Abstract
Equid herpesvirus 1 (EHV-1) can cause a wide spectrum of diseases ranging from inapparent respiratory infection to the induction of abortion and, in extreme cases, neurological disease resulting in paralysis and ultimately death. It has been suggested that distinct strains of EHV-1 that differ in pathogenic capacity circulate in the field. In order to investigate this hypothesis, it was necessary to identify genetic markers that allow subgroups of related strains to be identified. We have determined all of the genetic differences between a neuropathogenic strain (Ab4) and a nonneuropathogenic strain (V592) of EHV-1 and developed PCR/sequencing procedures enabling differentiation of EHV-1 strains circulating in the field. The results indicate the occurrence of several major genetic subgroups of EHV-1 among isolates recovered from outbreaks over the course of 30 years, consistent with the proposal that distinct strains of EHV-1 circulate in the field. Moreover, there is evidence that certain strain groups are geographically restricted, being recovered predominantly from outbreaks occurring in either North America or Europe. Significantly, variation of a single amino acid of the DNA polymerase is strongly associated with neurological versus nonneurological disease outbreaks. Strikingly, this variant amino acid occurs at a highly conserved position for herpesvirus DNA polymerases, suggesting an important functional role.
Equid herpesvirus 1 (EHV-1), a member of the subfamily Alphaherpesvirinae, is a highly prevalent equine pathogen that can cause a range of clinical signs, from respiratory distress to the induction of abortion, neonatal foal death, and occasionally neurological damage resulting in paralysis (9, 14, 19, 33, 37, 61). The severity of disease resulting from EHV-1 infection is likely to be influenced by a number of factors, including the age and physical condition of the host; whether the infection is primary, a reinfection, or a reactivation of latent virus; the immune status of the host; and the pathogenic potential of the strain involved. In order to assess the relative importance of EHV-1 strain variation regarding disease outcome, it is necessary to develop methods enabling precise discrimination between genetic subgroups of interrelated strains. Previous studies have utilized DNA restriction fragment length polymorphism (RFLP) to separate field isolates of EHV-1 into subgroups according to characteristic restriction enzyme site changes and the presence of variable numbers of copies of short sequence repeats. These studies demonstrated a relatively low frequency of genetic polymorphism for EHV-1 and suggested that distinct strains of EHV-1 do exist in the field (3, 4, 8, 25, 32, 41, 54, 57). However, the relative lack of variation of EHV-1 sequences between strains has resulted in too few RFLP variants to be identified for detailed epidemiological studies. Furthermore, although such analyses may be used for tracing the genetic relatedness of strains, they allow identification only of those genetic changes resulting in restriction fragment variation, rather than the majority of changes that do not affect restriction cleavage sites.
Studies of the pathogenesis of EHV-1 infection, in particular relating to induction of abortion and neurological damage, have demonstrated differences in pathogenic potential correlated with differences in the ability to disseminate to and establish infection at vascular endothelial sites, in particular within the endometrium and central nervous system (CNS) (1, 17, 38, 42, 44, 60). Infection of endothelial cells is associated with vasculitis and thrombosis, with the resulting restricted blood flow leading to ischemic damage at the utero-placental junction and the CNS. Two field isolates of EHV-1 which show consistent differences in pathogenicity have been extensively characterized. Ab4, a neuropathogenic strain, was originally isolated from a case of paralytic disease (14) and V592, a nonneuropathogenic strain, from a multicase outbreak of abortion, during which no neurological disease was reported (39). In experimental infections of horses, Ab4 infection resulted in severe clinical disease, including a high frequency of abortion and a less frequent occurrence of paralysis. In contrast, disease following experimental V592 infection was largely restricted to a short-lived fever and mild respiratory signs, with infrequent induction of abortion and no neurological damage (38, 51, 52). These differences in clinical outcome may be related to observed differences in replication of these strains in vivo in leukocytes and endothelial cells: Ab4 typically caused a prolonged high level of viremia and widespread endothelial cell infection in experimental challenge, whereas V592 typically caused a more short-lived, low-level viremia and restricted or absent endothelial cell infection (38, 52).
In order to identify a genetic basis for the observed differences in pathogenic potential of the prototypic neuropathogenic and nonneuropathogenic isolates described above, we have determined the complete genomic sequence of V592 (16) (GenBank accession number AY464052). The information generated, in conjunction with the previously determined sequence of Ab4 (56) (GenBank accession number AY665713), has provided a valuable resource for identification of multiple loci of sequence variation between two EHV-1 pathotypes, which may be useful for subtyping of EHV-1 field isolates. In this paper, we report on amino acid coding variation identified between EHV-1 strains V592 and Ab4. We further describe the testing of selected genetic markers, comprising either variable-repeat-length regions or nucleic acid point mutations, against a panel of EHV-1 field isolates from various European countries, North and South America, and Australia. The sequence results for the selected markers were compared and analyzed according to the geographical origins and disease classifications (either neuropathogenic or nonneuropathogenic) of the outbreaks from which the field isolates were derived. The most significant finding is that variation of a single amino acid of the DNA polymerase is strongly associated with neuropathogenic versus nonneuropathogenic disease outbreaks.
MATERIALS AND METHODS
Viruses and cells.
The majority of EHV-1 field isolates were obtained from archive material held at the Animal Health Trust (United Kingdom isolates) and the Gluck Equine Research Center (U.S. and Canadian isolates). Additional isolates, or purified DNA from isolates, originating from outbreaks in other countries were provided by G. Fortier (Laboratoire Frank Duncombe, France), M. Studdert (University of Melbourne, Australia), S. Raidal (Murdoch University, Australia), C. van Maanen (Animal Health Service, The Netherlands), C. Galosi (Cátedra de Virología, UNLP, Argentina), and K. van der Meulen (University of Ghent, Belgium). The majority of isolates were obtained by culture of diagnostic material (nasal swab or blood or tissue samples) on rabbit kidney (RK-13) or equine dermal (NBL-6) cell lines. Some isolates were obtained by culture on primary equine (equine embryonic lung [EEL] or equine fetal kidney) cell types.
In order to provide a consistent nomenclature, isolates were coded according to their country of origin (C), year of outbreak (Y), a unique identifier for the outbreak (x: 1, 2, 3, etc.), and the pathogenic features of the outbreak (p: 0, attenuated vaccine strain; 1, nonneuropathogenic; 2, neuropathogenic) according to the scheme CCYY_x_p. Countries of origin are designated AR (Argentina), AU (Australia), BE (Belgium), CA (Canada), FR (France), GB (Great Britain), NL (The Netherlands), PL (Poland), and US (United States).
Preparation and shotgun cloning of purified virion DNA from V592.
DNA was prepared from purified virions essentially as previously described (49, 56). Equine fibroblasts (EEL) were infected at low multiplicity and incubated at 37°C until the complete cytopathic effect had developed. The tissue culture medium was harvested and centrifuged at low speed (1,500 × g, 5 min) to remove cell debris, and the supernatant was centrifuged at high speed (17,000 × g, 200 min) to pellet virions. Pellets were resuspended in 5 ml minimum essential medium (Eagle)-2% fetal calf serum and then purified via a sucrose gradient (40% to 60% sucrose in phosphate-buffered saline), with high-speed centrifugation (69,000 × g, 120 min). The virion material present at the sucrose gradient interface was collected and pelleted by centrifugation (17,000 × g, 180 min), and the pellet was resuspended in 1 ml Tris-EDTA buffer (10 mM Tris, 0.1 mM EDTA, pH 8). Virion DNA was prepared by proteinase K-sodium dodecyl sulfate digestion followed by phenol-chloroform extraction and ethanol precipitation. The DNA (redissolved in Tris-EDTA buffer) was self-ligated (T4 DNA ligase) to remove free ends and then sonicated to generate randomly sheared fragments. Sonicated DNA was gel purified, and fragments with an approximate size range of 300 to 1,000 bp were selected. Fragments were cloned into M13mp19 via blunt-end cloning with a Perfectly Blunt cloning kit (Novagen, Madison, WI). In addition, a panel of semirandom clones was generated by digestion of viral DNA with frequent cutting restriction enzymes (AluI, PvuII, and BalI), followed by cloning into M13mp19 as described above.
Determination of V592 complete genomic sequence.
Single-stranded M13 templates were prepared and sequenced using proprietary sequencing reagents and methods of the ABI Prism DNA sequencing guide (Applied Biosystems), with sequencing reactions analyzed with either an ABI 377 or an ABI 3700 automated sequencer. Sequence reads were assembled using the Staden sequence analysis programs PREGAP4 and GAP4 (53), run using the LINUX operating system. In the latter stages of assembly, sequence data for the gaps between contigs were obtained via PCR amplification from virion DNA of fragments spanning the gap regions that were then cloned into M13mp19 and sequenced as described above (a minimum of three independent M13 clones were sequenced for each PCR-amplified region). In the final assembly, each nucleotide of the V592 sequence was determined, on average, 5.27 times. The sequences of the genomic termini of V592 were not specifically determined in this study but rather were assumed to be identical to those of the previously sequenced Ab4 strain.
Preparation of DNA from field isolates.
DNA samples were prepared from a panel of isolates originating from a number of different countries, recovered from outbreaks of either neurological or nonneurological disease. DNA was prepared from either a pooled tissue sample or infected cells (rabbit kidney [RK-13], equine dermal [NBL-6], or primary equine [EEL or equine fetal kidney] cells) by standard proteolytic digestion using a High Pure PCR template preparation kit (Roche Diagnostics GmbH, Mannheim, Germany). Briefly, for DNA extraction from pooled tissue samples, approximately 25 mg of tissue was ground in 2 ml of a 2% minimum essential medium (Eagle) solution by using a glass homogenizer; to 200 μl of this solution 200 μl lysis buffer and 40 μl proteinase K were added, and this mixture was incubated for 1 h at 55°C. Binding buffer (200 μl) was added, and the mixture was further incubated for 10 min at 72°C. Isopropanol (100 μl) was added, and the mixture was transferred to the upper reservoir of a combined High Pure filter tube-collection tube assembly. The DNA sample was then purified by a series of rapid wash and spin steps and eluted by the addition of 200 μl prewarmed elution buffer. For DNA extraction from either EEL or RK-13 cells, a 1-ml cell suspension (approximately 106 cells/ml) was centrifuged for 5 min at 8,000 rpm and the cells were resuspended in 200 μl phosphate-buffered saline. Binding buffer (200 μl) and proteinase K (40 μl) were added to the resuspended cell pellet, and the mixture was incubated for 10 min at 72°C. Isopropanol (100 μl) was added, and DNA was purified as described above.
