8, 16, 24, and 48 h p.i. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to nitrocellulose membranes (A, B, and C) or resolved on sodium dodecyl sulfate-15% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (D). (A) LEF-4 was stained with rabbit anti-Lef-4 antiserum. (B) Early gene expression was analyzed with rabbit antisera raised against IE2 (16), LEF-3 (3), DBP (22), or P35 (11) or with mouse monoclonal anti-GP64 (AcV5) (12). (C) Late gene expression was analyzed with mouse monoclonal anti-P39 (P10C6) (30). (D) Expression of the very late protein P10 was detectable with rabbit anti-P10 serum (29). Expression of LEF-4, IE2, LEF-3, DBP, P39, and P10 was analyzed on samples of nuclear protein fractions, and P35 and GP64 expression was detected in cytoplasmic fractions as described previously (21). Protein size markers are shown on the left, and the identities of the viral proteins are indicated on the right.
To study the effect of LEF-4 suppression on viral gene expression, we analyzed proteins expressed early, late, and very late during infection. IE2, an immediate-early protein, is detectable from 2 until 12 to 24 h p.i. in S. frugiperda cells (16). The loss of LEF-4 had no effect on IE2 levels, consistent with the assumption that the ie2 promoter is transcribed by host RNA polymerase II (Fig. 2B). We also found no effect on expression patterns of LEF-3 (3), DBP (22), or P35 (11) in infected cells, consistent with the idea that LEF-4 is not involved in early gene expression (Fig. 2B). Surprisingly, we observed comparable levels of the membrane protein GP64 in cells treated with LEF-4 dsRNA or GFP dsRNA (Fig. 2B). Since GP64 is known to be regulated by both early and late promoters, we expected that protein levels would be lower in the lef-4-silenced cells. The similar amounts of protein might be coincidental, because accumulation of GP64 in normal infection is a function of synthesis combined with the loss of protein due to budding of virus particles. In the case of lef-4-silenced cells, less budding presumably occurs, due to lower levels of synthesis of viral structural proteins (see below).
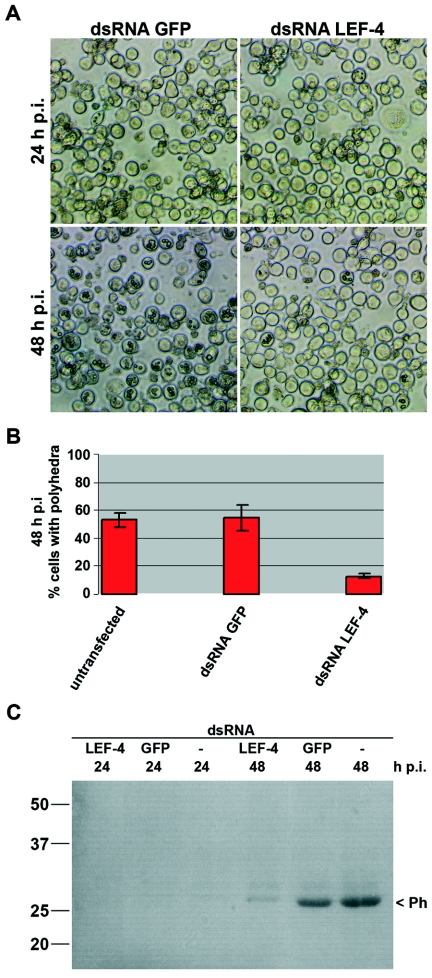
To examine the effect of LEF-4 on late genes, protein extracts were probed with antibody against the capsid protein P39 (30). Strong expression of P39 was observed at 24 and 48 h p.i. in cells treated with GFP dsRNA, while only a weak P39 signal was detected in cells with suppressed LEF-4 (Fig. 2C). The very late protein P10 was also shown to be dependent upon LEF-4 synthesis. P10 levels were undetectable in LEF-4 suppressed cells, while P10 was evident in the control cells at 48 h p.i. (Fig. 2D). Furthermore, polyhedrin levels were reduced. Immunoblot analysis showed a significant reduction in accumulation of polyhedrin protein in cells treated with LEF-4 dsRNA, compared to cells treated with GFP dsRNA or untreated cells (Fig. 3C). Microscopic analysis revealed the presence of polyhedron-positive cells at 24 and 48 h p.i. in the LEF-4-silenced cells, although the number of cells with polyhedra was about fivefold lower than that of the control GFP-treated cells (Fig. 3A and B). The few cells that were polyhedron positive had equivalent numbers of polyhedra per cell, and those polyhedra formed with the same kinetics as in the GFP dsRNA-treated and untreated controls (Fig. 3A and data not shown). This suggests that cells with polyhedra likely represent the population of cells that did not take up LEF-4 dsRNA.