PCR amplification and sequencing of selected variable loci of field isolates.
PCR primers used for amplification and sequencing of selected loci are detailed in Table 1. The PCR mix (50 μl) consisted of 0.3 μM of each primer (Proligo, France), 0.2 mM of each deoxynucleoside triphosphate (Applied Biosystems), 3× PCRx Enhancer solution (Invitrogen, United Kingdom), and 1.25 U AmpliTaq DNA polymerase (Applied Biosystems) in 10 mM Tris-HCl (pH 8.3) solution containing 1.5 mM MgCl2 (Applied Biosystems). The PCR was denatured for 4 min at 94°C and then cycled for 35 cycles for 30 s at 94°C, 1 min at the annealing temperature of the primers used, and 2 min at 72°C, followed by a final step of 10 min at 72°C. After cycling, 10 μl of each PCR product was analyzed on a 1.0% agarose gel containing ethidium bromide. Following product identification, the PCR products were purified using a QIAquick PCR purification kit (QIAGEN Ltd., Sussex, United Kingdom) and quantified on a 2% agarose gel containing ethidium bromide, with DNA quantification standards (Gibco BRL, Life Technologies, United Kingdom).
TABLE 1.
PCR amplification and sequencing primers
| ORF | Amplification primers
|
Sequencing primer(s)
|
||
|---|---|---|---|---|
| Name | Sequence (5′-3′) | Name | Sequence (5′-3′) | |
| 8 | 8f | AATTATTGGTGGGCAGAACC | 8s | CCATCACCACGCTATTCTC |
| 8r | TTCTCCAGCACAGCAACG | |||
| 11 | 11f | CTGTAACGATGCTAACGCTACG | 11sf1 | TAGCCCAAACGACGACAAAG |
| 11r | TTCAGAAACCTGTCCAGGTCC | |||
| 15 | 15f | CAAAGCGCAAAGCAAAATGC | 15sf1 | ATCCAACACACGCAACATC |
| 15r | GGGCCTTGTAGATCAGCTCTCC | |||
| 30 | 30f1 | GCGCTACTTCTGAAAACG | 30-1s | TACCCAAGCATTATCCAG |
| 30r1 | CCACAAACTTGATAAACACG | 30-1sr1 | GATAACCCTGACGGAGTAAG | |
| 30f2 | AAACCACAAGCCTTACTCC | 30-2s | GAGAAGACCTTTCAGCGAC | |
| 30r2 | GCGCAACTATCACATACG | 30-2sr1 | CTCAGCAGTCATAACGAAC | |
| 31 | 31f | GGACTTACCGTCGAACAGACC | 31sr1 | GGAGACAGGTTGAGAGAAG |
| 31r | ACCCAGTACGTAGCCATGAAGC | 31sr | CAGCCTCGCAAGCGCGGGTGAGGT | |
| 32 | 32f | TGATTACTACTACCCCCCTCC | 32sf1 | TATTTCTGACACCCACCAC |
| 32r | AGCCAGATTCACGTCATCC | |||
| 37 | 37f | ACATACTCCGCCATCTCC | 37s | CCATCTATAACGCCAGCC |
| 37r | ACAGCTACATGACTCGTGC | |||
| 40 | 40f | ATCACAAACGCATCCTCG | 40s | ACTCGCCGAATCCCAGATG |
| 40r | CACAGCTCACCCAAAATACC | |||
| 42 | 42f1 | ATCCCAGCGCAACTCTTAC | 42fs1 | GGCAAGTATAACCTGAACGG |
| 42r1 | GTCCGTAGAGCTGAGTAGAG | 42rs1 | TGAAACAGCGAACACGGGTC | |
| 45 | 45f | TGGCACGAAAGCACTACG | 45sr1 | TGATCTATCAGAGCCAAC |
| 45r | AAACCTGCACGAATGTTGG | |||
| 50 | 50f | ACTCGACTCAGACTACATTGG | 50sr1 | GCTCACTAAATAGCCAAAC |
| 50r | ATAGCAGAATGGGGATAGC | |||
| 52 | 52f | CCAACAGCTTTCACATCACC | 52s | GCTACTTCGTTCACTTCC |
| 52r | TTCAAATTCCTCGCTCACC | |||
| 57 | 57f1 | GGTGCTGAACGTAAACCTG | 57fs1 | CGTAAACCTGAGCGAG |
| 57r1 | CTCAGGGCTGCTAATATATGC | 57rs1 | GTAGAAACCTTGACGACACGG | |
| 67 | 67f | CAAAACTCCAACACCACG | 67s | GAAGAAGCATTCGCCAAG |
| 67r | GAGAAGAGGTTCTACAAGCAGC | |||
| 68 | 68f | AGCATTGCCAAACAGTTCC | 68s1 | GAAGATAGAATGGGTGTGAG |
| 68r | CAAGAAACCACTGCTCAACC | 68s3 | ACCGTTGAGCATAATCATCC | |
| 68r2 | GTCCCCTACCTTTTAACG | |||
| 68r3 | CGGCGGCCGTGGACCGA | |||
| 73 | 73f | GAACATATGAGCATGTACC | 73s | ACGGACAACGCTTACCCAC |
| 73r | GACCAGTAGAACATCTTCC | |||
Sequencing reactions and sequence analysis.
The purified PCR products were directly sequenced using ABI sequencing reagents (dRhodamine or Big Dye) according to the manufacturer's instructions and EHV-1 specific sequencing primers (Table 1). The extension products were purified by a series of ethanol washes, and the purified products were analyzed using either an ABI 3700 or an ABI 3100 automated sequencer. Sequences were assembled using the DNASTAR program SeqManII (version 4.03; DNASTAR, Inc.). Following sequence assembly, further analysis, in particular, identification of coding changes and multiple alignment of homologous sequences, was carried out using the OMIGA software package (version 2.0; Oxford Molecular Ltd., United Kingdom), BioEdit (version 7.0.1.4 [21]), or MEGA2 (26).
Statistical methods.
Fisher's exact test was used to test the null hypotheses that there were no differences in the relative proportions of open reading frame 68 (ORF68) groups between (i) isolates originating from either Europe or North America and (ii) isolates that were from outbreaks characterized clinically as either neurological or nonneurological. The same test was also used to test the null hypothesis that there was no difference in the relative proportions of specific single nucleotide polymorphisms (SNPs) between isolates that were from neurological or nonneurological outbreaks.
Nucleotide sequence accession numbers.
The nucleotide sequences determined in this study were submitted to GenBank and assigned accession numbers as follows: for the EHV-1 strain V592 complete genome, AY464052; for the EHV-1 isolate ORF30 sequences, DQ180606 to DQ180738; and for the EHV-1 isolate ORF68 sequences, DQ172308 to DQ172415.
RESULTS
Comparison between the genomic sequences of Ab4 and V592.
The V592 genomic sequence (149,430 bp) was determined via shotgun cloning and sequencing of DNA prepared from purified virions. The V592 stock used to generate DNA for sequencing had not been plaque purified and contained a mixture of DNA populations at certain sites. It should be noted that the determined sequence is the consensus for the majority of sequence reads. Sites with heterogeneity between individual sequencing templates are shown in Table 2. These sites are all regions of variable repeat length, apart from one position of single-nucleotide heterogeneity.
TABLE 2.
Sequence heterogeneity between individual sequence templates of V592
| Positiona | ORF | Variation |
|---|---|---|
| 105-308 | NAb | 10-13 copies of 17-bp repeat element |
| 366-707 | NA | 14-19 copies of 18-bp repeat element |
| 1959 | 2 | Substitution of A to G in minority of reads (2/9) |
| 73803-74090 | NA | 8-9 copies of 32-bp repeat element |
| 108895-108984 | NA | 4-5 copies of 18-bp repeat element |
| 112316-112419 | NA | 8-9 copies of 13-bp repeat element |
| 123549-123656 | NA | 2-4 copies of 27-bp repeat element |
| 129328-129552 | 71 | 17-18 copies of 15-bp repeat element |
Nucleotide positions are numbered according to the V592 genomic sequence.
NA, not applicable.
The V592 sequence was compared with that of Ab4 (56) to identify all positions of sequence variation and to determine amino acid coding changes for known ORFs. Overall, there was a nucleotide variation rate of approximately 0.1%, with 50 regions of insertion or deletion of 1 or more nucleotides and 110 single nucleotide substitutions, of which 43 resulted in amino acid coding changes. A total of 31 of 76 ORFs had amino acid variations between the two strains, as shown in Table 3. Two ORFs (ORF24 and ORF71) showed variable copy numbers of short nucleotide direct repeat elements, in agreement with previous studies that had identified variation in repeat element copy numbers for ORF24 and ORF71 among EHV-1 field isolates (8, 22, 25, 32). Such regions are relatively unstable and therefore of limited use for epidemiological studies. This instability is apparent from the variable copy numbers of several of these repeat regions identified during sequencing of V592 (Table 2). Furthermore, PCR amplification of several regions of variable lengths, including those present in ORF24, demonstrated variable lengths of PCR products upon comparison of individual isolates derived from the same outbreak (data not shown).
TABLE 3.