FIG. 3.
Polyhedron formation and polyhedrin expression upon lef-4 inhibition in AcMNPV-infected S. frugiperda cells. Cells were transfected with either LEF-4 dsRNA or GFP dsRNA, infected with AcMNPV (10 PFU/cells) at 20 h posttransfection, and analyzed at 24 and 48 h p.i. (A) Phase-contrast images are shown, and (B) polyhedron-containing cells were quantitated at 48 h p.i. (C) Cells transfected with LEF-4 dsRNA, GFP dsRNA, or untransfected cells were infected, and detergent-based nuclear extracts were prepared at 24 and 48 h p.i. Proteins were resolved on sodium dodecyl sulfate-10% polyacrylamide gels and transferred to nitrocellulose, and polyhedrin was viewed by Ponceau staining. Numbers at left are molecular masses in kilodaltons.
Our results strengthen the identification of LEF-4 as an essential component of the viral RNA polymerase that is responsible for late and very late transcription. The role of LEF-4, however, remains open. Previous results showed that the lef-4 ts mutant L104F failed to produce infectious virus at the nonpermissive temperature (2). Interestingly, a mutant version of LEF-4 with this substitution was not impaired for guanyltransferase activity (15). To address whether LEF-4 is essential for its capping activity, we constructed a virus with an interrupted lef-4 gene.
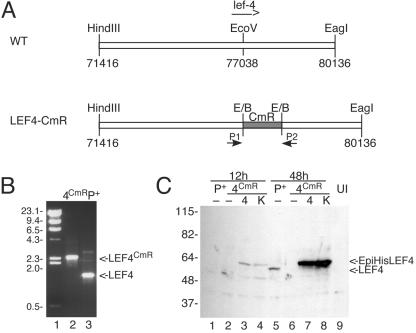
The bacmid BacP+ is derived from the Bac-to-Bac cloning vector (Invitrogen), which has the AcMNPV genome cloned into a single-copy plasmid (1). BacP+ has a reconstructed polyhedrin gene and lacks transposition sites for cloning into the polyhedrin locus. The lef-4 gene was inactivated by inserting a 1-kb BstBI fragment of pBR325, containing the chloramphenicol acetyltransferase (CAT) resistance marker under the control of its own promoter, into the EcoRV site of an 8.7-kb genome fragment containing lef-4 (Fig. 4A). The resulting plasmid was digested to excise the viral DNA that was used to transform BacP+-containing BJ5183 cells by electroporation. Recombinants were selected by plating on chloramphenicol, and interruption of the lef-4 open reading frame with CAT was confirmed by PCR using plasmids that flank the lef-4 gene (Fig. 4B).
FIG. 4.
Construction of BacP+/LEF-4CmR. (A) Cloning strategy. A plasmid containing the left 7 kb of the HindIII-C fragment of AcMNPV was digested with EcoRV, and a 1-kb fragment of pBR325 containing the CAT resistance marker was inserted. The resultant plasmid was transformed into BJ5183 cells containing a modified version of the AcMNPV genome, followed by selection on chloramphenicol, as previously described (1). (B) PCR screening. Correct insertion of CAT into lef-4 was verified by PCR using primers that flank the lef-4 open reading frame. Lane 2, BacP+/LEF-4CmR; lane 3, BacP+. The positions of molecular mass markers (lane 1) are indicated on the left in kilodaltons, and the migration of lef-4 and lef-4 with the CAT insertion is shown on the right. (C) Immunoblot analysis of LEF-4 expression in transfected cells. Cells were transfected with BacP+ (P+, lanes 1 and 5) or BacP+/LEF-4CmR (4CmR, lanes 2 to 4 and 6 to 8) and cotransfected where indicated with pHSEpiHisLEF-4 (4, lanes 3 and 7) or pHSEpiHisLEF-4(K255A) (K, lanes 4 and 8). Cells were harvested at 12 h (lanes 1 to 4) or 48 h (lanes 5 to 8) posttransfection. The blot was probed with rabbit LEF-4 antiserum. Untransfected cells were analyzed as a negative control (lane 9). The positions of molecular mass markers are indicated on the left in kilodaltons, and the migration of LEF-4 and EpiHisLEF-4 is indicated on the right.
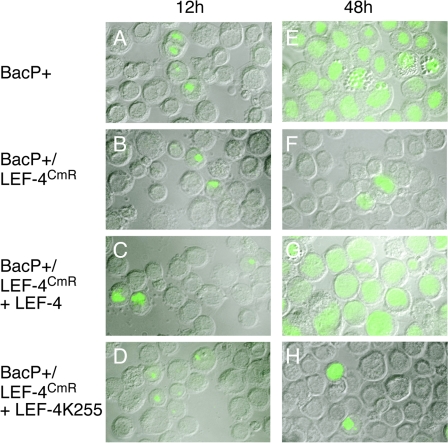
Purified bacmid DNA was transfected into Sf9 cells, which were analyzed at 12 h p.i. by immunofluorescence using antibody against the immediate-early IE1. Approximately 500 total cells were scored for IE1 expression. We found 4 to 12% positive cells, indicating similar transfection efficiencies for the two DNAs (Fig. 5A and B). At 48 h posttransfection, virtually all cells transfected with BacP+ were positive for IE1 (Fig. 5E), indicating that viral infection had spread to adjacent cells, but the percentage of IE1-positive cells in BacP+/LEF-4CmR had not significantly increased (Fig. 5F). The lack of replication was verified by plaque assay of the culture medium at 48 h posttransfection (Table 1).
FIG. 5.
Infectivity of lef-4 mutant viruses. S. frugiperda cells were transfected with BacP+ (A and E), BacP+/LEF-4CmR (B and F), or BacP+/LEF-4CmR in the presence of plasmids encoding wild-type LEF-4 (C and G) or a mutant version of LEF-4 that lacks guanyltransferase activity (D and H). Cells were harvested at 12 or 48 h p.i. and processed for immunofluorescence using a mouse monoclonal antibody raised against IE1. Images were collected using a Zeiss ApoTome/Axioplan 2 microscope. Images shown represent merged images of phase contrast and green fluorescence to visualize percentages of cells expressing IE1.
TABLE 1.
Infectious virus produced from transfection of bacmids
| Bacmid | Plasmid | Titer at 48 h posttransfection |
|---|---|---|
| BacP+ | None | 2 × 107 |
| BacP+/LEF-4CmR | None | 0 |
| pHSEpiHisLEF-4 | 5.3 × 106 | |
| pHSEpiHisLEF-4(K255A) | 2 × 102a |
All clones recovered were reversions.
To determine whether capping enzyme activity was necessary for viral infection, we cotransfected BacP+/LEF-4CmR and pHSEpiHisLEF-4 (27), encoding an epitope-tagged version of lef-4 or a mutant version of the same plasmid encoding an alanine substitution at lysine 255 (K255A). We have previously shown that this mutant is not active in guanyltransferase assays (15). Immunoblot analysis showed that the two proteins were expressed at equivalent levels by 12 h posttransfection (Fig. 4C). The wild-type lef-4 plasmid rescued the mutant bacmid DNA. The pattern of IE1 immunofluorescence was similar to that seen with the parental bacmid (Fig. 5C and G), and the yield of infectious virus was approximately 25% of that obtained with the parental clone (Table 1), indicating that the level of recombination between the bacmid and plasmid was very high. Cotransfection with the K255A mutant, however, did not significantly change the pattern of IE1 immunofluorescence (Fig. 5D and H). Some progeny virus was obtained (Table 1), but direct sequencing of four clones revealed that their genomes contained wild-type lef-4. This was possible because the BacP+/LEF-4CmR construct was made by inserting the CAT gene into the LEF-4 open reading frame without deleting any lef-4 sequences, including the K255 locus. The fact that only wild-type mutants were obtained with the K255A plasmid indicates that guanyltransferase activity is required for late gene expression and production of progeny virions.
The strategy used in these studies differs somewhat from previous protocols relying on bacmid systems (18, 23, 28). First, we did not construct a “repaired” virus control by inserting the lef-4 gene into the polyhedrin locus. Instead, we relied on the efficient recombination ability of baculoviruses to produce viable virus. The fact that the yield of infectious virus obtained from cotransfection of a wild-type lef-4 plasmid and BacP+/LEF-4CmR was 25% of that obtained with the parental bacmid indicates that in vivo recombination is very efficient. This was also evident in the high spread of IE1 fluorescence at 48 h posttransfection. Second, we did not construct bacmids that expressed reporter genes in order to follow infection. Instead we visualized expression of a viral gene.