Nonsynonymous differences between V592 and Ab4
| ORF | Positiona | Variation(s) in:
|
|
|---|---|---|---|
| Ab4 sequence | V592 sequence | ||
| 2 | 59 | G | D |
| 5 | 114 | G | V |
| 8 | 114 | D | N |
| 11 | 189 | Q | K |
| 13 | 305 | S | L |
| 460 | A | T | |
| 14 | 619-621 | *c | PSR (9-bp duplication resulting in 3-amino-acid insertion) |
| 15 | 166 | D | N |
| 22 | 430 | S | P |
| 24 | 2567-2568 | (GPPPTLPPAPPL PQSTSKAAS)2 | *(126-bp deletion [2 copies of 63-bp repeat element] resulting in 42-amino-acid deletion) |
| 2586 | G | S | |
| 2855-2862 | * | DQAKDQAK (24-bp insertion [2 copies of 12-bp repeat element] resulting in 8-amino-acid insertion) | |
| 2904 | E | K | |
| 2914-2928 | * | TGAVPENTPLPDDSP (45-bp insertion resulting in 15-amino-acid insertion) | |
| 3099 | T | A | |
| 29 | 12 | T | K |
| 30 | 752 | D | N |
| 990 | E | K | |
| 31 | 90 | N | S |
| 32 | 42 | S | L |
| 33 | 15 | N | H |
| 976 | N | D | |
| 34 | 66 | D | G |
| 36 | 47 | S | R |
| 37 | 265 | A | V |
| 39 | 440 | S | L |
| 40 | 196 | R | H |
| 42 | 1275 | K | R |
| 45 | 427 | E | G |
| 46 | 140 | F | S |
| 50 | 367 | P | S |
| 52 | 386 | A | V |
| 57 | 804 | K | R |
| 64 | 73 | T | A |
| 648 | T | S | |
| 67 | 261 | S | F |
| 68 | 210 | R | H |
| 247 | D | Md | |
| 71 | 226-227 | SS | TA |
| 231-299 | * | TAATTTAATTSSATTAATTSS (TTTAA)9TTT (additional copies of 15-bp repeat elements and nucleotide substitutions, resulting in 69-amino-acid insertionb | |
| 73 | 122 | A | V |
| 76 | 128 | F | S |
ORF amino acid positions are numbered according to the V592 amino acid sequence.
Repeat element with variable copy number in V592; sequence present in majority of reads is shown.
*, gap in sequence.
Due to a single C deletion resulting in frameshift relative to Ab4. The resulting polypeptide is 303 amino acids long (cf. 418 for Ab4).
ORF14 of V592 had a 9-bp duplication, resulting in the insertion of three amino acids. The majority of field isolates tested encoded an equivalent 9-bp repeat (J. Nugent and N. Davis-Poynter, unpublished data). ORF68 (homologous to herpes simplex virus type 1 [HSV-1] Us2 and related genes of other alphaherpesviruses) displayed the most significant coding change, with a single nucleotide deletion in V592 (G8 in Ab4, G7 in V592) resulting in a frameshift and hence multiple coding changes and premature truncation compared with Ab4. Such a frameshift had previously been noted from analysis of several other EHV-1 isolates (34). The other variable ORFs possessed minor coding changes between Ab4 and V592, usually comprising a single amino acid substitution.
Evaluation of sequence variation of multiple loci for a panel of EHV-1 field isolates.
As noted above, EHV-1 infection results in outbreaks of various levels of severity, which may either be restricted to respiratory disease or include abortion and/or neurological disease. Previous evidence had indicated genetic variability of EHV-1 isolates but had not demonstrated conclusively the cocirculation of genetically distinct strains of EHV-1. We assembled DNA samples prepared from a panel of field isolates recovered from outbreaks that occurred in various countries, collected over the course of approximately 30 years. PCR procedures to amplify selected regions of ORFs found to vary in predicted amino acid sequence between Ab4 and V592 were developed by using the amplification primers listed in Table 1. The purified PCR products were then sequenced using the respective sequencing primers (Table 1).
ORF68 as the primary strain grouping marker.
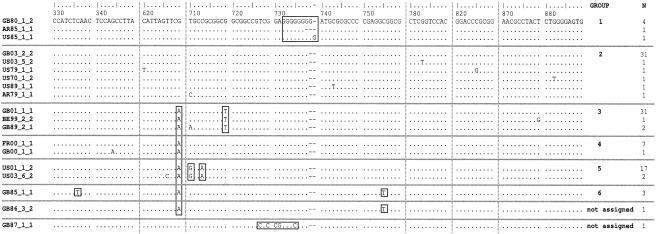
A region of ∼600 bp in ORF68 was particularly polymorphic and so was tentatively adopted as a marker system for efficiently distinguishing isolates without having to type multiple loci. ORF68 sequences from 106 field isolates (each from a different outbreak) and two tissue culture attenuated vaccine strains (RacH [PL68_1_0] [30, 31, 35] and the component of the vaccine Rhinoquin [US72_1_0] [Jensen-Salsbury] [45, 46, 47]) could be categorized into six common strain groups (Table 4). A sequence alignment of representative isolates for the various ORF68 sequences determined is shown in Fig. 1. Ab4 (GB80_1_2) is located in group 1, and V592 (GB85_1_1) is located in group 6. Group 1 is distinguished from group 2 only on the basis of the number of G residues in a homopolymeric tract (nucleotides [nt] 732 to 739). Within group 1, isolates encode G6 (AR85_1_1), G8 (Ab4 [GB80_1_2], GB83_1_1, GB83_3_1, and GB93_4_2), or G9 (US85_1_1) in this region, whereas group 2 and all other groups encode G7. Polymorphism in this region was originally identified by Meindl and Osterrieder (34), who demonstrated that Ab4 (G8) expressed a longer form of the protein than isolates with G7. Our data indicate that the ORF68 sequence for Ab4 is atypical and that the majority of EHV-1 isolates encode a shorter form of the protein than that expressed by Ab4. Isolates in groups 3 to 6 all encode A629, compared with G629 for groups 1 and 2. Additional SNPs characteristic of each group are indicated in Fig. 1, namely, group 3 (T719), group 5 (G710 and A713), and group 6 (T336 and T755). Isolate GB86_3_2 encodes A629 and T755 and has not been assigned a group, since it may be considered a member of either group 4 or group 6, differing by a single substitution from the consensus sequence for either group. Isolate GB87_1_1 is similar to isolates from group 2 but has five substitutions in the region from nt 727 to 735, none of which are seen with any other isolate. This isolate may represent a group distinct from groups 1 to 6; however, in the absence of other isolates sharing the signature 5 substitutions it has not been assigned a group.
TABLE 4.
EHV-1 isolate groups according to ORF68 sequence
| Group no. | Neurological isolate(s) | Nonneurological isolate(s) |
|---|---|---|
| 1 | GB80_1_2, GB83_4_2 | GB83_1_1, GB83_3_1, US85_1_1, AR85_1_1 |
| 2 | AR91_2_2, AU82_1_2, GB79_1_2, GB99_1_2, GB03_2_2, US85_2_2, US86_2_2, US86_3_2, US03_1_2, US03_2_2, US03_3_2, US03_4_2, US03_7_2, US70_1_2, US03_5_2 | AR79_1_1, AR90_1_1, AR90_2_1, AR91_1_1, AR91_3_1, AR96_1_1, AR00_1_1, FR97_1_1, FR99_1_1, FR99_2_1, FR99_5_1, GB86_2_1, US76_1_1, US79_1_1, US83_2_1, US84_1_1, US85_4_1, US89_1_1, US90_1_1, US99_1_1, US00_1_1 |
| 3 | BE95_1_2, BE99_2_2, BE03_1_2, FR02_1_2, GB88_2_2, GB03_3_2, GB03_4_2, GB03_5_2, GB04_10_2, GB04_11_2, NL95_1_2, NL99_1_2 | AU02_1_1, BE94_1_1, BE97_1_1, BE99_1_1, GB83_2_1, GB89_1_1, GB89_2_1, GB91_2_1, GB93_2_1, GB00_2_1, GB01_1_1, GB03_1_1, GB02_1_1, GB04_1_1, GB04_3_1, GB04_5_1, GB04_14_1, GB05_1_1, US86_1_1, US90_2_1, US90_3_1, US92_1_1 |
| 4 | GB04_12_2, GB04_13_2, GB04_15_2, US86_4_2 | FR00_1_1, GB81_2_1, GB81_3_1, GB00_1_1 |
| 5 | CA89_1_2, US41_1_2, US82_1_2, US83_3_2, US99_2_2, US99_3_2, US01_1_2, US02_1_2, US03_6_2, US03_9_2, US03_8_2, US03_10_2, US03_11_2 | PL68_1_0, US72_1_0, US75_1_1, US81_1_1, US85_3_1, US01_2_1 |
| 6 | GB85_1_1, GB91_1_1, GB01_2_1 | |
| Not assigned | GB86_3_2 | GB87_1_1 |
FIG. 1.
Regions of sequence variation for ORF68 are shown, with a representative isolate for each different sequence type displayed. N indicates the number of independent isolates identified with each particular sequence. Sequences are aligned with reference to the prototype Ab4 (GB80_1_2) sequence (GenBank accession number AY665713), with dots indicating sequence identity and dashes indicating gaps in the alignment. Nucleotide positions are indicated above the alignment, in accordance with the Ab4 (GB80_1_2) sequence. Vertical dotted lines indicate breaks between contiguous blocks of sequence. Nucleotide positions characteristic of each ORF68 group are indicated by boxes (further details in text). Isolate GB86_3_2 has one of the two nucleotides (T755) which distinguish between group 4 and group 6 and cannot be assigned to either of these groups. Isolate GB87_1_1 has a stretch of five nucleotide substitutions (boxed), none of which are seen with any other isolate. This may be a representative of a separate group, but in the absence of other isolates sharing the signature five substitutions, this isolate has not been assigned to a group.
Analysis of the ORF68 strain distribution according to geographical origin of the isolates provided evidence for geographical restriction of certain strain groups (Table 5). Isolates from 104 disease outbreaks from each of the six major strain groups were analyzed; the tissue culture attenuated vaccine strains RacH (PL68_1_0) and the component of the vaccine Rhinoquin (US72_1_0) and the two isolates not assigned to a group (GB86_3_2 and GB87_1_1) were excluded from the analysis. There was a statistically significant heterogeneity between North America and Europe in the assignment of outbreaks to groups (P < 0.001 by Fisher's exact test): all 17 group 5 field isolates came from outbreaks in North America, whereas the majority (36/42) of group 3 and group 4 isolates were from outbreaks in Europe. No single strain group was found to be responsible for outbreaks of neurological disease, with neurological disease represented in five of the six groups. Overall, 45% of the outbreaks presented neurological disease, and this is reflected in the proportion of neurological outbreaks in most of the groups (30 to 50% for groups 1 to 4). Group 5 had the highest proportion of neurological outbreaks (76%), while group 6 (the smallest group, with three outbreaks) included only nonneurological outbreaks.
TABLE 5.
ORF68 group distribution according to outbreak origin and disease characteristics
| ORF68 group | No. of isolates
|
|||||
|---|---|---|---|---|---|---|
| Geographical origin
|
Outbreak disease characteristics
|
|||||
| North America | South America | Australia | Europe | Neurological | Nonneurological | |
| 1 | 1 | 1 | 0 | 4 | 2 | 4 |
| 2 | 19 | 8 | 1 | 8 | 15 | 21 |
| 3 | 4 | 0 | 1 | 29 | 12 | 22 |
| 4 | 1 | 0 | 0 | 7 | 4 | 4 |
| 5 | 17 | 0 | 0 | 0 | 13 | 4 |
| 6 | 0 | 0 | 0 | 3 | 0 | 3 |
Comparison of ORF68 grouping with SNPs of other ORFs.
A panel of field isolates recovered from 12 neurological and 13 nonneurological disease outbreaks, and with representatives from each major strain group, was analyzed for SNPs for a number of ORFs distributed across the genome (Table 6). Each ORF was sequenced across the region with a coding difference between Ab4 and V592 by use of the primers detailed in Table 1. Extra SNPs were detected in some ORFs, and these are shown if present in more than one isolate. Notably, where multiple isolates from the same outbreak were analyzed, consistent sequences were found for all markers tested (data not shown).
TABLE 6.
Analysis of SNPs of 15 ORFs for a panel of EHV-1 field isolates
| Isolate | Group | SNP of ORF at nt positiona
|
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ORF8 | ORF11 | ORF15 | ORF30
|
ORF31
|
ORF32
|
ORF37
|
ORF40
|
ORF42
|
ORF45
|
ORF50
|
ORF52
|
ORF57
|
ORF67
|
ORF73
|
|||||||
| 340 | 565 | 496 | 2254 | 2279 | 2968 | 269 | 125 | 794 | 498c | 587 | 3824 | 1280 | 1099 | 1157 | 2411 | 2426c | 759c | 782 | 365 | ||
| GB80_1_2 | 1 | G | C | G | G | A | G | A | C | C | C | G | A | A | C | C | A | T | G | C | C |
| GB93_4_2 | G | C | G | G | A | G | A | C | C | C | G | A | ND | ND | C | A | T | G | C | C | |
| GB83_1_1 | G | C | G | A | A | G | A | C | C | C | G | A | A | C | C | A | T | G | C | C | |
| US85_1_1 | G | C | G | A | A | G | A | C | NDb | C | G | A | A | C | C | A | T | G | C | C | |
| US70_1_2 | 2 | G | C | G | G | A | G | A | C | C | C | G | A | A | C | C | A | T | ND | ND | C |
| US85_2_2 | G | C | G | A | A | G | A | C | C | C | G | A | A | ND | C | A | T | G | C | C | |
| GB99_1_2 | G | C | A | G | A | ND | ND | C | ND | ND | ND | A | G | C | ND | A | T | ND | ND | C | |
| US99_1_1 | ND | C | ND | A | A | G | A | ND | T | C | G | A | A | C | ND | ND | ND | G | C | C | |
| GB81_4_1 | G | C | G | A | A | G | A | C | C | C | G | A | A | ND | C | A | T | G | C | C | |
| NL95_1_2 | 3 | G | C | A | G | A | G | G | C | T | C | G | A | G | C | T | G | T | ND | ND | C |
| GB88_2_2 | G | C | A | G | A | G | G | C | T | C | G | A | G | C | T | G | T | G | C | C | |
| GB89_1_1 | G | C | A | A | A | G | G | C | T | C | G | A | ND | ND | T | G | T | G | C | C | |
| GB91_2_1 | G | C | ND | A | A | G | ND | ND | T | C | G | ND | ND | ND | ND | ND | ND | G | C | ND | |
| US86_4_2 | 4 | G | C | A | G | A | G | A | C | T | C | G | A | G | C | T | A | C | G | C | C |
| FR00_1_1 | G | ND | ND | G | A | ND | A | C | ND | ND | ND | A | G | C | ND | A | C | ND | ND | C | |
| GB81_3_1 | G | C | A | A | A | G | A | C | T | C | G | A | G | C | T | A | C | G | C | C | |
| GB00_1_1 | G | C | A | A | A | G | A | C | T | C | G | A | G | C | ND | A | C | ND | ND | C | |
| US99_2_2 | 5 | G | C | A | G | A | G | A | C | T | C | G | A | G | C | T | A | T | A | C | C |
| US99_3_2 | G | C | A | A | G | G | A | C | T | C | G | A | G | ND | T | A | T | A | C | C | |
| US02_1_2 | G | C | A | A | G | G | A | C | T | C | G | A | G | C | T | A | T | A | C | C | |
| US75_1_1 | G | C | A | A | A | G | A | C | T | C | G | A | G | C | T | A | T | ND | ND | C | |
| GB91_1_1 | 6 | A | A | A | A | A | A | G | T | T | T | A | ND | G | C | T | G | T | G | T | T |
| GB01_2_1 | A | A | A | A | A | A | G | ND | T | T | A | G | G | C | T | G | T | ND | ND | T | |
| GB85_1_1 | A | A | A | A | A | A | G | T | T | T | A | G | G | T | T | G | T | G | T | T | |
| GB86_3_2 | Not assigned | G | C | A | ND | ND | G | G | C | T | C | G | A | G | C | T | G | T | G | C | C |
For each ORF, the nucleotide position of the SNP is indicated relative to the start of the ORF, according to the Ab4 (GB80_1_2) sequence (56). Nucleotides differing from the Ab4 sequence are highlighted in bold.
ND indicates the sequence was not determined.
Three SNPs (nt 498 of ORF40, nt 2426 of ORF57, and nt 759 of ORF67) did not result in amino acid coding changes.
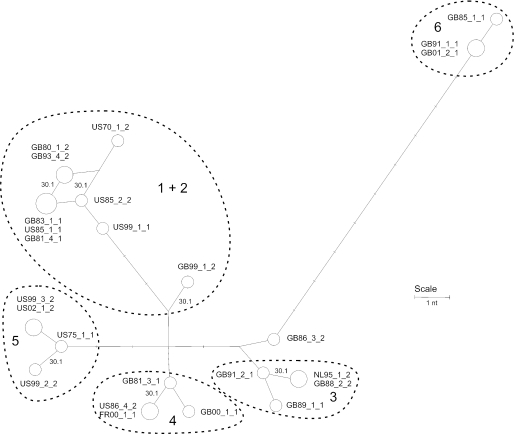
Data from ORF68 and the genome-wide SNP typing is presented as a network (Fig. 2) (5). The available data set was incomplete, and sequencing of further isolates and SNPs is under way to conduct a more thorough analysis, but the following preliminary inferences about the evolution and epidemiology of EHV-1 may be drawn. First, the pooled data are consistent with a tree-like genealogy (with the exception of a single repeated substitution of ORF30 [see below]), indicating that there is currently no evidence for recombination among the isolates in the representative panel. Second, the genome-wide data substantiate the ORF68-based classification, except for groups 1 and 2, which are not distinguished by any further SNPs and are superimposed on the network. Third, group 6 is quite divergent from the other strains analyzed, with nine further distinctive substitutions, namely, with ORF8 (A340), ORF11 (A565), ORF30 (A2968), ORF32 (T125), ORF40 (T498 and A587), ORF42 (G3824), ORF67 (T782), and ORF73 (T365). Other groups had their characteristic SNPs, but none was as divergent as group 6. V592 (GB85_1_1), a member of group 6, had an extra substitution in ORF50 (T1099) that was not observed with any other isolate. Analysis of two independent isolates from the same outbreak confirmed that this SNP is characteristic of the outbreak, rather than being an adventitious mutation of V592 (data not shown). Three isolates could not be reliably assigned to groups or were reassigned based on combined data. Based on ORF68, isolate GB86_3_2 had differed by a single substitution from the consensus sequence of either group 4 or group 6 and by two substitutions from that of group 3. Interestingly, genome-wide data placed this isolate nearest to group 3 isolates. Using SNP data, two group 2 isolates (GB99_1_2 and US99_1_1) could be distinguished from the rest of groups 1 and 2.
FIG. 2.
Reduced median network for panel of EHV-1 isolates. Polymorphism data were derived from a set of ORFs containing at least one nonsynonymous substitution and from ORF68 sequences in a panel of 25 isolates representing neuropathogenic and nonneuropathogenic outbreaks and all six ORF68 groupings. Isolates are labeled as shown in Table 6, and the area of each circle is in proportion to the frequency of isolates sharing the same genotype. The positions of substitutions at ORF30 nt 2254 are shown (30.1’), and other substitutions are to scale but unlabeled. The parallelogram at top left indicates the alternative mutational paths involving two variable sites. Groupings based on ORF68 are indicated by dashed lines.
ORF30 sequence variation is associated with pathogenic potential.