Taken together, our results further characterize the essential nature of LEF-4 and demonstrate that the guanyltransferase function of LEF-4 is essential for productive infection. In addition, LEF-4 may also be required for its RNA triphosphatase activity and another function that was disrupted by the L105F ts substitution (2). Biochemical assays of a protein with an L105F substitution revealed that it was normal with respect to guanyltranferase and had only a modest decrease in RNA triphosphatase activity, which is probably insignificant (15). Since these are the only two enzymatic activities that the protein is known to possess, it is possible that L105 is important for structural integrity of the polymerase complex, and high temperature inhibits replication because the polymerase dissociated although the enzymatic functions of LEF-4 are unaffected. Analysis of additional LEF-4 mutants should help to define roles of LEF-4 in viral infection.
Acknowledgments
We thank Keiju Okano for the antiserum against Bombyx mori nucleopolyhedrovirus DBP, George F. Rohrmann for anti-LEF-3 antiserum, Paul D. Friesen for anti-p35 antiserum, Gary W. Blissard for monoclonal antibody gp64 (AcV5), Loy Volkman for monoclonal antibody p39 (P10C6), and Monique van Oers for anti-p10 antiserum. We are grateful to Ute Schepers for helpful comments on RNA silencing experiments.
Research was supported by grant KN536/11-1 from the Deutsche Forschungsgemeinschaft and the Köln Fortune Program/Faculty of Medicine, University of Cologne, and by grant MCB-0416484 from the National Science Foundation.
REFERENCES
- 1.Bideshi, D. K., and B. A. Federici. 2000. The Trichoplusia ni granulovirus helicase is unable to support replication of Autographa californica multicapsid nucleopolyhedrovirus in cells and larvae of T. ni. J. Gen. Virol. 81:1593-1599. [DOI] [PubMed] [Google Scholar]
- 2.Carstens, E. B., H. Chan, H. Yu, G. V. Williams, and R. Casselman. 1994. Genetic analyses of temperature-sensitive mutations in baculovirus late expression factors. Virology 204:323-337. [DOI] [PubMed] [Google Scholar]
- 3.Evans, J. T., and G. F. Rohrmann. 1997. The baculovirus single-stranded DNA binding protein, LEF-3, forms a homotrimer in solution. J. Virol. 71:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Flores-Jasso, C. F., V. J. Valdes, A. Sampieri, V. Valadez-Graham, F. Recillas-Targa, and L. Vaca. 2004. Silencing structural and nonstructural genes in baculovirus by RNA interference. Virus Res. 102:75-84. [DOI] [PubMed] [Google Scholar]
- 5.Fuchs, L. Y., M. S. Woods, and R. F. Weaver. 1983. Viral transcription during Autographa californica nuclear polyhedrosis virus infection: a novel RNA polymerase induced in Spodoptera frugiperda cells. J. Virol. 48:641-646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Glocker, B., R. R. Hoopes, Jr., L. Hodges, and G. F. Rohrmann. 1993. In vitro transcription from baculovirus late gene promoters: accurate mRNA initiation by nuclear extracts prepared from infected Spodoptera frugiperda cells. J. Virol. 67:3771-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross, C. H., and S. Shuman. 1998. RNA 5′-triphosphatase, nucleoside triphosphatase, and guanylyltransferase activities of baculovirus LEF-4 protein. J. Virol. 72:10020-10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grula, M. A., P. L. Buller, and R. F. Weaver. 1981. α-Amanitin-resistant viral RNA synthesis in nuclei isolated from nuclear polyhedrosis virus-infected Heliothis zea larvae and Spodoptera frugiperda cells. J. Virol. 38:916-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guarino, L. A., J. Jin, and W. Dong. 1998. Guanylyltransferase activity of the LEF-4 subunit of baculovirus RNA polymerase. J. Virol. 72:10003-10010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guarino, L. A., B. Xu, J. Jin, and W. Dong. 1998. A virus-encoded RNA polymerase purified from baculovirus-infected cells. J. Virol. 72:7985-7991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hershberger, P. A., D. J. LaCount, and P. D. Friesen. 1994. The apoptotic suppressor P35 is required early during baculovirus replication and is targeted to the cytosol of infected cells. J. Virol. 68:3467-3477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hohmann, A. W., and P. Faulkner. 1983. Monoclonal antibodies to baculovirus structural proteins: determination of specificities by Western blot analysis. Virology 125:432-444. [DOI] [PubMed] [Google Scholar]