Strikingly, variation at a single nucleotide position among the typed SNPs was inconsistent with the simple pattern of rare unique mutations seen across the EHV-1 genome. Using the network analysis described above, approximately six distinct, independent mutations can be inferred at nt 2254 of ORF30 (encoding the catalytic subunit of the DNA polymerase). In contrast to all of the other SNPs detected, disease severity was more closely related to the genotype of an isolate at this position than to its genetically more stable ORF68 group membership. Fisher's exact test was used to assess the statistical association with neurological disease of a total of 51 SNPs from 22 ORFs (including the ORFs specified in Table 6 and seven additional ORFs: ORF2, -5, -13, -14, -29, -34, and -36) and of the six ORF68 groups. Each SNP had data available from at least 10 neuropathogenic and 10 nonneuropathogenic outbreaks. Only ORF30 at nt 2254 had a significant association (P < 0.01) with neurological/nonneurological outbreak status. ORF30 sequences (nt 2200 to 2450) were analyzed for a panel of 131 field isolates, comprising 82 from outbreaks where no neurological disease was reported and 49 from outbreaks including at least one case of neurological disease (Table 7). Seventy-eight (95%) of the nonneurological isolates encoded A2254 (amino acid N752), whereas 42 (86%) of the neurological isolates encoded G2254 (amino acid D752). This distribution was found to be highly significant statistically (P < 0.0001 by Fisher's exact test). Although we cannot exclude the possibility that other SNPs (alone or in combination) may show an association with disease severity, it is clear that alleles at the SNP of ORF30 at nt 2254 are strongly associated with disease phenotype but are not associated with group membership defined by ORF68 and supported by other SNPs.
TABLE 7.
Change in amino acid coding identified for ORF30 that correlates with distribution of isolates according to EHV-1 disease severity
| Significant amino acid in ORF30 sequence at position 752a (D/N) | Amino acid encoded at positionb:
|
No. of isolates from outbreaks of EHV-1 disease severity
|
||||
|---|---|---|---|---|---|---|
| 753 (Y/S) | 760 (D/G) | 769 (H/N) | Neurological | Nonneurological | Attenuated | |
| D | Y | D | H | 42 | 4 | 1 |
| D | S | D | H | 0 | 0 | 1 |
| N | Y | D | H | 4 | 76 | 0 |
| N | Y | G | H | 3 | 1 | 0 |
| N | Y | D | N | 0 | 1 | 0 |
Amino acid coding change D/N752 corresponds to nucleotide change G/A2254.
Amino acid coding change Y/S753 corresponds to nucleotide change A/C2258, D/G760 corresponds to nucleotide change A/G2279, and H/N769 corresponds to nucleotide change C/A2305.
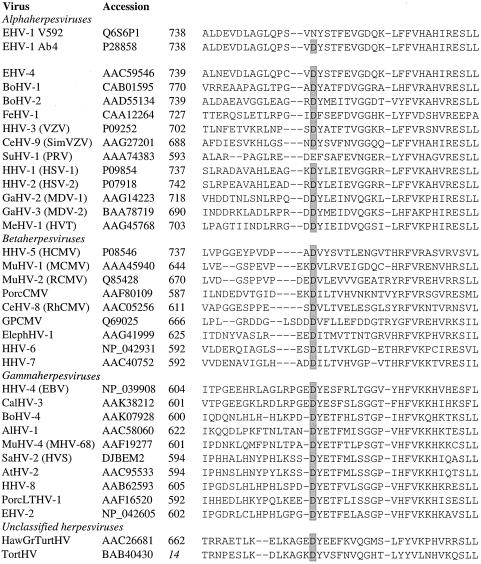
Interestingly, EHV-1 ORF30 amino acid 752 was found to correspond to a highly conserved position for herpesvirus DNA polymerases, as shown in Fig. 3. Remarkably, this position was found to be conserved as an acidic residue (aspartate, with the exception of pseudorabies virus [glutamate]), followed by a hydrophobic residue, for all mammalian herpesvirus polymerases evaluated, including representatives of alpha-, beta-, and gammaherpesviruses. This conservation also extended to unclassified herpesviruses from nonmammalian species (tortoise and turtle). The presence of a polar, neutral residue (asparagine) at this position for the majority of EHV-1 isolates tested is therefore highly unusual.
FIG. 3.
An alignment of amino acid sequences for various herpesvirus DNA polymerases is shown, generated using ClustalW (run using the BioEdit software [21]). For each sequence, the name of the virus, database accession number, and residue number of the first amino acid are shown (the residue number in italics refers to a partial polymerase sequence). The conserved aspartate residue, corresponding to D/N752 of the EHV-1 polymerase, is highlighted by gray shading. BoHV-1, bovine herpesvirus 1; FeHV-1, felid herpesvirus 1; HHV-3, human herpesvirus 3; CeHV-9, cercopithecine herpesvirus 9; SimVZV, simian VZV; SuHV-1, suid herpesvirus 1; PRV, pseudorabies virus; GaHV-2, gallid herpesvirus 2; MDV-1, Marek's disease virus type 1; MeHV-1, meleagrid herpesvirus 1; HVT, herpesvirus of turkeys; MuHV-1, murid herpesvirus 1; MCMV, mouse cytomegalovirus; RCMV, rat cytomegalovirus; PorcCMV, porcine cytomegalovirus; RhCMV, rhesus monkey cytomegalovirus; GPCMV, guinea pig cytomegalovirus; ElephHV-1, elephantid herpesvirus 1; EBV, Epstein-Barr virus; CalHV-3, callitrichine herpesvirus 3; AlHV-1, alcelaphine herpesvirus 1; MHV-68, mouse herpesvirus strain 68; SaHV-2, saimiriine herpesvirus 2; HVS, herpesvirus saimiri; AtHV-2, ateline herpesvirus 2; PorcLTHV-1, porcine lymphotropic herpesvirus 1; EHV-2, equine herpesvirus 2; HawGrTurtHV, Hawaiian green turtle herpesvirus; and TortHV, tortoise herpesvirus.
Two attenuated vaccine strains were also analyzed for their ORF30 sequences (Table 7). The component of the vaccine Rhinoquin (US72_1_0) was found to have an ORF30 sequence in this region that was identical to that of Ab4 and the majority of isolates from neurological disease outbreaks. The second vaccine strain, RacH (PL68_1_0), was found to encode a sequence not seen in any other field isolate tested (D752 S753). Additional sequence variation was noted to occur at nt 2279 (A/G) and 2305 (C/A) in a minority of the isolates tested, corresponding to amino acid coding changes G760 and N769, respectively (Table 7). The four isolates encoding G760 were from different ORF68 strain groups, two from group 3 (BE95_1_2 and GB04_8_1) and two from group 5 (US99_3_2 and US02_1_2). Three of these isolates were from outbreaks of neurological disease, and one was from a single case of abortion without neurological signs. The variant N769 was identified for isolates from an outbreak of abortion without neurological signs (GB01_2_1, group 6).
DISCUSSION
EHV-1 is associated predominantly with outbreaks of abortigenic disease and more rarely with outbreaks of neurological disease. For many years, it has been suspected that distinct strains of EHV-1 may be responsible for outbreaks of neurological disease. However, previous molecular epidemiology studies, although demonstrating genetic heterogeneity (e.g., via RFLP analyses), had failed to identify genetic markers associated with neuropathogenic disease outbreaks. The starting point for this study was the determination of all nucleotide differences between two strains of EHV-1, Ab4 (high frequency of abortion, neuropathogenic) and V592 (low frequency of abortion, nonneuropathogenic), which have been shown by experimental infection studies with ponies to exhibit markedly different pathogenicities. Following determination of the complete genomic sequence of V592 and comparison with the previously determined sequence of Ab4 (56), we sought to test the following hypotheses. (i) Positions of sequence variation between Ab4 and V592 are indicative of regions of sequence variability among EHV-1 field isolates and will therefore provide markers enabling discrimination between EHV-1 strains. (ii) Ab4 is representative of a distinct group of genetically closely related EHV-1 strains capable of causing neurological disease. (iii) One or more specific sequence markers that vary between Ab4 and V592 are indicative of strains capable of causing neurological disease.
Overall, there was relatively low variation between the Ab4 and V592 sequences, with approximately one variable position per 1,000 bp. The majority of changes were point mutations, involving single nucleotide substitutions. There were also several small deletions or insertions, involving 1 or 2 nucleotides, together with larger deletions and insertions involving a contraction or expansion of short direct repeat elements. Analysis of the larger regions of variable repeat length were of limited value for typing of strains, due to their inherent instability, agreeing with previous studies (2, 8, 22, 25, 32).
On the assumption that the pathogenic differences between Ab4 and V592 are the result of sequence changes in one or more viral proteins, we evaluated SNPs for several ORFs, found to vary in coding sequence between Ab4 and V592, for a panel of EHV-1 isolates. A region of ORF68 was found to be the most variable, with 19 distinct sequences observed, and was used as the primary marker for grouping isolates into six major strain groups (Table 4; Fig. 1). A frameshift in ORF68 had previously been identified (34), due to the presence of either 7 or 8 G residues in a homopolymeric tract, with Ab4 (G8) expressing a longer form of the protein (418 amino acids) than other isolates (303 amino acids). In this study, additional variants were identified with either G6 or G9 in this region. Isolates encoding G6, G8, or G9 were all assigned to ORF68 group 1 and were closely related in ORF68 sequence (and other SNPs [see below]) to group 2 isolates. The majority of field isolates were found to encode G7 and are therefore predicted to express the 303-amino-acid form of the polypeptide. ORF68 is homologous to Us2 of HSV-1 and related genes in other alphaherpesviruses (56). The region of homology between ORF68 and Us2 lies upstream of the frameshift region, suggesting that all of the forms expressed by EHV-1 isolates are likely to retain important functional domains.
Analysis of SNPs for several other ORFs (Table 6; Fig. 2) supported the groupings assigned according to ORF68 sequence variation. Groups 1 and 2 appear to be closely related, with characteristic SNPs for ORF15, -37, -45, and -52, which were not observed with other strain groups. Group 1 is distinguished from group 2 solely on the basis of the number of Gs in a homopolymeric run, likely to be a relatively unstable marker. Groups 3, 4, and 5 shared SNPs for most of the ORFs analyzed, but SNPs distinguishing between these groups were also observed. Group 6 has unusual SNPs in several ORFs and, on the basis of the ORFs analyzed, is the most highly diverged from the other groups. Since the ORFs analyzed were selected on the basis of differences between Ab4 (group 1) and V592 (group 6), it is uncertain whether the high proportion of SNPs specific to group 6 reflects relatively early divergence of group 6 from the other groups or is due to sampling bias. These findings support the first hypothesis presented above and suggest that genetically distinct strains of EHV-1 are in circulation.
Studies of other herpesviruses have demonstrated differing degrees of sequence diversity. Clinical isolates of human cytomegalovirus (HCMV) show significant sequence diversity by RFLP analysis and sequencing of selected ORFs (10, 13, 15, 43). Furthermore, it is difficult to assign HCMV isolates into strain groups, due to a relatively high frequency of recombination, indicated by variation of gB via intragenic recombination and relative rarity of genetic linkages between nine separate variable loci (20, 48). Similarly, a study of sequence variation between clinical isolates for genes within the Us region of HSV-1 (gG, gI, and gE) concluded that homologous recombination (intra- and intergenic) was a common event (40). In contrast, varicella-zoster virus (VZV) is relatively genetically stable and has been estimated to have a sequence diversity approximately 10 times lower than that of HSV and 40 times lower than that of HCMV (36, 50, 62). Analysis of VZV strains indicates significant geographical/ethnic restriction of the circulation of strain groups and shows that recombination between different strain groups is a relatively rare event (28, 36, 58). These differences in the various herpesviruses are likely to reflect differences in their biological properties, in particular relating to their modes of transmission and frequency of reactivation.
The low level of sequence divergence (0.1%) between the Ab4 and V592 genomes and the generally good agreement between the ORF68 grouping and SNPs in other ORFs suggest that EHV-1 genomes are relatively stable, in agreement with previous assessments of EHV-1 sequence diversity (4, 32). The current study has uncovered no evidence for recombination in the isolates and SNPs so far, although more data would be required to reach a definitive conclusion about the potential significance of recombination in the virus' life cycle. The relatively low degree of sequence diversity observed for EHV-1 is similar to that of VZV. EHV-1 is also similar to VZV in showing geographical restriction of certain strain groups. Thus, EHV-1 group 5 isolates were isolated predominantly from outbreaks in North America and group 3 isolates predominantly from outbreaks in Europe. Notably, there was no obvious association between any particular ORF68 strain group and outbreaks of neurological disease, with five of the six major groups including both neuropathogenic and nonneuropathogenic isolates. It is clear that Ab4 is not representative of a group of related strains capable of causing neurological disease, thereby disproving the second hypothesis presented above.
In contrast to all of the other markers tested, an SNP in ORF30 (G/A2254) did not cosegregate according to the ORF68 strain grouping (Table 6 and unpublished data). Both forms were present in five of the six major strain groups (the exception being group 6, which comprises only three members, all of which encoded A2254). The ORF30 SNP (G/A2254), corresponding to amino acid variation D/N752 of the DNA polymerase (GAC-AAC is the only single nucleotide substitution that will result in a D-to-N change), instead showed a very strong association with neuropathogenic/nonneuropathogenic isolates, respectively. This result is in agreement with the third hypothesis presented above and demonstrates that one of the Ab4 specific alleles (ORF30 G2254) shows strong predictive value as a marker of isolates with a propensity for causing neurological disease. It was also noted that three of the seven isolates from neurological outbreaks that encoded ORF30 A2254 encoded an additional nucleotide change, A/G2279, resulting in the coding change D/G760. The single outbreak involving an isolate encoding G2279 that did not include neurological disease occurred on premises with only two pregnant mares (of which one aborted), where there were no other horses in contact with the infected horses. There was no statistically significant association of the G2279 SNP with neuropathogenicity (P = 0.066), although considering this allele occurred infrequently (4/131 isolates), analysis of additional isolates encoding this allele is required to determine whether this may be an uncommon ORF30 variant associated with increased pathogenic potential.
It is important to consider whether differences in the techniques used to generate DNA from the two different outbreak types (neuropathogenic and nonneuropathogenic) may have affected the results, in particular, the potential selection of ORF30 sequence variants in tissue culture. In this study, the majority of DNA samples were analyzed from isolates which had been prepared by limited passage (typically two to five passages) in tissue culture, with only 8/131 samples (all from abortigenic outbreaks) prepared directly from tissue samples without cell culture. There was not, therefore, a consistent difference in the techniques used to generate the panels of neuropathogenic and nonneuropathogenic samples within each laboratory submitting the samples (although the choices of cells used for virus isolation differed between the various laboratories). Where ORF30 sequences were compared between DNA samples prepared directly from tissues and samples prepared from tissue culture isolates (two neuropathogenic outbreaks), the sequences were found to be identical. Combined with the observation that where samples from more than one horse in a given outbreak were analyzed (seven neuropathogenic and five nonneuropathogenic outbreaks), all samples yielded the same ORF30 sequence, we are confident that the ORF30 D/N752 sequence variation is not an artifact of tissue culture selection.
Two tissue culture attenuated vaccine strains were characterized for their DNA polymerase sequence. One of these strains, the component of the vaccine Rhinoquin (47), was found to encode the DNA polymerase D752 sequence typical of neuropathogenic strains. Although found to be attenuated in safety and efficacy studies (45, 46), Rhinoquin was associated with paralytic disease when used as a vaccine in the United States and as a consequence was withdrawn from the market (27). It is difficult to verify whether the cases of neurological disease were directly related to administration of Rhinoquin or coincidental to the vaccine trial, but the cases showed a strong temporal link, having occurred 8 to 11 days after administration of the vaccine. It can be speculated that either Rhinoquin reverted the attenuating mutations and was then able to express neuropathogenicity or the Rhinoquin DNA polymerase sequence was introduced into wild-type field strains via recombination, resulting in neurological disease. A second tissue culture attenuated strain used as the progenitor of live vaccines (RacH [30, 31, 35]) also encoded DNA polymerase D752. In contrast to Rhinoquin, vaccines derived from RacH have not been associated with neurological disease, and such vaccines are in current use in the United States (Rhinomune; Pfizer Animal Health [6, 7]) and in Europe (Prevaccinol; Intervet [30]). It may be significant, therefore, that RacH encodes a second amino acid coding change, Y/S753, that was not identified in any of the field isolates. In this regard, it should be noted that the position corresponding to Y/S753 is conserved in other herpesviruses as a hydrophobic residue (Fig. 3); hence, the S753 mutation of RacH is a nonconservative change.
The strong association of DNA polymerase D/N752 with neurological versus nonneurological disease outbreaks suggests that the viral genotype is a major contributory factor in determining whether a disease outbreak will include animals presenting neurological signs. This does not mean that infection with neuropathogenic strains inevitably results in clinical signs of neurological damage, since the clinical outcome of infection for individual animals within an outbreak will be modulated by other factors, which may include age, host immunity, challenge dose, and hormonal status. Consequently, the proportions of animals that ultimately show neurological signs when infected with a neuropathogenic strain vary for each outbreak. For example, during a large outbreak of neurological EHV-1 disease in the United Kingdom, the neurological attack rate (number of cases presenting neurological signs/number of horses confirmed to be infected) was 20% (33). In four recent, large outbreaks of EHV-1 neurological disease in horses in the United States, for which complete records were kept (G. Allen, unpublished data), the mean neurological attack rate was 33% (range of 22% to 50%) and the mean neurological case fatality rate was 40% (range of 20% to 50%). All five outbreaks described above involved EHV-1 strains carrying the neuropathogenic-associated form of the DNA polymerase (D752).
Notably, the D/N752 variable position of EHV-1 DNA polymerase corresponds to a highly conserved position in other herpesviruses. Alignment of herpesvirus DNA polymerase sequences from diverse host species indicated that the equivalent position was conserved as an acidic residue (almost invariably D) in other herpesviruses (Fig. 3). This suggests that the ancestral EHV-1 virus probably encoded D752 and that variants expressing N752 arose subsequently, possibly due to a selective advantage. The observation that the D/N752 SNP is unlinked to any of the other SNPs tested suggests that either the D/N752 mutation has arisen relatively recently and independently in five of the six major strain groups or this position is relatively unstable. Outbreaks of EHV-1 neurological disease are relatively rare compared with outbreaks of nonneurological disease, which suggests that the majority of EHV-1 strains in circulation encode N752. It may further be proposed that D752 variants arise spontaneously but may have a selective disadvantage for long-term maintenance in the population. Comparison between herpesvirus DNA polymerase sequences and those of other related polymerases (from organisms as diverse as bacteriophages and mammals) has revealed a number of conserved domains (23, 59). The variable D/N752 residue lies between the conserved domains designated II and VI, which comprise regions essential for DNA polymerase activity. Mutations proximal to this position (mostly within domains II and VI), which influence antiviral drug sensitivity for herpes simplex virus, varicella-zoster virus, and human cytomegalovirus, have been noted previously (11, 12, 18, 24, 29). Thus, although the variable D/N752 residue is not within one of the domains known to be critical for function, it does lie within the core, catalytic region of the enzyme and may therefore influence enzymatic activity.
Considering the strong association of D/N752 with neuropathogenic potential, it is reasonable to speculate that the D/N752 coding change affects functional properties of the DNA polymerase that play a direct role in the etiology of EHV-1 neurological disease. Replication studies with various cell types in tissue culture (fibroblast and endothelial) have shown no consistent differences between V592 and Ab4 (55; R. Chiam, K. Smith, and N. Davis-Poynter, unpublished data). It is possible, however, that alterations of DNA polymerase activity affecting replication may be apparent only in specific cell types or in the context of tissues in vivo. Other possible (albeit speculative) modes of action may include variation in the efficiency with which different forms of the DNA polymerase stimulate immune responses (e.g., cytotoxic T lymphocyte) against EHV-1-infected target cells, thereby affecting the efficiency of clearance of the infection. Alternatively, the neuropathogenic form of EHV-1 DNA polymerase may promote damage to blood vessels supplying the CNS, either directly via effects upon infected endothelial cells or indirectly via immunopathology. In order to test the hypothesis that the D/N752 substitution of the DNA polymerase affects neuropathogenicity, recombinant EHV-1 with the substitution at nt 752 engineered on a syngeneic background has been generated (L. Goodman and N. Osterrieder, unpublished data) and will be used in future in vivo studies to determine the influence of DNA polymerase sequence variation upon EHV-1 disease outcome.