- 13.Hoopes, R. R., Jr., and G. F. Rohrmann. 1991. In vitro transcription of baculovirus immediate early genes: accurate mRNA initiation by nuclear extracts from both insect and human cells. Proc. Natl. Acad. Sci. USA 88:4513-4517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hou, S., X. Chen, H. Wang, M. Tao, and Z. Hu. 2002. Efficient method to generate homologous recombinant baculovirus genomes in E. coli. BioTechniques 32:783-784, 786, 788. [DOI] [PubMed] [Google Scholar]
- 15.Jin, J., W. Dong, and L. A. Guarino. 1998. The LEF-4 subunit of baculovirus RNA polymerase has RNA 5′-triphosphatase and ATPase activities. J. Virol. 72:10011-10019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krappa, R., R. Roncarati, and D. Knebel-Morsdorf. 1995. Expression of PE38 and IE2, viral members of the C3HC4 finger family, during baculovirus infection: PE38 and IE2 localize to distinct nuclear regions. J. Virol. 69:5287-5293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, Y., J. Wang, R. Deng, Q. Zhang, K. Yang, and X. Wang. 2005. vlf-1 deletion brought AcMNPV to defect in nucleocapsid formation. Virus Genes 31:275-284. [DOI] [PubMed] [Google Scholar]
- 18.Lin, G., and G. W. Blissard. 2002. Analysis of an Autographa californica nucleopolyhedrovirus lef-11 knockout: LEF-11 is essential for viral DNA replication. J. Virol. 76:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu, A., and L. K. Miller. 1994. Identification of three late expression factor genes within the 33.8- to 43.4-map-unit region of Autographa californica nuclear polyhedrosis virus. J. Virol. 68:6710-6718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Means, J. C., I. Muro, and R. J. Clem. 2003. Silencing of the baculovirus Op-iap3 gene by RNA interference reveals that it is required for prevention of apoptosis during Orgyia pseudotsugata M nucleopolyhedrovirus infection of Ld652Y cells. J. Virol. 77:4481-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murges, D., I. Quadt, J. Schroer, and D. Knebel-Morsdorf. 2001. Dynamic nuclear localization of the baculovirus proteins IE2 and PE38 during the infection cycle: the promyelocytic leukemia protein colocalizes with IE2. Exp. Cell Res. 264:219-232. [DOI] [PubMed] [Google Scholar]
- 22.Okano, K., V. S. Mikhailov, and S. Maeda. 1999. Colocalization of baculovirus IE-1 and two DNA-binding proteins, DBP and LEF-3, to viral replication factories. J. Virol. 73:110-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Okano, K., A. L. Vanarsdall, and G. F. Rohrmann. 2004. Characterization of a baculovirus lacking the alkaline nuclease gene. J. Virol. 78:10650-10656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Partington, S., H. Yu, A. Lu, and E. B. Carstens. 1990. Isolation of temperature sensitive mutants of Autographa californica nuclear polyhedrosis virus: phenotype characterization of baculovirus mutants defective in very late gene expression. Virology 175:91-102. [DOI] [PubMed] [Google Scholar]
- 25.Passarelli, A. L., and L. K. Miller. 1993. Identification and characterization of lef-1, a baculovirus gene involved in late and very late gene expression. J. Virol. 67:3481-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passarelli, A. L., J. W. Todd, and L. K. Miller. 1994. A baculovirus gene involved in late gene expression predicts a large polypeptide with a conserved motif of RNA polymerases. J. Virol. 68:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rapp, J. C., J. A. Wilson, and L. K. Miller. 1998. Nineteen baculovirus open reading frames, including LEF-12, support late gene expression. J. Virol. 72:10197-10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stewart, T. M., I. Huijskens, L. G. Willis, and D. A. Theilmann. 2005. The Autographa californica multiple nucleopolyhedrovirus ie0-ie1 gene complex is essential for wild-type virus replication, but either IE0 or IE1 can support virus growth. J. Virol. 79:4619-4629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Oers, M. M., J. T. Flipsen, C. B. Reusken, and J. M. Vlak. 1994. Specificity of baculovirus p10 functions. Virology 200:513-523. [DOI] [PubMed] [Google Scholar]
- 30.Whitt, M. A., and J. S. Manning. 1988. A phosphorylated 34-kDa protein and a subpopulation of polyhedrin are thiol linked to the carbohydrate layer surrounding a baculovirus occlusion body. Virology 163:33-42. [DOI] [PubMed] [Google Scholar]