The strong association of a naturally occurring SNP of a single gene with pathogenic potential, observed for EHV-1 DNA polymerase in this study, is to our knowledge unprecedented for a herpesvirus. The implication is that mutation of a single nucleotide has a significant effect upon pathogenicity, although this hypothesis has not yet been tested. This raises an important question concerning the origin of EHV-1 strains giving rise to neurological disease outbreaks, namely, whether they usually originate from nonneuropathogenic strains, via spontaneous mutation of the DNA polymerase (from N752 to D), or whether they are usually the result of reactivation from animals previously exposed to neuropathogenic strains. Currently, we have no data regarding the relative frequency of latent carriage of EHV-1 strains encoding D/N752. DNA polymerase activity may conceivably influence the establishment of, or reactivation from, latency. Thus, establishment of latency may be promoted by abortive replication, if DNA polymerase activity is insufficient to promote full late gene expression prior to the genome entering a quiescent state; conversely, during the early stages of reactivation, DNA polymerase activity may be critical in triggering full lytic cycle gene expression. If EHV-1 DNA polymerase sequence variation does have an impact upon latency/reactivation, this may provide a source of selective pressure acting upon the DNA polymerase D/N752 sequence. In the absence of a reliable system for evaluating the efficiency of EHV-1 latency establishment and reactivation, it will be difficult to test the abovementioned hypothesis.
In conclusion, determination of the complete genomic sequence of a nonneuropathogenic strain of EHV-1 (V592) and comparison with the previously published genomic sequence of a neuropathogenic strain (Ab4) has led to two important findings. First, a method for subtyping EHV-1 isolates recovered from the field via multilocus typing, which will be useful in tracing the transmission of virus strains between outbreaks, has been developed. Second, an SNP of ORF30 (G/A2254), resulting in an amino acid coding change of the DNA polymerase (D/N752), shows strong predictive value as a marker of isolates with a propensity to cause neurological disease. If a causal link between DNA polymerase sequence variation and neuropathogenic potential is established, determination of the molecular mechanisms underlying the link will assist future diagnostic, prophylactic, and therapeutic strategies designed to reduce the incidence and severity of neurological EHV-1 disease. These findings may also have implications for molecular epidemiological studies of other herpesviruses. In the case of EHV-1, variation of a single gene was found to be associated with differences in pathogenic potential, whereas none of the other SNPs tested showed significant association with disease. If single gene polymorphisms similarly have a significant association with disease manifestation for other herpesviruses, such strain-related differences may not have been apparent from previous analyses of a limited subset of variable gene sequences. The availability of methods for whole-genome SNP analysis will facilitate more-comprehensive studies and may reveal previously unrecognized associations between herpesvirus strain variation and disease.
Acknowledgments
This work was supported by grants from the Home of Rest for Horses, the Horserace Betting Levy Board, the Grayson Jockey Club Research Foundation, and the European Breeders Fund.
We are grateful to the following contributors for generous provision of EHV-1 isolates/DNA and information concerning the relevant outbreaks: G. Fortier (Laboratoire Frank Duncombe, France), M. Studdert (University of Melbourne, Australia), S. Raidal (Murdoch University, Australia), K. van Maanen (Animal Health Service, The Netherlands), C. Galosi (Cátedra de Virología, UNLP, Argentina), and K. van der Meulen (University of Ghent, Belgium).
REFERENCES
- 1.Allen, G. P., J. H. Kydd, J. D. Slater, and K. C. Smith. 1999. Advances in understanding the pathogenesis, epidemiology and immunological control of equid herpes-1 (EHV-1) abortion, p. 129-146. In U. Wernery, J. F. Wade, J. A. Mumford, and O. R. Kaaden (ed.), Equine infectious diseases VIII. Proceedings of the Eighth International Conference on Equine Infectious Diseases. R&W Publications Limited, Newmarket, United Kingdom.
- 2.Allen, G. P., M. Yeargan, and J. Bryans. 1983. Alterations in the equine herpesvirus 1 genome after in vitro and in vivo virus passage. Infect. Immun. 40:436-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allen, G. P., M. R. Yeargan, L. W. Turtinen, and J. T. Bryans. 1985. A new field isolate of equine abortion virus (equine herpesvirus-1) among Kentucky horses. Am. J. Vet. Res. 46:138-140. [PubMed] [Google Scholar]
- 4.Allen, G. P., M. R. Yeargan, L. W. Turtinen, J. T. Bryans, and W. H. McCollum. 1983. Molecular epizootiologic studies of equine herpesvirus-1 infections by restriction endonuclease fingerprinting of viral DNA. Am. J. Vet. Res. 44:263-271. [PubMed] [Google Scholar]
- 5.Bandelt, H.-J., P. Forster, B. C. Sykes, and M. B. Richards. 1995. Mitochondrial portraits of human populations using median networks. Genetics 141:743-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bass, E. P. 1979. Immunization with a modified live virus equine rhinopneumonitis vaccine and an aluminum hydroxide absorbed equine influenza vaccine. Proc. Am. Assoc. Equine Pract. 24:65-74. [Google Scholar]
- 7.Beckenaur, W. H., and E. P. Bass. 1973. A modified live virus vaccine for equine rhinopneumonitis. J. Am. Vet. Med. Assoc. 163:1182. [Google Scholar]
- 8.Binns, M. M., S. McCann, L. Zhang, J. Wood, and J. A. Mumford. 1994. Molecular epidemiology of EHV-1 and EHV-4: a search for variable restriction sites, p. 237-241. In H. Nakajima and W. Plowright (ed.), Equine infectious diseases VII. Proceedings of the Seventh International Conference on Equine Infectious Diseases. R&W Publications, Newmarket, United Kingdom.
- 9.Blunden, A. S., K. E. Whitwell, and K. M. Pegler. 1993. An outbreak of paralysis associated with equine herpes virus type 1 infection in a livery stable. Prog. Vet. Neurol. 3:95-100. [Google Scholar]
- 10.Chou, S. 1992. Comparative analysis of sequence variation in gp116 and gp55 components of glycoprotein B of human cytomegalovirus. Virology 188:388-390. [DOI] [PubMed] [Google Scholar]
- 11.Chou, S., N. S. Lurain, A. Weinberg, G.-Y. Cai, P. L. Sharma, C. S. Crumpacker, and Adult AIDS Clinical Trials Group CMV Laboratories. 1999. Interstrain variation in the human cytomegalovirus DNA polymerase sequence and its effect on genotypic diagnosis of antiviral drug resistance. Antimicrob. Agents Chemother. 43:1500-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chou, S., N. S. Lurain, K. D. Thompson, R. C. Miner, and W. L. Drew. 2003. Viral DNA polymerase mutations associated with drug resistance in human cytomegalovirus. J. Infect. Dis. 188:32-39. [DOI] [PubMed] [Google Scholar]
- 13.Chou, S. W., and K. M. Dennison. 1991. Analysis of interstrain variation in cytomegalovirus glycoprotein B sequences encoding neutralization-related epitopes. J. Infect. Dis. 163:1229-1234. [DOI] [PubMed] [Google Scholar]
- 14.Crowhurst, F. A., G. Dickinson, and R. Burrows. 1981. An outbreak of paresis in mares and geldings associated with equid herpesvirus 1. Vet. Rec. 109:527-528. [PubMed] [Google Scholar]
- 15.Darlington, J., M. Super, K. Patel, J. Grundy, P. Griffiths, and V. C. Emery. 1991. Use of the polymerase chain reaction to analyse sequence variation within a major neutralizing epitope of glycoprotein B (gp58) in clinical isolates of human cytomegalovirus. J. Gen. Virol. 72:1985-1989. [DOI] [PubMed] [Google Scholar]
- 16.Davis-Poynter, N. J., J. Nugent, I. Birch-Machin, and G. P. Allen. Viral marker. U.S. patent application 20,050,003,342.
- 17.Edington, N., C. G. Bridges, and J. R. Patel. 1986. Endothelial cell infection and thrombosis in paralysis caused by equid herpesvirus-1: equine stroke. Arch. Virol. 90:111-124. [DOI] [PubMed] [Google Scholar]
- 18.Gibbs, J. S., H. C. Chiou, K. F. Bastow, Y.-C. Cheng, and D. M. Coen. 1988. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc. Natl. Acad. Sci. USA 85:6672-6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Greenwood, R. E. S., and A. R. B. Simson. 1980. Clinical report of a paralytic syndrome affecting stallions, mares and foals on a thoroughbred studfarm. Equine Vet. J. 12:113-117. [DOI] [PubMed] [Google Scholar]
- 20.Haberland, M., U. Meyer-Konig, and F. T. Hufert. 1999. Variation within the glycoprotein B gene of human cytomegalovirus is due to homologous recombination. J. Gen. Virol. 80:1495-1500. [DOI] [PubMed] [Google Scholar]
- 21.Hall, T. A. 1999. Bioedit: a userfriendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 41:95-98. [Google Scholar]
- 22.Huang, J., N. Ficorilli, C. A. Hartley, G. P. Allen, and M. J. Studdert. 2002. Polymorphism of open reading frame 71 of equine herpesvirus-4 (EHV-4) and EHV-1. J. Gen. Virol. 83:525-531. [DOI] [PubMed] [Google Scholar]
- 23.Hwang, C. B., K. L. Ruffner, and D. M. Coen. 1992. A point mutation within a distinct conserved region of the herpes simplex virus DNA polymerase gene confers drug resistance. J. Virol. 66:1774-1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamiyama, T., M. Kurokawa, and K. Shiraki. 2001. Characterisation of the DNA polymerase gene of varicella-zoster viruses resistant to acyclovir. J. Gen. Virol. 82:2761-2765. [DOI] [PubMed] [Google Scholar]
- 25.Kirisawa, R., H. Ohmori, H. Iwai, and Y. Kawakami. 1993. The genomic diversity among equine herpesvirus-1 strains isolated in Japan. Arch. Virol. 129:11-22. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 27.Liu, I. K. M., and W. Castleman. 1977. Equine posterior paresis associated with equine herpesvirus 1 vaccine in California: a preliminary report. J. Eqine Med. Surg. 12:397-401. [Google Scholar]
- 28.Loparev, V. N., A. Gonzalez, M. Deleon-Carnes, G. Tipples, H. Fikenscher, E. G. Torfason, and D. S. Schid. 2004. Global identification of three major genotypes of varicella-zoster virus: longitudinal clustering and strategies for genotyping. J. Virol. 78:8349-8358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews, J. T., B. J. Terry, and K. A. Field. 1993. The structure and function of the HSV DNA replication proteins: defining novel antiviral targets. Antivir. Res. 20:89-114. [DOI] [PubMed] [Google Scholar]
- 30.Mayr, A. 1970. Vaccination of horses against equine herpesvirus 1 infection, p. 41-45. In J. T. Bryans and H. Gerber (ed.), Equine infectious diseases II. Proceedings of the Second International Conference on Equine Infectious Diseases. S. Karger, Basel, Switzerland.
- 31.Mayr, A., J. Pette, K. Petzoldt, and K. Wagener. 1968. Studies on the development of a live vaccine against rhinopneumonitis (mare abortion) of horses. Zentbl. Vetmed. Reihe B 15:406-418. (In German.) [PubMed] [Google Scholar]
- 32.McCann, S. H. E., J. A. Mumford, and M. M. Binns. 1995. Development of PCR assays to detect genetic variation amongst equine herpesvirus-1 isolates as an aid to epidemiological investigation. J. Virol. Methods 52:183-194. [DOI] [PubMed] [Google Scholar]
- 33.McCartan, C. G., M. M. Russell, J. L. N. Wood, and J. A. Mumford. 1995. Clinical, serological and virological characteristics of an outbreak of paresis and neonatal foal disease to equine herpesvirus-1 on a stud farm. Vet. Rec. 136:7-12. [DOI] [PubMed] [Google Scholar]
- 34.Meindl, A., and N. Osterrieder. 1999. The equine herpesvirus 1 US2 homolog encodes a nonessential membrane-associated virion component. J. Virol. 73:3430-3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer, H., P. H. Hubert, and W. Eichhorn. 1987. Changes in restriction enzyme pattern of the equine herpes virus type 1 (EHV-1) strain RacH DNA during attenuation. Zentbl. Vetmed. Reihe B 34:310-313. [DOI] [PubMed] [Google Scholar]
- 36.Muir, W. B., R. Nichols, and J. Breuer. 2002. Phylogenetic analysis of varicella-zoster virus: evidence of intercontinental spread of genotypes and recombination. J. Virol. 76:1971-1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mumford, J. A., and N. Edington. 1980. EHV-1 and equine paresis. Vet. Rec. 106:277. [DOI] [PubMed] [Google Scholar]
- 38.Mumford, J. A., D. Hannant, D. M. Jessett, T. O'Neill, K. C. Smith, and E. N. Ostlund. 1994. Abortigenic and neurological disease caused by experimental infection with equid herpesvirus-1, p. 261-275. In H. Nakajima and W. Plowright (ed.), Equine infectious diseases VII. Proceedings of the Seventh International Conference on Equine Infectious Diseases. R&W Publications, Newmarket, United Kingdom.
- 39.Mumford, J. A., P. D. Rossdale, D. M. Jesset, S. J. Gann, J. Ousey, and R. F. Cook. 1987. Serological and virological investigations of an equid herpesvirus-1 (EHV-1) abortion storm on a stud farm in 1985. J. Reprod. Fertil. 35(Suppl.):509-518. [PubMed] [Google Scholar]
- 40.Norberg, P., T. Bergstrom, E. Rekabdar, M. Lindh, and J.-A. Liljeqvist. 2004. Phylogenetic analysis of clinical herpes simplex virus type 1 isolates identified three genetic groups and recombinant viruses. J. Virol. 78:10755-10764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Palfi, V., and L. S. Christensen. 1995. Analyses of restriction fragment patterns (RFPs) and pathogenicity in baby mice of equine herpesvirus 1 and 4 (EHV-1 and EHV-4) strains circulating in Danish horses. Vet. Microbiol. 47:199-204. [DOI] [PubMed] [Google Scholar]
- 42.Patel, J. R., N. Edington, and J. A. Mumford. 1982. Variation in cellular tropism between isolates of equine herpesvirus-1 in foals. Arch. Virol. 74:41-52. [DOI] [PubMed] [Google Scholar]
- 43.Pignatelli, S., P. Dal Monte, and M. P. Landini. 2001. gpUL73 (gN) genomic variants of human cytomegalovirus isolates are clustered into four distinct genotypes. J. Gen. Virol. 82:2777-2784. [DOI] [PubMed] [Google Scholar]
- 44.Platt, H., H. Singh, and K. E. Whitwell. 1980. Pathological observations on an outbreak of paralysis in broodmares. Equine Vet. J. 12:118-126. [DOI] [PubMed] [Google Scholar]
- 45.Purdy, C. W., R. C. Porter, and S. J. Ford. 1978. Equine rhinopneumonitis vaccine: immunogenicity and safety in foals. Am. J. Vet. Res. 39:745-752. [PubMed] [Google Scholar]
- 46.Purdy, C. W., S. J. Ford, and R. C. Porter. 1978. Equine rhinopneumonitis vaccine: immunogenicity and safety in adult horses, including pregnant mares. Am. J. Vet. Res. 39:377-383. [PubMed] [Google Scholar]
- 47.Purdy, C. W., S. J. Ford, and W. F. Grant. 1977. Equine rhinopneumonitis virus (equine herpesvirus type 1): attenuation in stable monkey cell line. Am. J. Vet. Res. 38:1211-1215. [PubMed] [Google Scholar]
- 48.Rasmussen, L., A. Geissler, and M. Winters. 2003. Inter- and intragenic variations complicate the molecular epidemiology of human cytomegalovirus. J. Infect. Dis. 187:809-819. [DOI] [PubMed] [Google Scholar]
- 49.Rawlinson, W. D., H. E. Farrell, and B. G. Barrell. 1996. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 70:8833-8849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sakaoka, H., K. Kurita, Y. Iida, S. Takada, K. Umene, Y. T. Kim, C. S. Ren, and A. J. Nahmias. 1994. Quantitative analysis of genomic polymorphism of herpes simplex virus type 1 strains from six countries: studies of molecular evolution and molecular epidemiology of the virus. J. Gen. Virol. 75:513-527. [DOI] [PubMed] [Google Scholar]
- 51.Smith, K. C., K. E. Whitwell, J. A. Mumford, D. Hannant, A. S. Blunden, and J. P. Tearle. 2000. Virulence of the V592 isolate of equid herpesvirus type 1 in ponies. J. Comp. Pathol. 122:288-297. [DOI] [PubMed] [Google Scholar]
- 52.Smith, K. C., K. E. Whitwell, M. M. Binns, C. A. Dolby, D. Hannant, and J. A. Mumford. 1992. Abortion of virologically negative foetuses following experimental challenge of pregnant pony mares with equid herpesvirus 1. Equine Vet. J. 24:256-259. [DOI] [PubMed] [Google Scholar]
- 53.Staden, R. 1996. The Staden sequence analysis package. Mol. Biotechnol. 5:233-241. [DOI] [PubMed] [Google Scholar]
- 54.Studdert, M. J., B. S. Crabb, and N. Ficorilli. 1992. The molecular epidemiology of equine herpesvirus 1 (equine abortion virus) in Australasia 1975-1989. Aust. Vet. J. 69:104-111. [DOI] [PubMed] [Google Scholar]
- 55.Tearle, J. P., K. C. Smith, A. Platt, D. Hannant, N. J. Davis-Poynter, and J. A. Mumford,. 2003. In vitro characterisation of high and low virulence isolates of equine herpesviruses-1 and -4. Res. Vet. Sci. 75:83-86. [DOI] [PubMed] [Google Scholar]
- 56.Telford, E. A. R., M. S. Watson, K. McBride, and A. J. Davison. 1992. The DNA sequence of equine herpesvirus-1. Virology 189:304-316. [DOI] [PubMed] [Google Scholar]
- 57.van Maanen, C., M. M. Sloet van Oldruitenborgh-Oosterbaan, E. A. Damen, and A. G. Derksen. 2001. Neurological disease associated with EHV-1-infection in a riding school: clinical and virological characteristics. Equine Vet. J. 33:191-196. [DOI] [PubMed] [Google Scholar]
- 58.Wagenaar, T. R., V. T. K. Chow, C. Buranathai, P. Thawatsupha, and C. Grose. 2003. The out of Africa model of varicella-zoster virus evolution: single nucleotide polymorphisms and private alleles distinguish Asian clades from European/North American clades. Vaccine 21:1072-1081. [DOI] [PubMed] [Google Scholar]
- 59.Wang, T. S.-F. 1991. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 60:513-552. [DOI] [PubMed] [Google Scholar]
- 60.Whitwell, K. E., and A. S. Blunden. 1992. Pathological findings in horses dying during an outbreak of the paralytic form of equid herpesvirus type 1 (EHV-1) infection. Equine Vet. J. 24:13-19. [DOI] [PubMed] [Google Scholar]
- 61.Wilson, D. 1997. Equine herpesvirus 1 myeloencephalopathy. Vet. Clin. N. Am. Equine Pract. 13:53-72. [DOI] [PubMed] [Google Scholar]
- 62.Zwegyberg, L., B. Wirgart, M. Brytting, A. Linde, B. Wahren, and L. Grillner. 1998. Sequence variation within three important cytomegalovirus gene regions in isolates from four different patient populations. J. Clin. Microbiol. 36:3662-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]